Insights into the Microbiome and Antibiotic Resistance Genes from Hospital Environmental Surfaces: A Prime Source of Antimicrobial Resistance
Abstract
1. Introduction
2. Results
2.1. Hospital Sample Collection
2.2. Alpha and Beta Diversity of the Samples
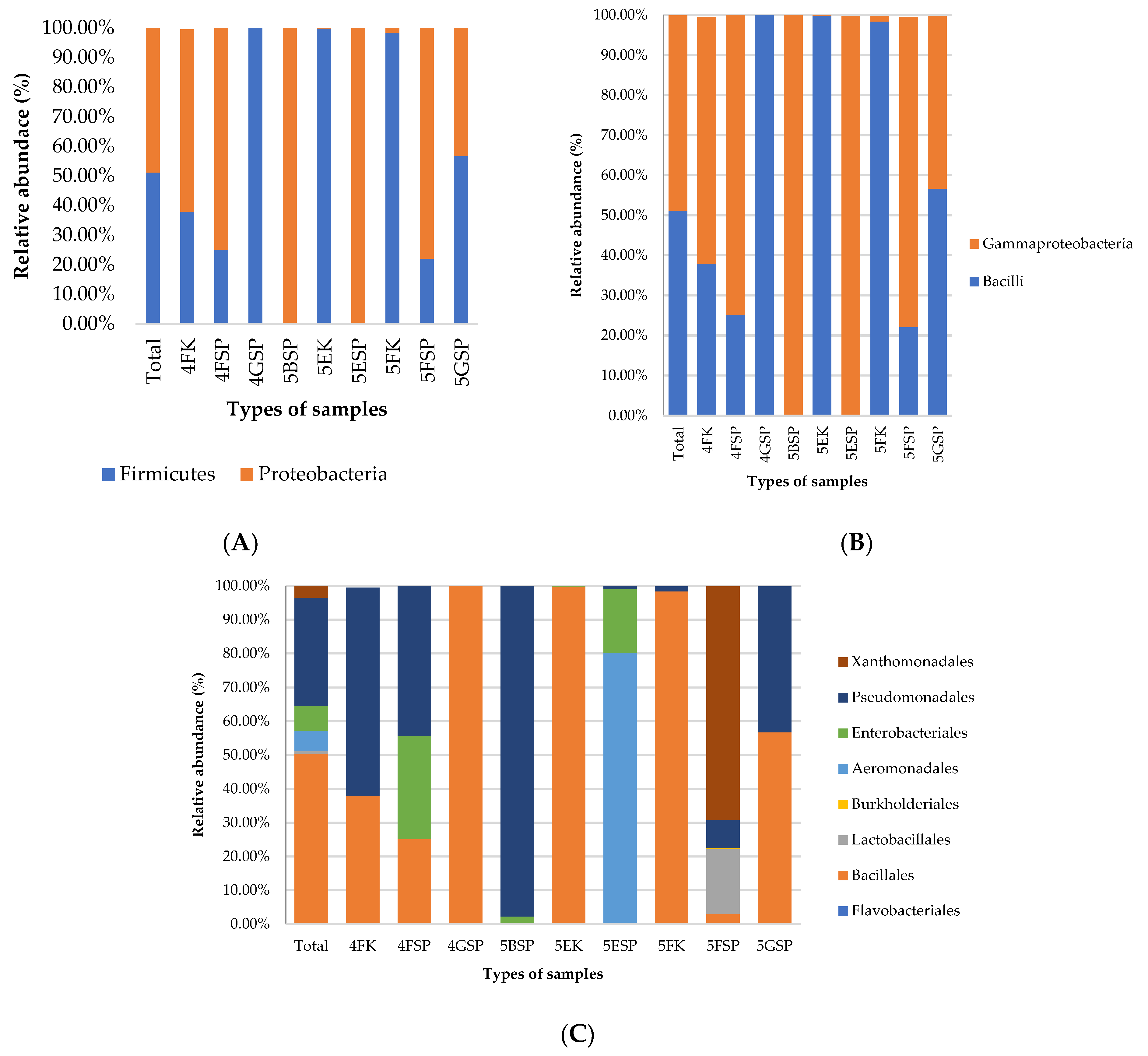
2.3. Taxonomic Profiles at Phylum, Class, and Order Levels
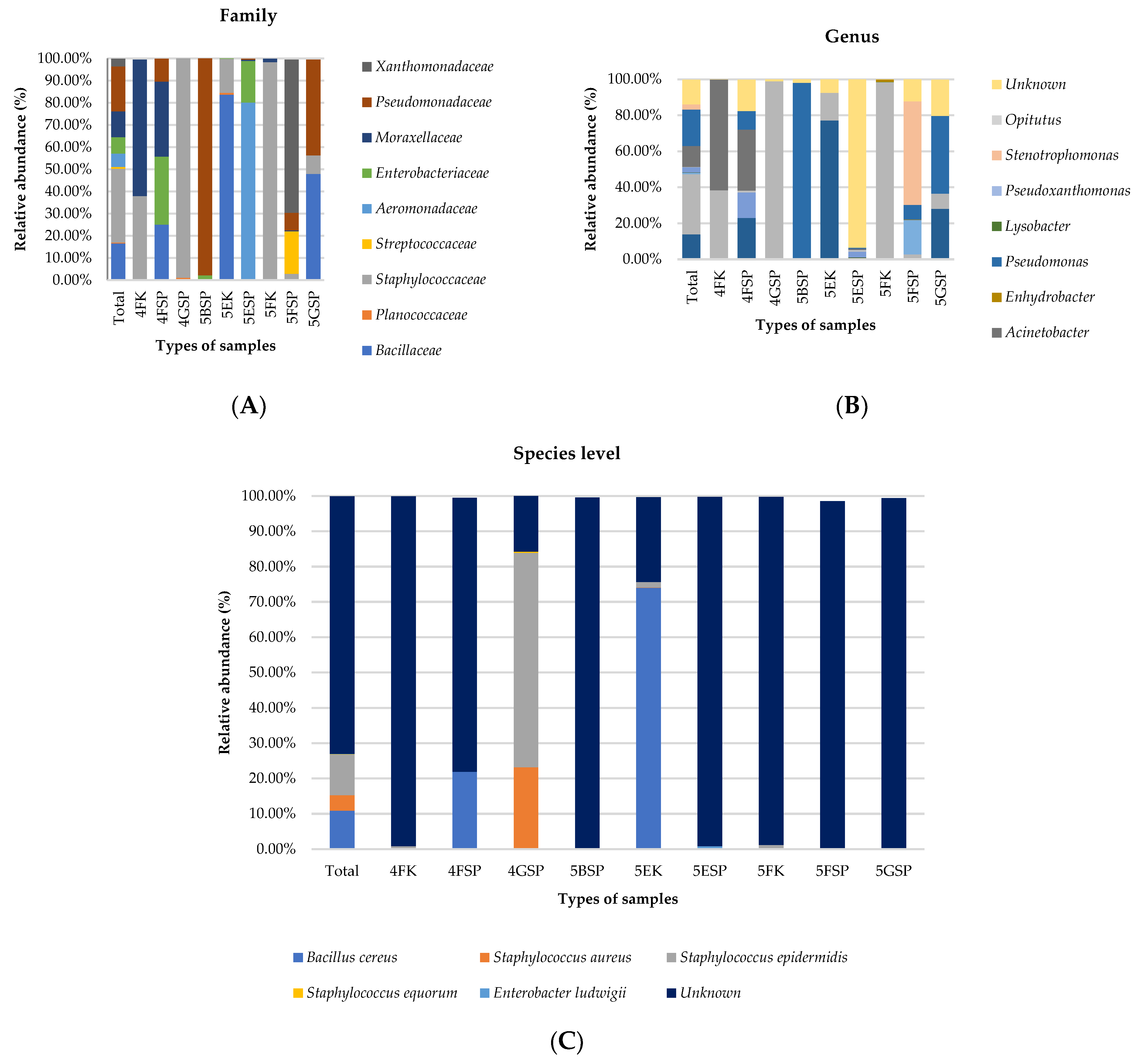
2.4. Taxonomic Profiles at Family, Genus, and Species Levels
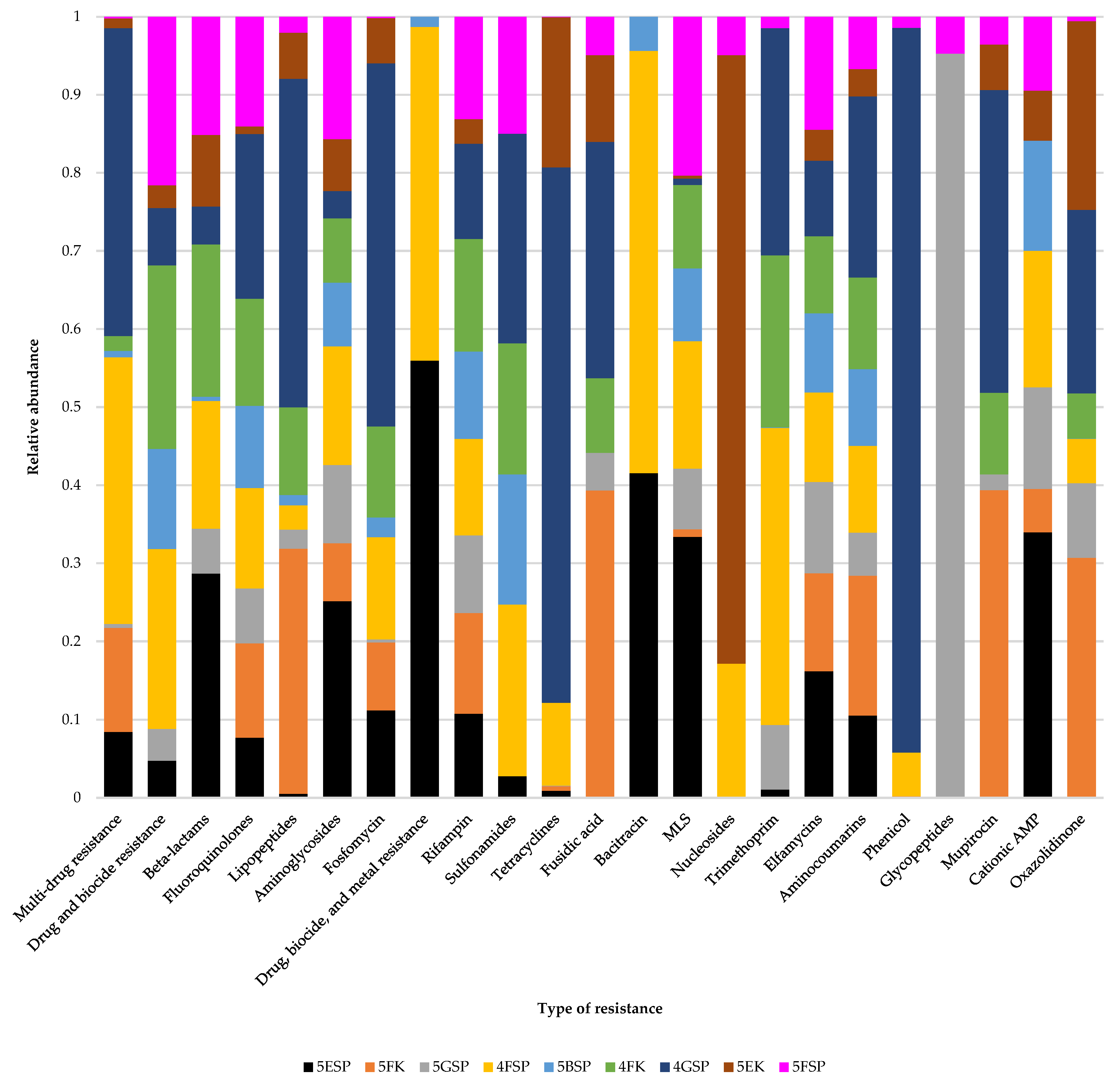
2.5. Antibiotic Resistance Gene Profiles
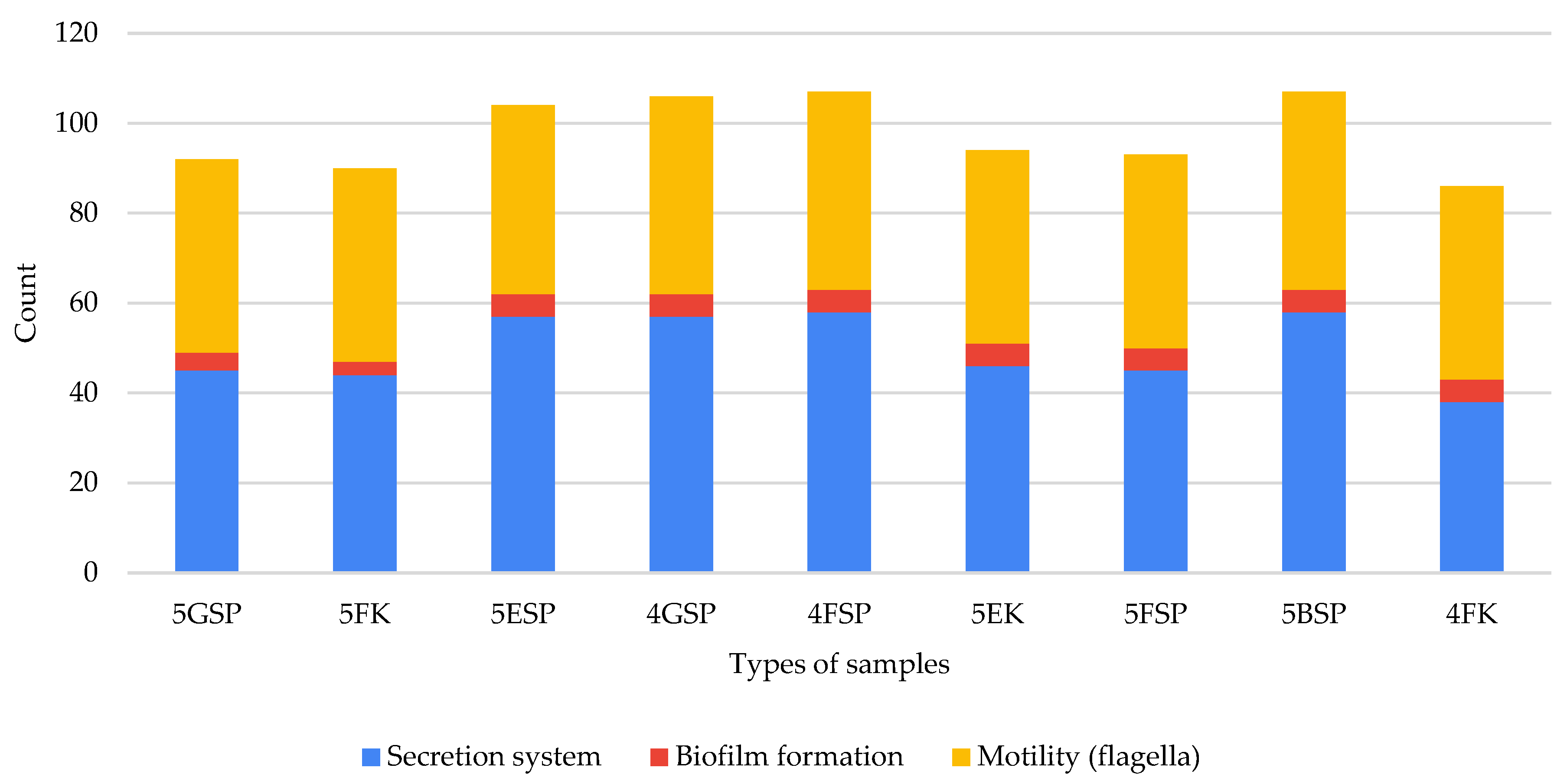
2.6. Virulence Pathways from Hospital Environmental Samples
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. 16S rRNA Amplicon Sequencing
4.3. Shotgun Sequencing to Identify Antibiotic Resistance Genes
4.4. Raw Read Processing
4.5. De Novo Assembly and Identification of AMR Genes
4.6. AMRplusplus2 Quantification of AMR Genes based on MEGARes Database
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haque, M.; McKimm, J.; Sartelli, M.; Dhingra, S.; Labricciosa, F.M.; Islam, S.; Jahan, D.; Nusrat, T.; Chowdhury, T.S.; Coccolini, F.; et al. Strategies to prevent healthcare-associated infections: A narrative overview. Risk Manag. Healthc. Policy 2020, 13, 1765–1780. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.G.; Fasugba, O.; Russo, P.L. Where is the strength of evidence? A review of infection prevention and control guidelines. J. Hosp. Infect. 2020, 105, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Facciolà, A.; Pellicanò, G.F.; Visalli, G.; Paolucci, I.A.; Venanzi Rullo, E.; Ceccarelli, M.; D’Aleo, F.; Di Pietro, A.; Squeri, R.; Nunnari, G.; et al. The role of the hospital environment in the healthcare-associated infections: A general review of the literature. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1266–1278. [Google Scholar] [PubMed]
- Alhumaid, S.; Al Mutair, A.; Al Alawi, Z.; Alsuliman, M.; Ahmed, G.Y.; Rabaan, A.A.; Al-Tawfiq, J.A.; Al-Omari, A. Knowledge of infection prevention and control among healthcare workers and factors influencing compliance: A systematic review. Antimicrob. Resist. Infect. Control 2021, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Knight, G.M.; Costelloe, C.; Deeny, S.R.; Moore, L.S.P.; Hopkins, S.; Johnson, A.P.; Robotham, J.V.; Holmes, A.H. Quantifying where human acquisition of antibiotic resistance occurs: A mathematical modelling study. BMC Med. 2018, 16, 137. [Google Scholar] [CrossRef] [PubMed]
- Chai, C.S.; Ng, D.L.; Chua, W.J.; Tung, Y.Z.; Sindeh, W.; Ibrahim, M.A.; Badlishah Sham, S.F.; Tan, S.B. Knowledge, attitude, and practices among the general population during the later stage of the COVID-19 pandemic in Malaysia: A cross-sectional study. Risk Manag. Healthc. Policy 2022, 15, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Sukhum, K.V.; Newcomer, E.P.; Cass, C.; Wallace, M.A.; Johnson, C.; Fine, J.; Sax, S.; Barlet, M.H.; Burnham, C.D.; Dantas, G.; et al. Antibiotic-resistant organisms establish reservoirs in new hospital-built environments and are related to patient blood infection isolates. Commun. Med. 2022, 2, 62. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Cave, R.; Cole, J.; Mkrtchyan, H.V. Surveillance and prevalence of antimicrobial resistant bacteria from public settings within urban built environments: Challenges and opportunities for hygiene and infection control. Environ. Int. 2021, 157, 106836. [Google Scholar] [CrossRef]
- Liu, S.; Moon, C.D.; Zheng, N.; Huws, S.; Zhao, S.; Wang, J. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 2022, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, F.X.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.; Hao, H.; et al. Advances in metagenomics and its application in environmental microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Li, Y.; Guo, H.; Zhou, J.; Wang, T. Metagenomic analysis revealed sources, transmission, and health risk of antibiotic resistance genes in confluence of Fenhe, Weihe, and Yellow Rivers. Sci. Total Environ. 2023, 858, 159913. [Google Scholar] [CrossRef]
- Hospital Canselor Tuanku Muhriz. Hospital Statistics. Available online: https://hctm.ukm.my/statistik-hospital/ (accessed on 27 July 2023).
- Mazloomirad, F.; Hasanzadeh, S.; Sharifi, A.; Nikbakht, G.; Roustaei, N.; Khoramrooz, S.S. Identification and detection of pathogenic bacteria from patients with hospital-acquired pneumonia in southwestern Iran; evaluation of biofilm production and molecular typing of bacterial isolates. BMC Pulm. Med. 2021, 21, 408. [Google Scholar] [CrossRef]
- Dougnon, V.T.; Sintondji, K.; Koudokpon, C.H.; Houéto, M.; Agbankpé, A.J.; Assogba, P.; Oussou, A.; Gnamy, A.; Legba, B.; Idrissou, A.; et al. Investigating catheter-related infections in Southern Benin Hospitals: Identification, susceptibility, and resistance genes of involved bacterial strains. Microorganisms 2023, 11, 617. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, P.; Delicati, A.; Caenazzo, L. Human microbiome and microbiota identification for preventing and controlling healthcare-associated infections: A systematic review. Front. Public Health 2022, 10, 989496. [Google Scholar] [CrossRef]
- Ismail, M.A.H.; Kamarudin, N.; Abdul Samat, M.N.; Raja Abdul Rahman, R.M.F.; Saimun, S.; Tan, T.L.; Neoh, H.M. Methicillin-resistant Staphylococcus aureus (MRSA) clonal replacement in a malaysian teaching hospital: Findings from an eight-year interval molecular surveillance. Antibiotics 2021, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Mohd Pauzi, Z.; Rasool Hassan, B.A.; Neo, C.F.; Mohammed, A.H.; Blebil, A.; Dujaili, J. Antibiotic use and resistance in a tertiary care hospital: Knowledge and attitude among patients of orthopaedic and surgical wards in Malaysia. J Pharm. Health Ser. Res. 2022, rmab068. [Google Scholar] [CrossRef]
- Firesbhat, A.; Tigabu, A.; Tegene, B.; Gelaw, B. Bacterial profile of high-touch surfaces, leftover drugs and antiseptics together with their antimicrobial susceptibility patterns at University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Microbiol. 2021, 21, 309. [Google Scholar] [CrossRef]
- Chaoui, L.; Mhand, R.; Mellouki, F.; Rhallabi, N. Contamination of the surfaces of a health care environment by multidrug-resistant (MDR) bacteria. Int. J. Microbiol. 2019, 2019, 3236526. [Google Scholar] [CrossRef]
- Kembel, S.W.; Jones, E.; Kline, J.; Northcutt, D.; Stenson, J.; Womack, A.M.; Bohannan, B.J.; Brown, G.Z.; Green, J.L. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012, 6, 1469–1479. [Google Scholar] [CrossRef]
- Ruiz-Gil, T.; Acuña, J.J.; Fujiyoshi, S.; Tanaka, D.; Noda, J.; Maruyama, F.; Jorquera, M.A. Airborne bacterial communities of outdoor environments and their associated influencing factors. Environ. Int. 2020, 145, 106156. [Google Scholar] [CrossRef]
- Gupta, S.; Mortensen, M.S.; Schjørring, S.; Trivedi, U.; Vestergaard, G.; Stokholm, J.; Bisgaard, H.; Krogfelt, K.A.; Sørensen, S.J. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun. Biol. 2019, 2, 291. [Google Scholar] [CrossRef] [PubMed]
- Withey, Z.; Goodall, T.; MacIntyre, S.; Gweon, H. Characterization of communal sink drain communities of a university campus. Environ. DNA 2021, 3, 901–911. [Google Scholar] [CrossRef]
- Silva, H.O.; Lima, J.A.S.; Aguilar, C.E.G.; Rossi, G.A.M.; Mathias, L.A.; Vidal, A.M.C. Efficiency of different disinfectants on Bacillus cereus Sensu Stricto biofilms on stainless-steel surfaces in contact with milk. Front. Microbiol. 2018, 9, 2934. [Google Scholar] [CrossRef] [PubMed]
- Betchen, M.; Giovinco, H.M.; Curry, M.; Luu, J.; Fraimow, H.; Carabetta, V.J.; Nahra, R. Evaluating the effectiveness of hospital antiseptics on multidrug-resistant Acinetobacter baumannii: Understanding the relationship between microbicide and antibiotic resistance. Antibiotics 2022, 11, 614. [Google Scholar] [CrossRef]
- Li, Y.; Song, Y.; Huang, Z.; Mei, L.; Jiang, M.; Wang, D.; Wei, Q. Screening of Staphylococcus aureus for disinfection evaluation and transcriptome analysis of high tolerance to chlorine-containing disinfectants. Microorganisms 2023, 11, 475. [Google Scholar] [CrossRef]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas aeruginosa biofilm: A review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Alajlan, A.A.; Mukhtar, L.E.; Almussallam, A.S.; Alnuqaydan, A.M.; Albakiri, N.S.; Almutari, T.F.; Bin Shehail, K.M.; Aldawsari, F.S.; Alajel, S.M. Assessment of disinfectant efficacy in reducing microbial growth. PLoS ONE 2022, 17, e0269850. [Google Scholar] [CrossRef]
- Jamaluddin, N.A.H.; Periyasamy, P.; Lau, C.L.; Ponnampalavanar, S.; Lai, P.S.M.; Ramli, R.; Tan, T.L.; Kori, N.; Yin, M.K.; Azman, N.J.; et al. Point prevalence survey of antimicrobial use in a Malaysian tertiary care university hospital. Antibiotics 2021, 10, 531. [Google Scholar] [CrossRef]
- Turner, J.; Muraoka, A.; Bedenbaugh, M.; Childress, B.; Pernot, L.; Wiencek, M.; Peterson, Y.K. The chemical relationship among beta-lactam antibiotics and potential impacts on reactivity and decomposition. Front. Microbiol. 2022, 13, 807955. [Google Scholar] [CrossRef]
- Madaha, E.L.; Mienie, C.; Gonsu, H.K.; Bughe, R.N.; Fonkoua, M.C.; Mbacham, W.F.; Alayande, K.A.; Bezuidenhout, C.C.; Ateba, C.N. Whole-genome sequence of multi-drug resistant Pseudomonas aeruginosa strains UY1PSABAL and UY1PSABAL2 isolated from human broncho-alveolar lavage, Yaoundé, Cameroon. PLoS ONE 2020, 15, e0238390. [Google Scholar] [CrossRef]
- Wareth, G.; Brandt, C.; Sprague, L.D.; Neubauer, H.; Pletz, M.W. WGS based analysis of acquired antimicrobial resistance in human and non-human Acinetobacter baumannii isolates from a German perspective. BMC Microbiol. 2021, 21, 210. [Google Scholar] [CrossRef]
- Carvalho, I.; Chenouf, N.S.; Carvalho, J.A.; Castro, A.P.; Silva, V.; Capita, R.; Alonso-Calleja, C.; Enes Dapkevicius, M.L.N.; Igrejas, G.; Torres, C.; et al. Multidrug-resistant Klebsiella pneumoniae harboring extended spectrum β-lactamase encoding genes isolated from human septicemias. PLoS ONE 2021, 16, e0250525. [Google Scholar] [CrossRef] [PubMed]
- Navaneethan, Y.; Effarizah, M.E. Prevalence, toxigenic profiles, multidrug resistance, and biofilm formation of Bacillus cereus isolated from ready-to eat cooked rice in Penang, Malaysia. Food Control 2021, 121, 107553. [Google Scholar] [CrossRef]
- Liu, C.; Yu, P.; Yu, S.; Wang, J.; Guo, H.; Zhang, Y.; Zhang, J.; Liao, X.; Li, C.; Wu, S.; et al. Assessment and molecular characterization of Bacillus cereus isolated from edible fungi in China. BMC Microbiol. 2020, 20, 310. [Google Scholar] [CrossRef] [PubMed]
- Torki Baghbadorani, S.; Rahimi, E.; Shakerian, A. Investigation of virulence and antibiotic-resistance of Bacillus cereus isolated from various spices. Can. J. Infect. Dis. Med. Microbiol. 2023, 2023, 8390778. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Institute for Medical Research. National Antibiotic Resistance Surveillance Report 2022. Available online: https://imr.nih.gov.my/images/uploads/NSAR/2022/NSAR-REPORT_2022_to-be-published.pdf (accessed on 1 September 2023).
- Devan, S.S.; Aklilu, E.; Hamdan, R.H.; Lemlem, M.; Zakaria, Z. Detection of colistin-resistant Escherichia coli isolated from broiler chickens in Kelantan, Malaysia. Trop. Biomed. 2022, 39, 197–202. [Google Scholar] [PubMed]
- Olaitan, A.O.; Dandachi, I.; Baron, S.A.; Daoud, Z.; Morand, S.; Rolain, J.M. Banning colistin in feed additives: A small step in the right direction. Lancet Infect. Dis. 2021, 21, 29–30. [Google Scholar] [CrossRef]
- Wang, C.; Feng, Y.; Liu, L.; Wei, L.; Kang, M.; Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg. Microbes Infect. 2020, 9, 508–516. [Google Scholar] [CrossRef]
- Xu, L.; Wan, F.; Fu, H.; Tang, B.; Ruan, Z.; Xiao, Y.; Luo, Q. Emergence of colistin resistance gene mcr-10 in Enterobacterales isolates recovered from fecal samples of chickens, slaughterhouse workers, and a nearby resident. Microbiol. Spec. 2022, 10, e0041822. [Google Scholar] [CrossRef]
- Khan, S.; Scholey, J.M. Assembly, functions and evolution of archaella, flagella and cilia. Curr. Biol. 2018, 28, R278–R292. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.J.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Cherrak, Y.; Flaugnatti, N.; Durand, E.; Journet, L.; Cascales, E. Structure and activity of the Type VI secretion system. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Sgro, G.G.; Oka, G.U.; Souza, D.P.; Cenens, W.; Bayer-Santos, E.; Matsuyama, B.Y.; Bueno, N.F.; Dos Santos, T.R.; Alvarez-Martinez, C.E.; Salinas, R.K.; et al. Bacteria-Killing type IV secretion systems. Front. Microbiol. 2019, 10, 1078. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, S.; Wang, J.; Al-Shamiri, M.M.; Han, B.; Chen, Y.; Han, S.; Han, L. Uncovering the secretion systems of Acinetobacter baumannii: Structures and functions in pathogenicity and antibiotic resistance. Antibiotics 2023, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.J.; Peri, A.M.; Lüftinger, L.; Beisken, S.; Bergh, H.; Forde, B.M.; Buckley, C.; Cuddihy, T.; Tan, P.; Paterson, D.L.; et al. Optimized method for bacterial nucleic acid extraction from positive blood culture broth for whole-genome sequencing, resistance phenotype prediction, and downstream molecular applications. J. Clin. Microbiol. 2022, 60, e0101222. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Seemann, T. Abricate, Github. Available online: https://github.com/tseemann/abricate (accessed on 1 September 2023).
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Lakin, S.M.; Dean, C.; Noyes, N.R.; Dettenwanger, A.; Ross, A.S.; Doster, E.; Rovira, P.; Abdo, Z.; Jones, K.L.; Ruiz, J.; et al. MEGARes: An antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res. 2017, 45, D574–D580. [Google Scholar] [CrossRef]

| No. | Ward | Location | Blood Agar | MacConkey Agar | Mannitol Salt Agar |
|---|---|---|---|---|---|
| 1. | 4F | SP | GNR | GNR | GNR in chains |
| SS | NG | GPR | NG | ||
| K | Mixed growth positive/negative cocci | GN Diplo | NG | ||
| 2. | 4G | SP | GPC in cluster | NG | Mixed growth positive/negative cocci |
| SS | NG | GNR | NG | ||
| K | NG | NG | NG | ||
| 3. | 4H | SP | NG | NG | NG |
| SS | NG | NG | NG | ||
| K | NG | NG | NG | ||
| 4. | 4J | SP | NG | NG | NG |
| SS | NG | NG | NG | ||
| K | NG | NG | NG | ||
| 5. | 5F | SP | GPC/GNR | GNR | NG |
| SS | GNC in pairs | NG | NG | ||
| K | GPC/GNC | GPR | GPC | ||
| 6. | 5G | SP | Growth | GNR | GNR |
| SS | NG | NG | NG | ||
| K | NG | NG | NG | ||
| 7. | 5E | SP | GNC | GNC | GNC |
| SS | NG | NG | NG | ||
| K | GPC | NG | GPC | ||
| 8. | 5B | SP | GNR | GNR | mixed growth |
| SS | NG | NG | NG | ||
| K | NG | NG | NG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanafiah, A.; Sukri, A.; Yusoff, H.; Chan, C.S.; Hazrin-Chong, N.H.; Salleh, S.A.; Neoh, H.-m. Insights into the Microbiome and Antibiotic Resistance Genes from Hospital Environmental Surfaces: A Prime Source of Antimicrobial Resistance. Antibiotics 2024, 13, 127. https://doi.org/10.3390/antibiotics13020127
Hanafiah A, Sukri A, Yusoff H, Chan CS, Hazrin-Chong NH, Salleh SA, Neoh H-m. Insights into the Microbiome and Antibiotic Resistance Genes from Hospital Environmental Surfaces: A Prime Source of Antimicrobial Resistance. Antibiotics. 2024; 13(2):127. https://doi.org/10.3390/antibiotics13020127
Chicago/Turabian StyleHanafiah, Alfizah, Asif Sukri, Hamidah Yusoff, Chia Sing Chan, Nur Hazlin Hazrin-Chong, Sharifah Azura Salleh, and Hui-min Neoh. 2024. "Insights into the Microbiome and Antibiotic Resistance Genes from Hospital Environmental Surfaces: A Prime Source of Antimicrobial Resistance" Antibiotics 13, no. 2: 127. https://doi.org/10.3390/antibiotics13020127
APA StyleHanafiah, A., Sukri, A., Yusoff, H., Chan, C. S., Hazrin-Chong, N. H., Salleh, S. A., & Neoh, H.-m. (2024). Insights into the Microbiome and Antibiotic Resistance Genes from Hospital Environmental Surfaces: A Prime Source of Antimicrobial Resistance. Antibiotics, 13(2), 127. https://doi.org/10.3390/antibiotics13020127






