A FtsZ Inhibitor That Can Utilize Siderophore-Ferric Iron Uptake Transporter Systems for Activity against Gram-Negative Bacterial Pathogens
Abstract
1. Introduction
2. Results
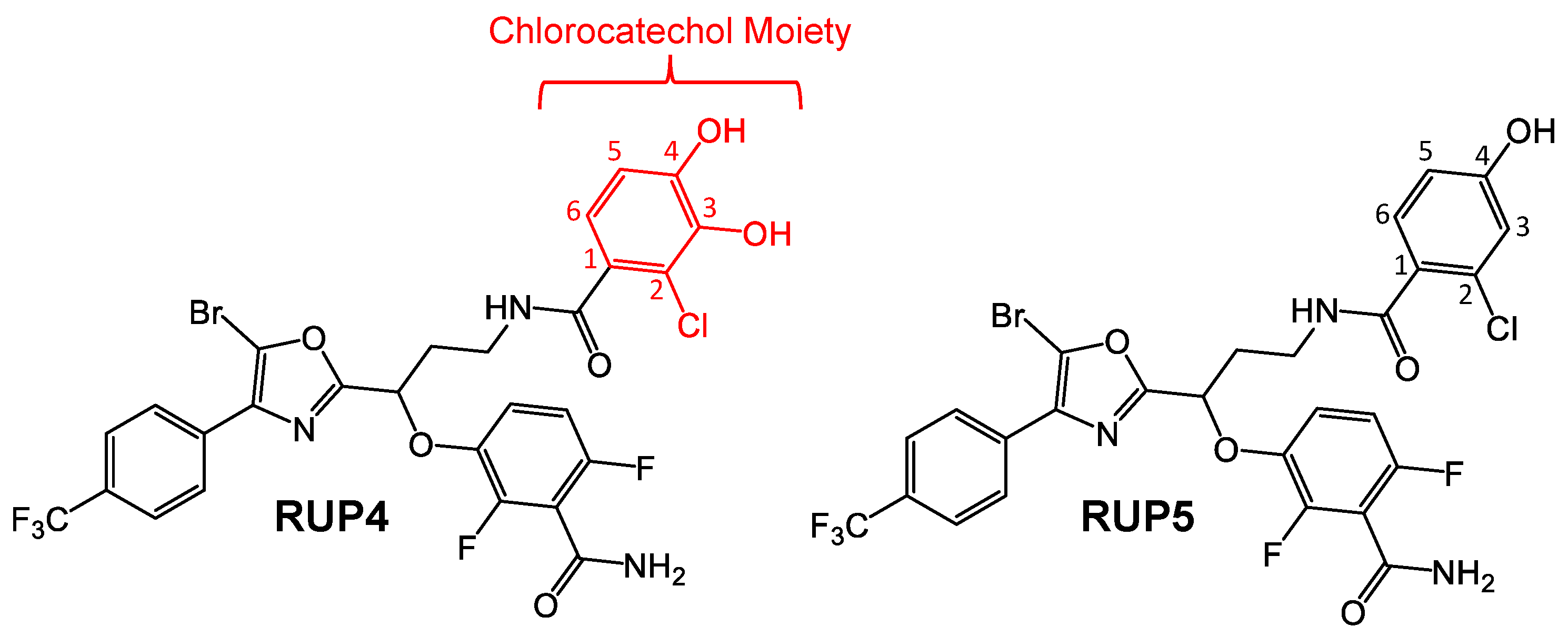
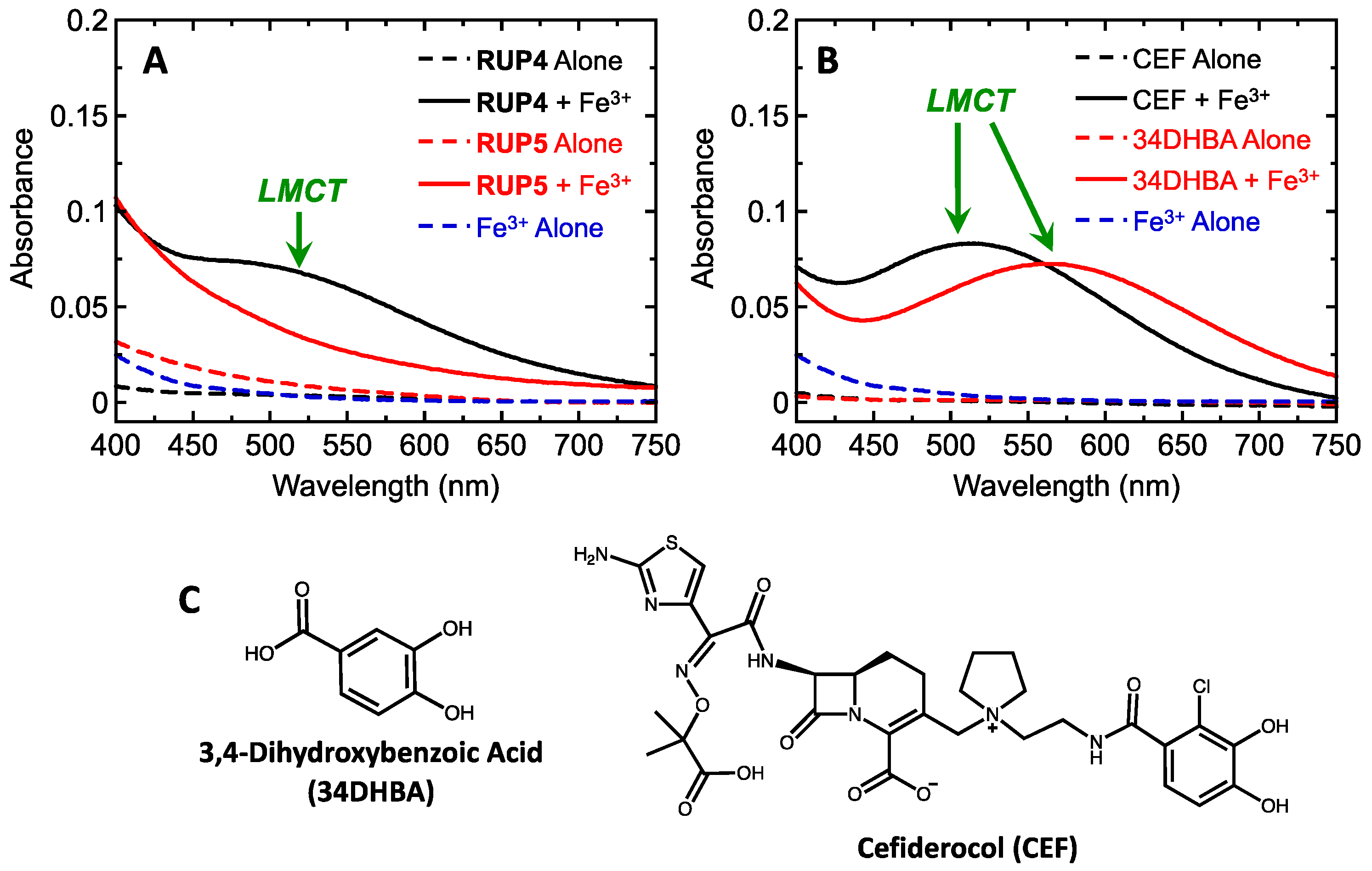
2.1. Evaluation of RUP4 and RUP5 for Their Ability to Chelate Fe3+
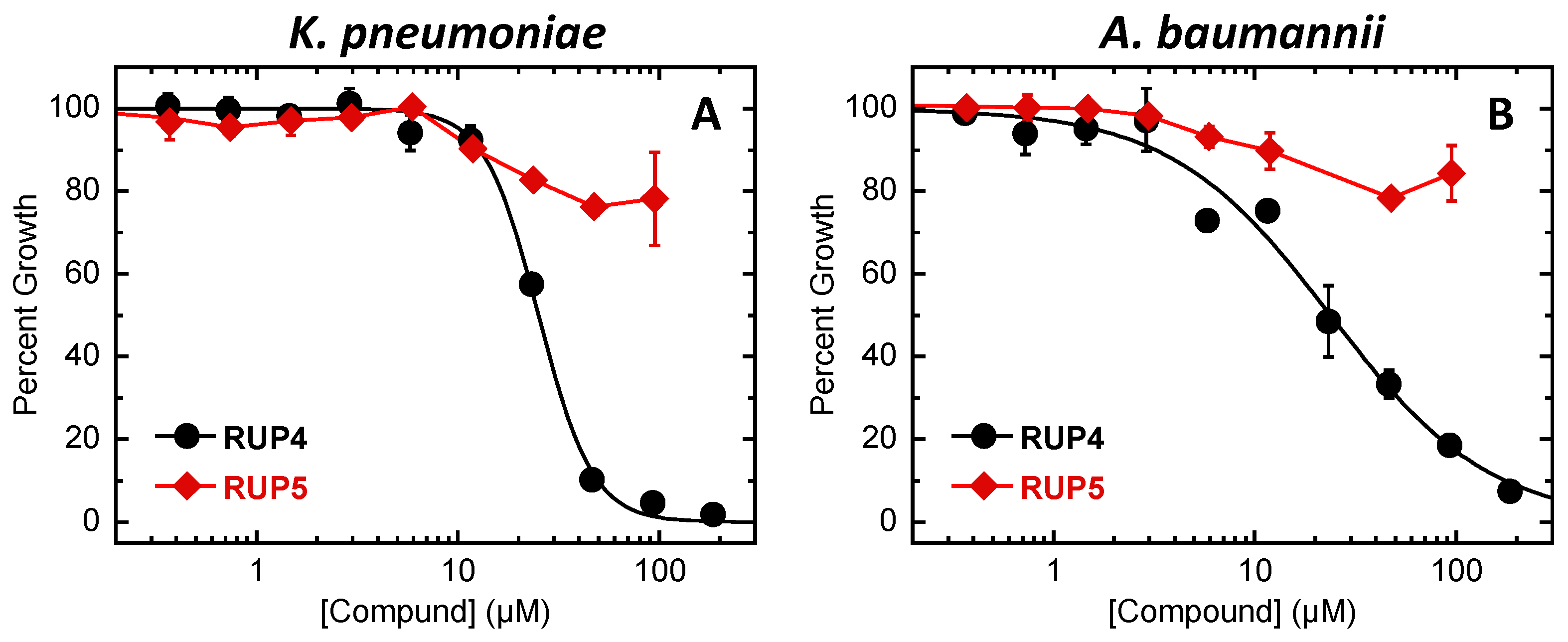
2.2. Evaluation of RUP4 and RUP5 for Antibacterial Activity against K. pneumoniae and A. baumannii
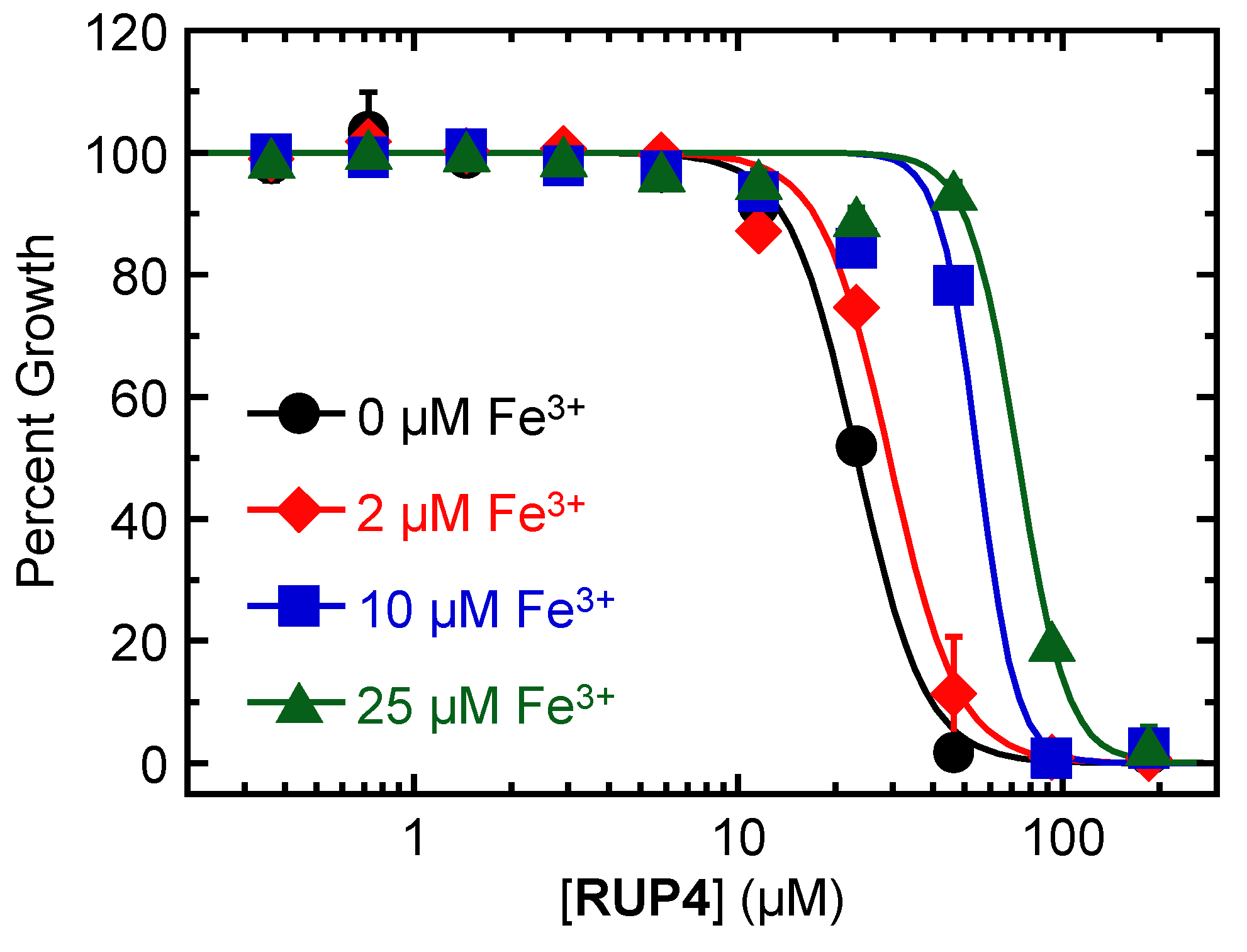
2.3. Impact of Added Exogenous Fe3+ on the Antibacterial Activity of RUP4 against K. pneumoniae
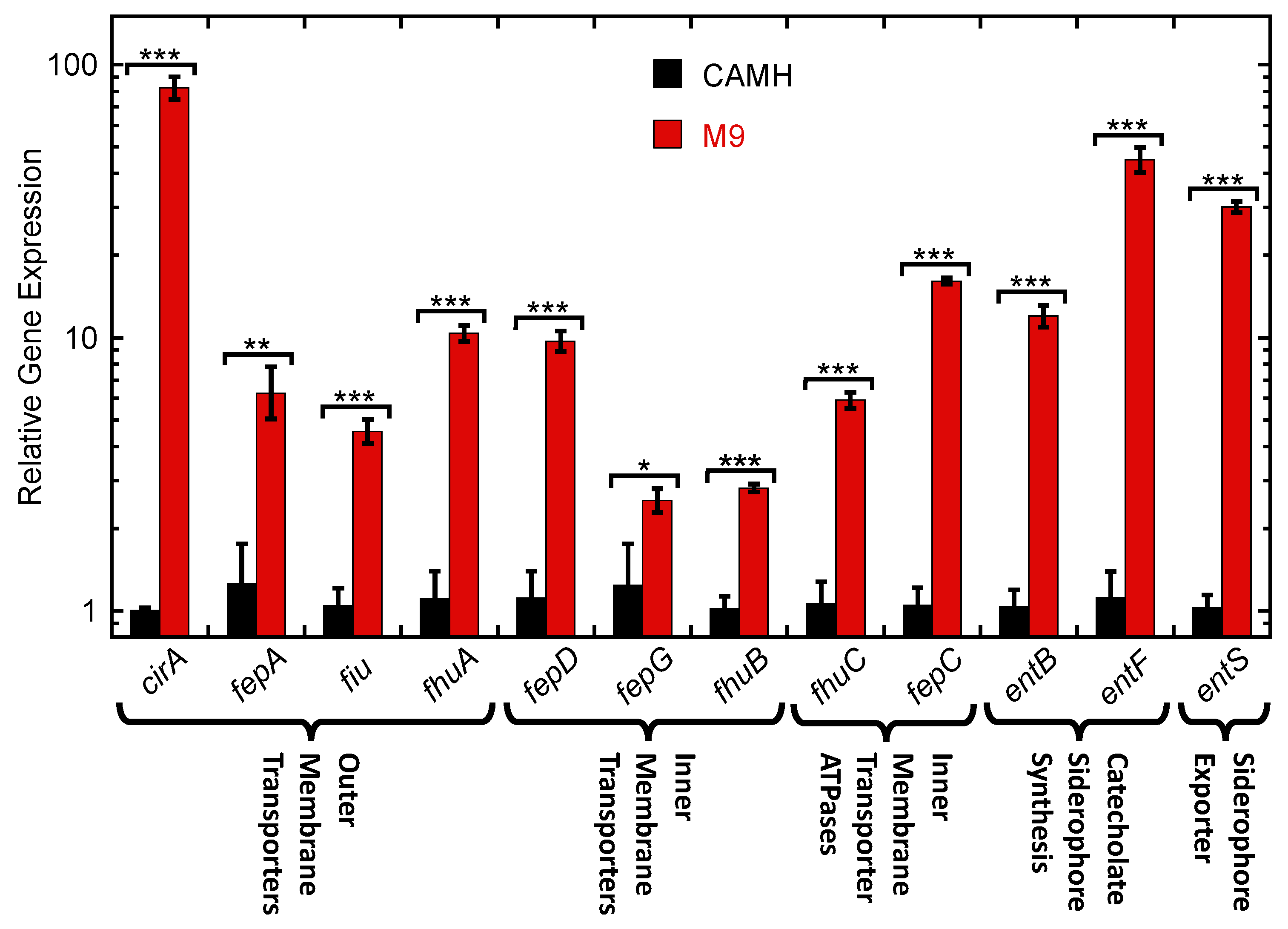
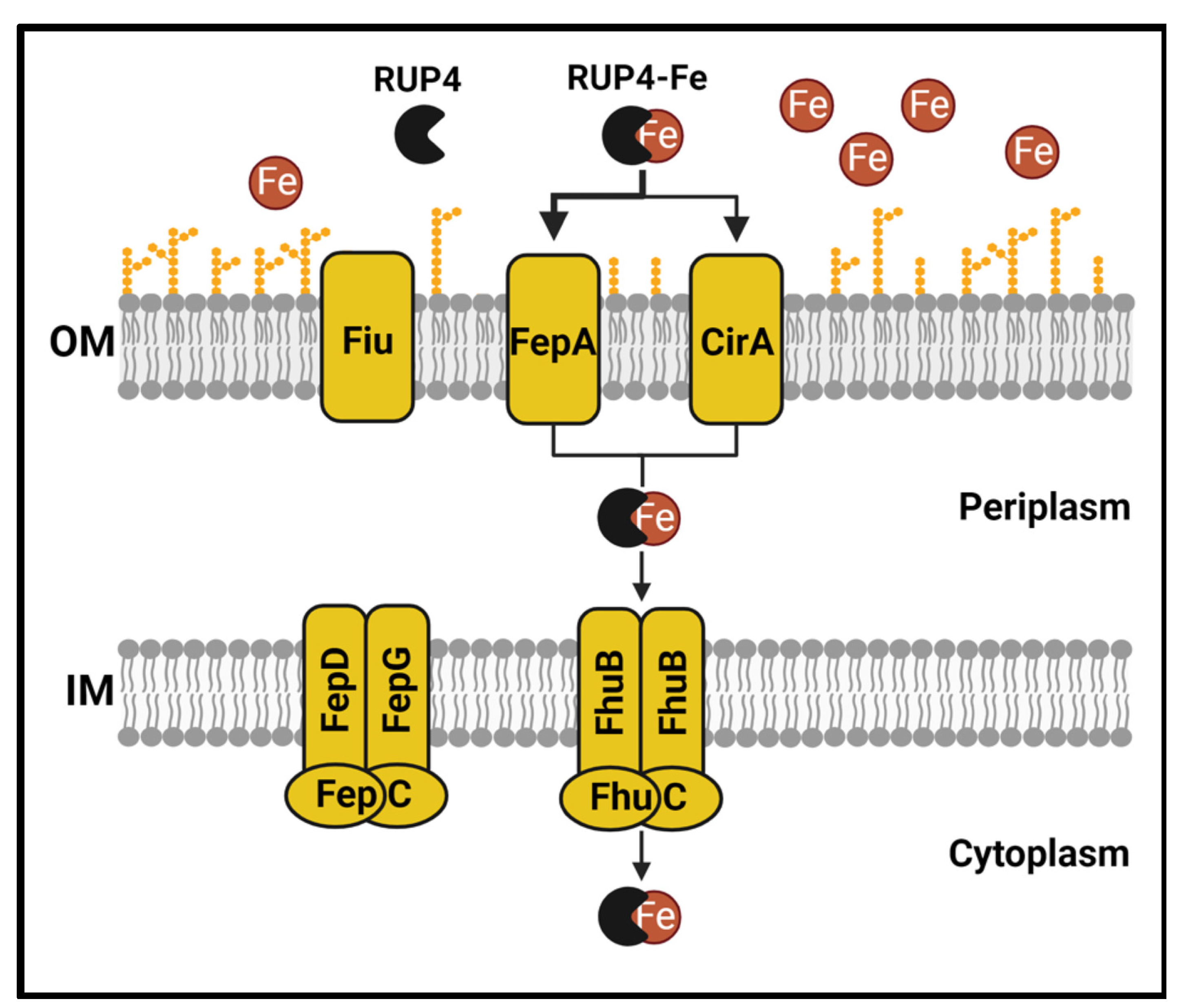
2.4. Expression of Siderophore Biosynthesis, Exporter, and Uptake Transporter Genes by K. pneumoniae in Nutrient-Rich CAMH Media Versus Fe3+-Limiting M9 Media
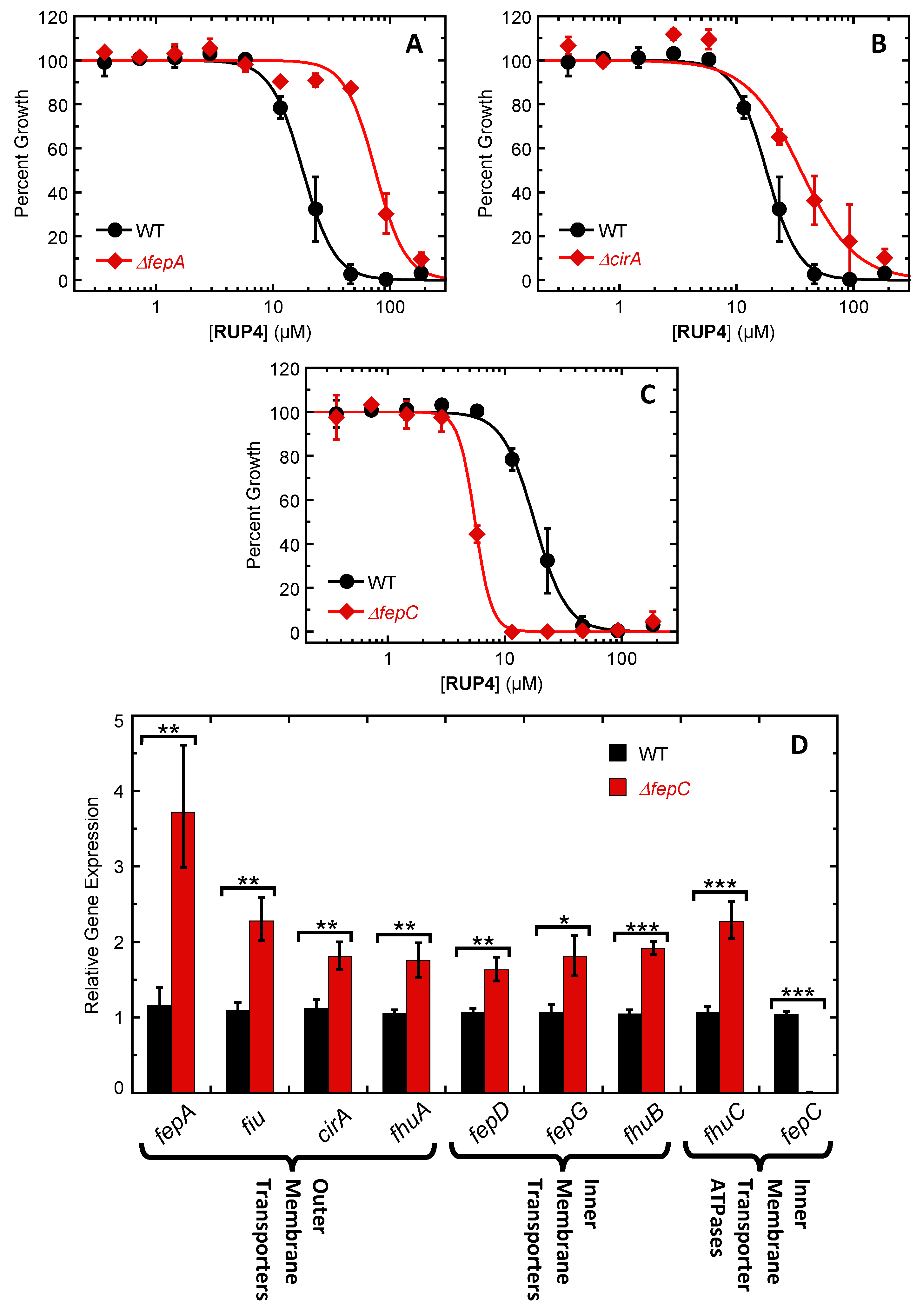
2.5. Impact of Deleting Select Siderophore-Fe3+ Uptake Transporter Genes on RUP4 Activity against K. pneumoniae
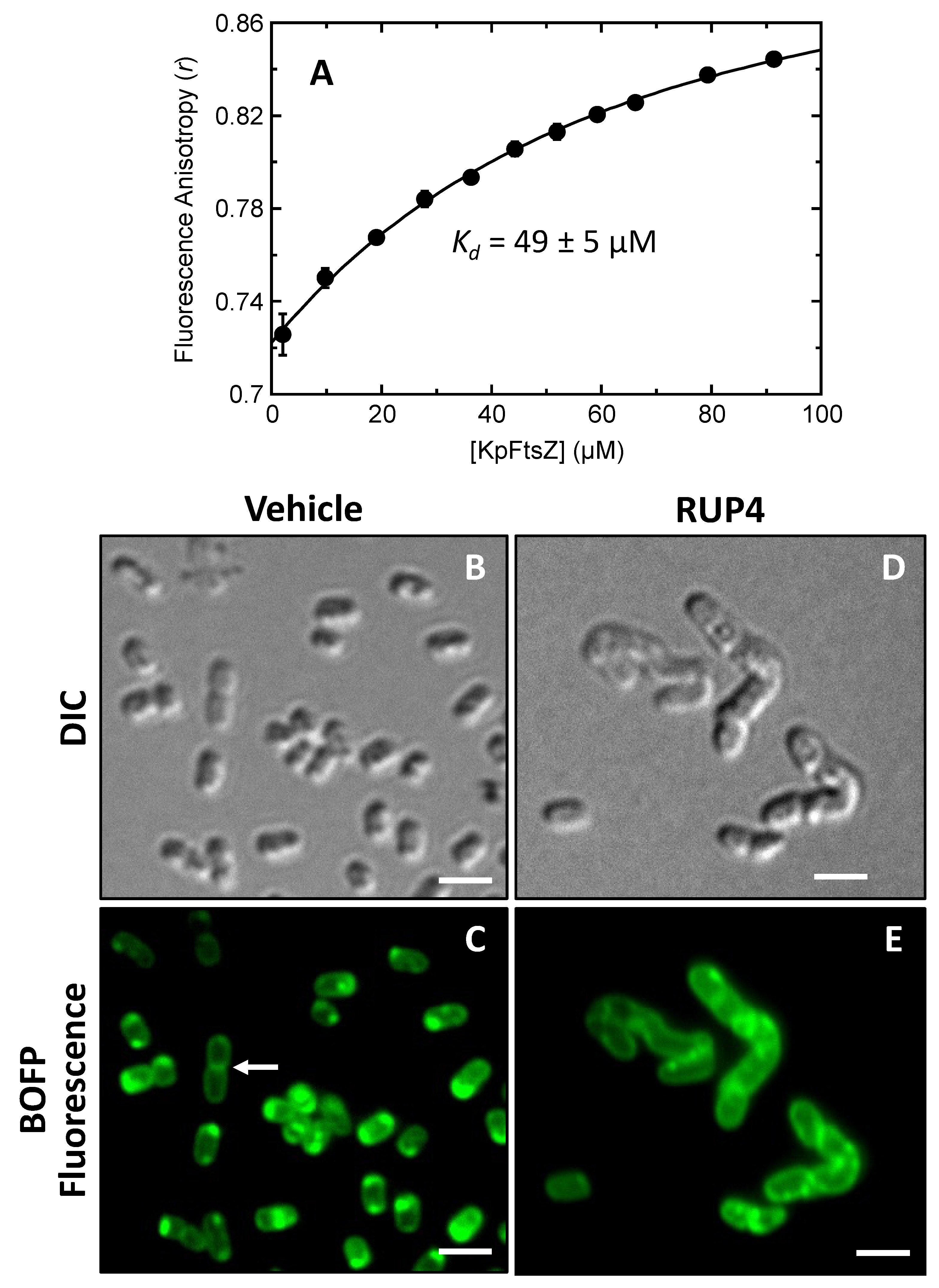
2.6. Direct Binding of RUP4 to Purified K. pneumoniae FtsZ (KpFtsZ)
2.7. Impact of RUP4 on Cell Division and FtsZ Localization in K. pneumoniae
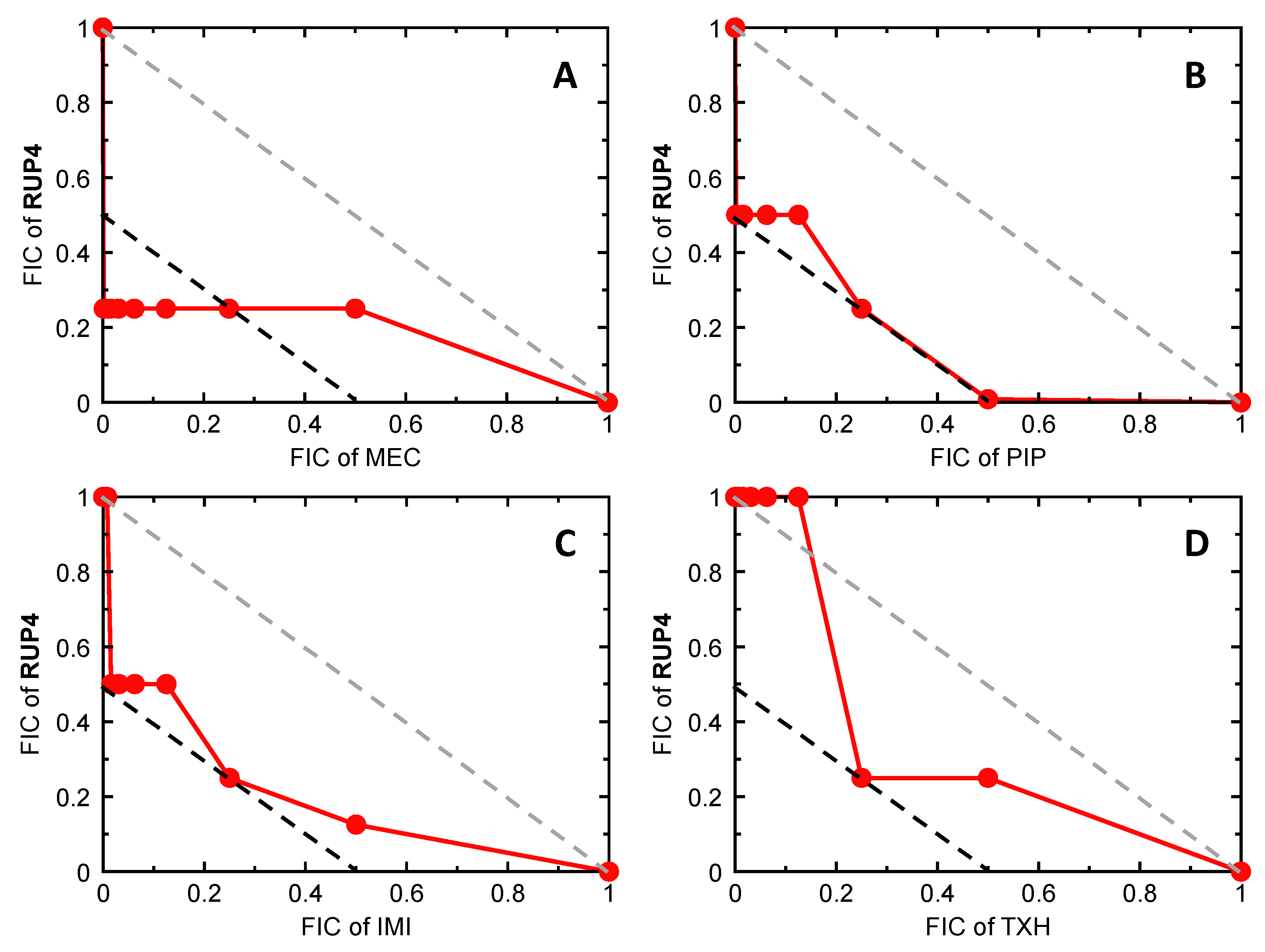
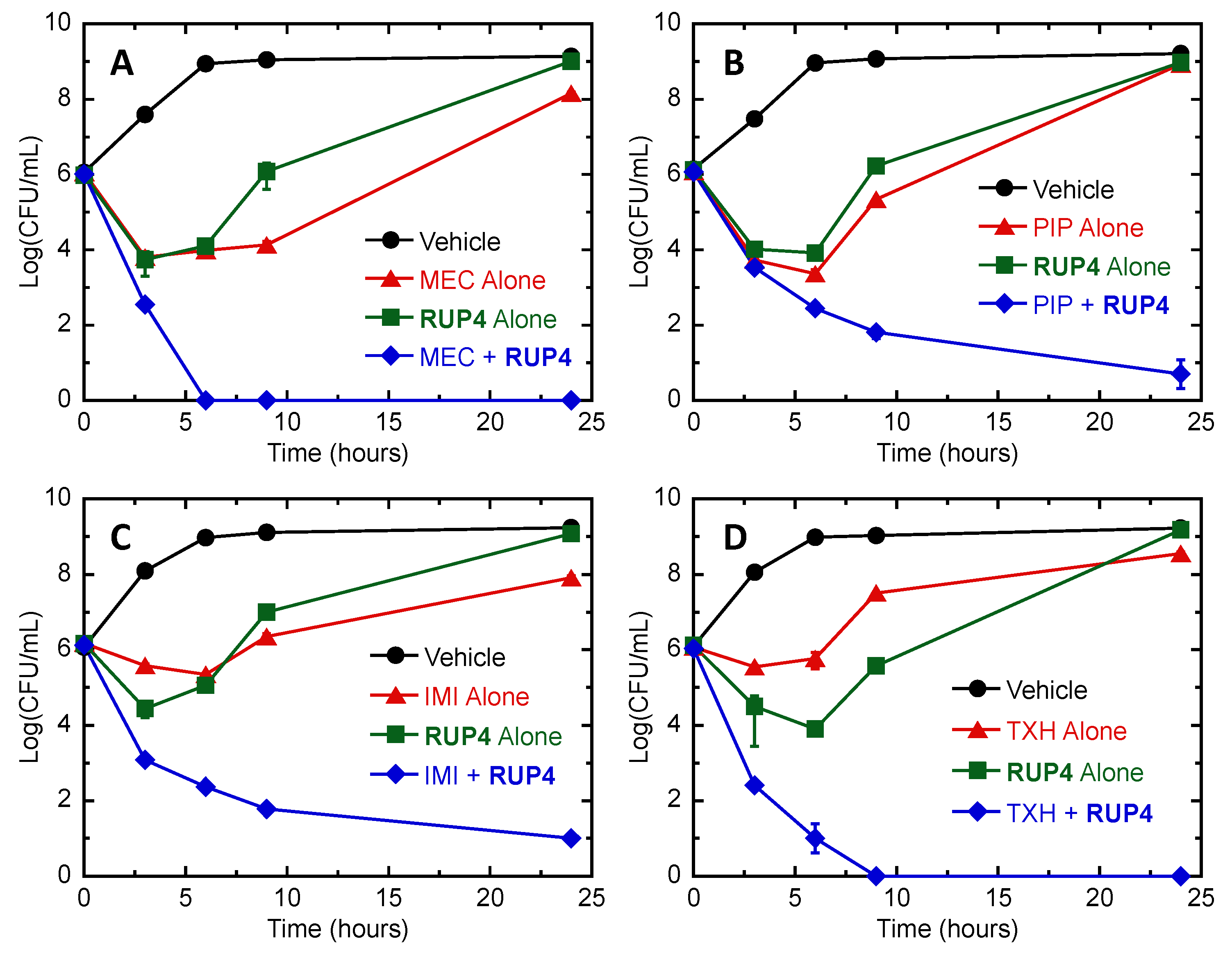
2.8. Bactericidal Synergy of RUP4 in Combination with Select PBP-Targeting β-Lactam Antibiotics and an MreB-Targeting Agent against K. pneumoniae
3. Discussion
3.1. Importance of the Catechol Siderophore Functionality for the Antibacterial Activity of RUP4
3.2. Validation of FtsZ as the Antibacterial Target of RUP4
3.3. Synergistic Drug Combinations with RUP4
4. Materials and Methods
4.1. Bacterial Strains, Growth Media, and Reagents
4.2. Compound Synthesis and K. pneumoniae FtsZ Protein Expression and Purification
4.3. Antibacterial Assays
4.4. Absorption Spectroscopy
4.5. Fluorescence Spectroscopy
4.6. Differential Interference Contrast (DIC) and Fluorescence Microscopy
4.7. RNA Extraction and Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR) Assays
4.8. Antibacterial Synergy Assays
4.8.1. Checkerboard Assays
4.8.2. Time–Kill Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015.
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States, 2019; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- O-Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Wellcome Collection: London, UK, 2014. [Google Scholar]
- Oliveira, D.M.P.D.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef]
- Morris, S.; Cerceo, E. Trends, Epidemiology, and Management of Multi-Drug Resistant Gram-Negative Bacterial Infections in the Hospitalized Setting. Antibiotics 2020, 9, 196. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A Major Worldwide Source and Shuttle for Antibiotic Resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef]
- Margolin, W. FtsZ and the Division of Prokaryotic Cells and Organelles. Nat. Rev. Mol. Cell Biol. 2005, 6, 862–871. [Google Scholar] [CrossRef] [PubMed]
- Silber, N.; Opitz, C.L.M.d.; Mayer, C.; Sass, P. Cell Division Protein FtsZ: From Structure and Mechanism to Antibiotic Target. Future Microbiol. 2020, 15, 801–831. [Google Scholar] [CrossRef]
- Adams, D.W.; Errington, J. Bacterial Cell Division: Assembly, Maintenance and Disassembly of the Z Ring. Nat. Rev. Microbiol. 2009, 7, 642–653. [Google Scholar] [CrossRef]
- Erickson, H.P.; Anderson, D.E.; Osawa, M. FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiol. Mol. Biol. Rev. 2010, 74, 504–528. [Google Scholar] [CrossRef]
- Fenton, A.K.; Gerdes, K. Direct Interaction of FtsZ and MreB is Required for Septum Synthesis and Cell Division in Escherichia coli. Embo J. 2013, 32, 1953–1965. [Google Scholar] [CrossRef]
- Varma, A.; Pedro, M.A.d.; Young, K.D. FtsZ Directs a Second Mode of Peptidoglycan Synthesis in Escherichia coli. J. Bacteriol. 2007, 189, 5692–5704. [Google Scholar] [CrossRef]
- Bryan, E.; Ferrer-González, E.; Sagong, H.Y.; Fujita, J.; Mark, L.; Kaul, M.; LaVoie, E.J.; Matsumura, H.; Pilch, D.S. Structural and Antibacterial Characterization of a New Benzamide FtsZ Inhibitor with Superior Bactericidal Activity and In Vivo Efficacy against Multidrug-Resistant Staphylococcus aureus. ACS Chem. Biol. 2023, 18, 629–642. [Google Scholar] [CrossRef]
- Chiodini, G.; Pallavicini, M.; Zanotto, C.; Bissa, M.; Radaelli, A.; Straniero, V.; Bolchi, C.; Fumagalli, L.; Ruggeri, P.; De Giuli Morghen, C.; et al. Benzodioxane–Benzamides as New Bacterial Cell Division Inhibitors. Eur. J. Med. Chem. 2015, 89, 252–265. [Google Scholar] [CrossRef]
- Deng, J.; Zhang, T.; Li, B.; Xu, M.; Wang, Y. Design, Synthesis and Biological Evaluation of Biphenyl-Benzamides as Potent FtsZ Inhibitors. Eur. J. Med. Chem. 2022, 239, 114553. [Google Scholar] [CrossRef]
- Haydon, D.J.; Stokes, N.R.; Ure, R.; Galbraith, G.; Bennett, J.M.; Brown, D.R.; Baker, P.J.; Barynin, V.V.; Rice, D.W.; Sedelnikova, S.E.; et al. An Inhibitor of FtsZ with Potent and Selective Anti-Staphylococcal Activity. Science 2008, 321, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Mark, L.; Zhang, Y.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S. An FtsZ-Targeting Prodrug with Oral Antistaphylococcal Efficacy In Vivo. Antimicrob. Agents Chemother. 2013, 57, 5860–5869. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Mark, L.; Zhang, Y.; Parhi, A.K.; Lyu, Y.L.; Pawlak, J.; Saravolatz, S.; Saravolatz, L.D.; Weinstein, M.P.; LaVoie, E.J.; et al. TXA709, an FtsZ-Targeting Benzamide Prodrug with Improved Pharmacokinetics and Enhanced In Vivo Efficacy against Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2015, 59, 4845–4855. [Google Scholar] [CrossRef] [PubMed]
- Stokes, N.R.; Baker, N.; Bennett, J.M.; Berry, J.; Collins, I.; Czaplewski, L.G.; Logan, A.; Macdonald, R.; Macleod, L.; Peasley, H.; et al. An Improved Small-Molecule Inhibitor of FtsZ with Superior In Vitro Potency, Drug-Like Properties, and In Vivo Efficacy. Antimicrob. Agents Chemother. 2013, 57, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Stokes, N.R.; Baker, N.; Bennett, J.M.; Chauhan, P.K.; Collins, I.; Davies, D.T.; Gavade, M.; Kumar, D.; Lancett, P.; Macdonald, R.; et al. Design, Synthesis and Structure–Activity Relationships of Substituted Oxazole–Benzamide Antibacterial Inhibitors of FtsZ. Bioorg. Med. Chem. Lett. 2014, 24, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Straniero, V.; Pallavicini, M.; Chiodini, G.; Zanotto, C.; Volontè, L.; Radaelli, A.; Bolchi, C.; Fumagalli, L.; Sanguinetti, M.; Menchinelli, G.; et al. 3-(Benzodioxan-2-ylmethoxy)-2,6-Difluorobenzamides Bearing Hydrophobic Substituents at the 7-Position of the Benzodioxane Nucleus Potently Inhibit Methicillin-Resistant Sa and Mtb Cell Division. Eur. J. Med. Chem. 2016, 120, 227–243. [Google Scholar] [CrossRef] [PubMed]
- Fujita, J.; Maeda, Y.; Mizohata, E.; Inoue, T.; Kaul, M.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S.; Matsumura, H. Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ. ACS Chem. Biol. 2017, 12, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Mark, L.; Zhang, Y.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S. Pharmacokinetics and In Vivo Antistaphylococcal Efficacy of TXY541, a 1-Methylpiperidine-4-Carboxamide Prodrug of PC190723. Biochem. Pharmacol. 2013, 86, 1699–1707. [Google Scholar] [CrossRef]
- Tan, C.M.; Therien, A.G.; Lu, J.; Lee, S.H.; Caron, A.; Gill, C.J.; Lebeau-Jacob, C.; Benton-Perdomo, L.; Monteiro, J.M.; Pereira, P.M.; et al. Restoring Methicillin-Resistant Staphylococcus aureus Susceptibility to β-Lactam Antibiotics. Sci. Transl. Med. 2012, 4, 126ra135. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Bi, F.; Zhang, N.; Qin, Y.; Liu, X.; Teng, Y.; Ma, S. Design, Synthesis of Novel 4,5-Dihydroisoxazole-Containing Benzamide Derivatives as Highly Potent FtsZ Inhibitors Capable of Killing a Variety of MDR Staphylococcus aureus. Bioorg. Med. Chem. 2020, 28, 115729. [Google Scholar] [CrossRef] [PubMed]
- Bi, F.; Song, D.; Qin, Y.; Liu, X.; Teng, Y.; Zhang, N.; Zhang, P.; Zhang, N.; Ma, S. Discovery of 1,3,4-Oxadiazol-2-One-Containing Benzamide Derivatives Targeting FtsZ as Highly Potent Agents of Killing a Variety of MDR Bacteria Strains. Bioorg. Med. Chem. 2019, 27, 3179–3193. [Google Scholar] [CrossRef]
- Suigo, L.; Margolin, W.; Ulzurrun, E.; Hrast Rambaher, M.; Zanotto, C.; Sebastián-Pérez, V.; Campillo, N.E.; Straniero, V.; Valoti, E. Benzodioxane–Benzamides as FtsZ Inhibitors: Effects of Linker’s Functionalization on Gram-Positive Antimicrobial Activity. Antibiotics 2023, 12, 1712. [Google Scholar] [CrossRef]
- Kaul, M.; Zhang, Y.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S. Inhibition of RND-Type Efflux Pumps Confers the FtsZ-Directed Prodrug TXY436 With Activity against Gram-Negative Bacteria. Biochem. Pharmacol. 2014, 89, 321–328. [Google Scholar] [CrossRef]
- Delcour, A.H. Outer Membrane Permeability and Antibiotic Resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef]
- Khare, S.; Hsin, J.; Sorto, N.A.; Nepomuceno, G.M.; Shaw, J.T.; Shi, H.; Huang, K.C. FtsZ-Independent Mechanism of Division Inhibition by the Small Molecule PC190723 in Escherichia coli. Adv. Biosyst. 2019, 3, 1900021. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.I. Antibiotic Resistance and Regulation of the Gram-Negative Bacterial Outer Membrane Barrier by Host Innate Immune Molecules. mBio 2016, 7, e01541-16. [Google Scholar] [CrossRef] [PubMed]
- Negash, K.H.; Norris, J.K.S.; Hodgkinson, J.T. Siderophore–Antibiotic Conjugate Design: New Drugs for Bad Bugs? Molecules 2019, 24, 3314. [Google Scholar] [CrossRef]
- Andrews, S.; Norton, I.; Salunkhe, A.S.; Goodluck, H.; Aly, W.S.M.; Mourad-Agha, H.; Cornelis, P. Control of Iron Metabolism in Bacteria. In Metallomics and the Cell; Banci, L., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 203–239. [Google Scholar]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial Iron Homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Finkelstein, R.A.; Sciortino, C.V.; McIntosh, M.A. Role of Iron in Microbe-Host Interactions. Rev. Infect. Dis. 1983, 5, S759–S777. [Google Scholar] [CrossRef]
- Schaible, U.E.; Kaufmann, S.H.E. Iron and Microbial Infection. Nat. Rev. Microbiol. 2004, 2, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.C.; Garcia-Herrero, A.; Johanson, T.H.; Krewulak, K.D.; Lau, C.K.; Peacock, R.S.; Slavinskaya, Z.; Vogel, H.J. Siderophore Uptake in Bacteria and the Battle for Iron with the Host; A Bird’s Eye View. BioMetals 2010, 23, 601–611. [Google Scholar] [CrossRef]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef]
- Sheldon, J.R.; Skaar, E.P. Acinetobacter baumannii Can Use Multiple Siderophores for Iron Acquisition, but Only Acinetobactin is Required for Virulence. PLoS Pathog. 2020, 16, e1008995. [Google Scholar] [CrossRef]
- Hackel, M.A.; Tsuji, M.; Yamano, Y.; Echols, R.; Karlowsky, J.A.; Sahm, D.F. In Vitro Activity of the Siderophore Cephalosporin, Cefiderocol, against Carbapenem-Nonsusceptible and Multidrug-Resistant Isolates of Gram-Negative Bacilli Collected Worldwide in 2014 to 2016. Antimicrob. Agents Chemother. 2018, 62, e01968-17. [Google Scholar] [CrossRef] [PubMed]
- Shortridge, D.; Streit, J.M.; Mendes, R.; Castanheira, M. In Vitro Activity of Cefiderocol against U.S. and European Gram-Negative Clinical Isolates Collected in 2020 as Part of the SENTRY Antimicrobial Surveillance Program. Microbiol. Spectr. 2022, 10, e0271221. [Google Scholar] [CrossRef] [PubMed]
- Yamano, Y. In Vitro Activity of Cefiderocol against a Broad Range of Clinically Important Gram-Negative Bacteria. Clin. Infect. Dis. 2019, 69, S544–S551. [Google Scholar] [CrossRef] [PubMed]
- Neumann, W.; Sassone-Corsi, M.; Raffatellu, M.; Nolan, E.M. Esterase-Catalyzed Siderophore Hydrolysis Activates an Enterobactin–Ciprofloxacin Conjugate and Confers Targeted Antibacterial Activity. J. Am. Chem. Soc. 2018, 140, 5193–5201. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-González, E.; Fujita, J.; Yoshizawa, T.; Nelson, J.M.; Pilch, A.J.; Hillman, E.; Ozawa, M.; Kuroda, N.; Al-Tameemi, H.M.; Boyd, J.M.; et al. Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens. Sci. Rep. 2019, 9, 20092. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Gorostiaga, A.; Palacios, P.; Martínez-Arteaga, R.; Sánchez, M.; Casanova, M.; Vicente, M. Life without Division: Physiology of Escherichia coli FtsZ-Deprived Filaments. mBio 2016, 7, e01620-16. [Google Scholar] [CrossRef]
- Bryan, E.J.; Sagong, H.Y.; Parhi, A.K.; Grier, M.C.; Roberge, J.Y.; LaVoie, E.J.; Pilch, D.S. TXH11106: A Third-Generation MreB Inhibitor with Enhanced Activity against a Broad Range of Gram-Negative Bacterial Pathogens. Antibiotics 2022, 11, 693. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Orhan, G.; Bayram, A.; Zer, Y.; Balci, I. Synergy Tests by E test and Checkerboard Methods of Antimicrobial Combinations against Brucella melitensis. J. Clin. Microbiol. 2005, 43, 140–143. [Google Scholar] [CrossRef]
- Kaul, M.; Mark, L.; Parhi, A.K.; LaVoie, E.J.; Pilch, D.S. Combining the FtsZ-Targeting Prodrug TXA709 and the Cephalosporin Cefdinir Confers Synergy and Reduces the Frequency of Resistance in Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2016, 60, 4290–4296. [Google Scholar] [CrossRef]
- Roemer, T.; Boone, C. Systems-Level Antimicrobial Drug and Drug Synergy Discovery. Nat. Chem. Biol. 2013, 9, 222–231. [Google Scholar] [CrossRef]
- Figge, R.M.; Divakaruni, A.V.; Gober, J.W. MreB, the Cell Shape-Determining Bacterial Actin Homologue, Co-ordinates Cell Wall Morphogenesis in Caulobacter crescentus. Mol. Microbiol. 2004, 51, 1321–1332. [Google Scholar] [CrossRef] [PubMed]
- Den Blaauwen, T.; De Pedro, M.A.; Nguyen-Distèche, M.; Ayala, J.A. Morphogenesis of Rod-Shaped Sacculi. FEMS Microbiol. Rev. 2008, 32, 321–344. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Fisher, Z.B.; Edgar, K.J. Azide Reduction by DTT or Thioacetic Acid Provides Access to Amino and Amido Polysaccharides. Cellulose 2019, 26, 445–462. [Google Scholar] [CrossRef]
- Huang, T.-W.; Lam, I.; Chang, H.-Y.; Tsai, S.-F.; Palsson, B.O.; Charusanti, P. Capsule Deletion Via a λ-Red Knockout System Perturbs Biofilm Formation and Fimbriae Expression in Klebsiella pneumoniae MGH 78578. BMC Res. Notes 2014, 7, 13. [Google Scholar] [CrossRef] [PubMed]
| Strain | MIC (µM) | MBC (µM) | IC50 (µM) | ||
|---|---|---|---|---|---|
| RUP4 | RUP5 | RUP4 | RUP4 * | RUP5 | |
| K. pneumoniae 10031 | 46 | >95 | 92 | 25 ± 1 | >95 |
| A. baumannii 19606 | 185 | >95 | >185 | 24 ± 2 | >95 |
| Added [Fe3+] (µM) | IC50 (µM) |
|---|---|
| 0 | 23 ± 1 |
| 2 | 29 ± 1 |
| 10 | 55 ± 6 |
| 25 | 73 ± 4 |
| Strain | IC50 (µM) |
|---|---|
| WT | 18 ± 1 |
| ΔfepA | 75 ± 5 |
| ΔcirA | 35 ± 5 |
| Δfiu | 22 ± 4 |
| ΔfepC | 5.6 ± 0.1 |
| Test Agent (TA) | FICTA | FICRUP4 | FICI |
|---|---|---|---|
| MEC | 0.0005 | 0.25 | 0.2505 |
| PIP | 0.25 | 0.25 | 0.5 |
| IMI | 0.25 | 0.25 | 0.5 |
| TXH | 0.25 | 0.25 | 0.5 |
| CFZ | 0.008 | 0.5 | 0.508 |
| AMX | 0.031 | 0.5 | 0.531 |
| MER | 0.031 | 0.5 | 0.531 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bryan, E.J.; Qiao, Q.; Wang, Y.; Roberge, J.Y.; LaVoie, E.J.; Pilch, D.S. A FtsZ Inhibitor That Can Utilize Siderophore-Ferric Iron Uptake Transporter Systems for Activity against Gram-Negative Bacterial Pathogens. Antibiotics 2024, 13, 209. https://doi.org/10.3390/antibiotics13030209
Bryan EJ, Qiao Q, Wang Y, Roberge JY, LaVoie EJ, Pilch DS. A FtsZ Inhibitor That Can Utilize Siderophore-Ferric Iron Uptake Transporter Systems for Activity against Gram-Negative Bacterial Pathogens. Antibiotics. 2024; 13(3):209. https://doi.org/10.3390/antibiotics13030209
Chicago/Turabian StyleBryan, Eric J., Qi Qiao, Yuxuan Wang, Jacques Y. Roberge, Edmond J. LaVoie, and Daniel S. Pilch. 2024. "A FtsZ Inhibitor That Can Utilize Siderophore-Ferric Iron Uptake Transporter Systems for Activity against Gram-Negative Bacterial Pathogens" Antibiotics 13, no. 3: 209. https://doi.org/10.3390/antibiotics13030209
APA StyleBryan, E. J., Qiao, Q., Wang, Y., Roberge, J. Y., LaVoie, E. J., & Pilch, D. S. (2024). A FtsZ Inhibitor That Can Utilize Siderophore-Ferric Iron Uptake Transporter Systems for Activity against Gram-Negative Bacterial Pathogens. Antibiotics, 13(3), 209. https://doi.org/10.3390/antibiotics13030209








