General and Specific Cytotoxicity of Chimeric Antisense Oligonucleotides in Bacterial Cells and Human Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Bioinformatical Analyses
2.2.2. Bacterial Cultivation
2.2.3. Cell Cultures and MTT Assay
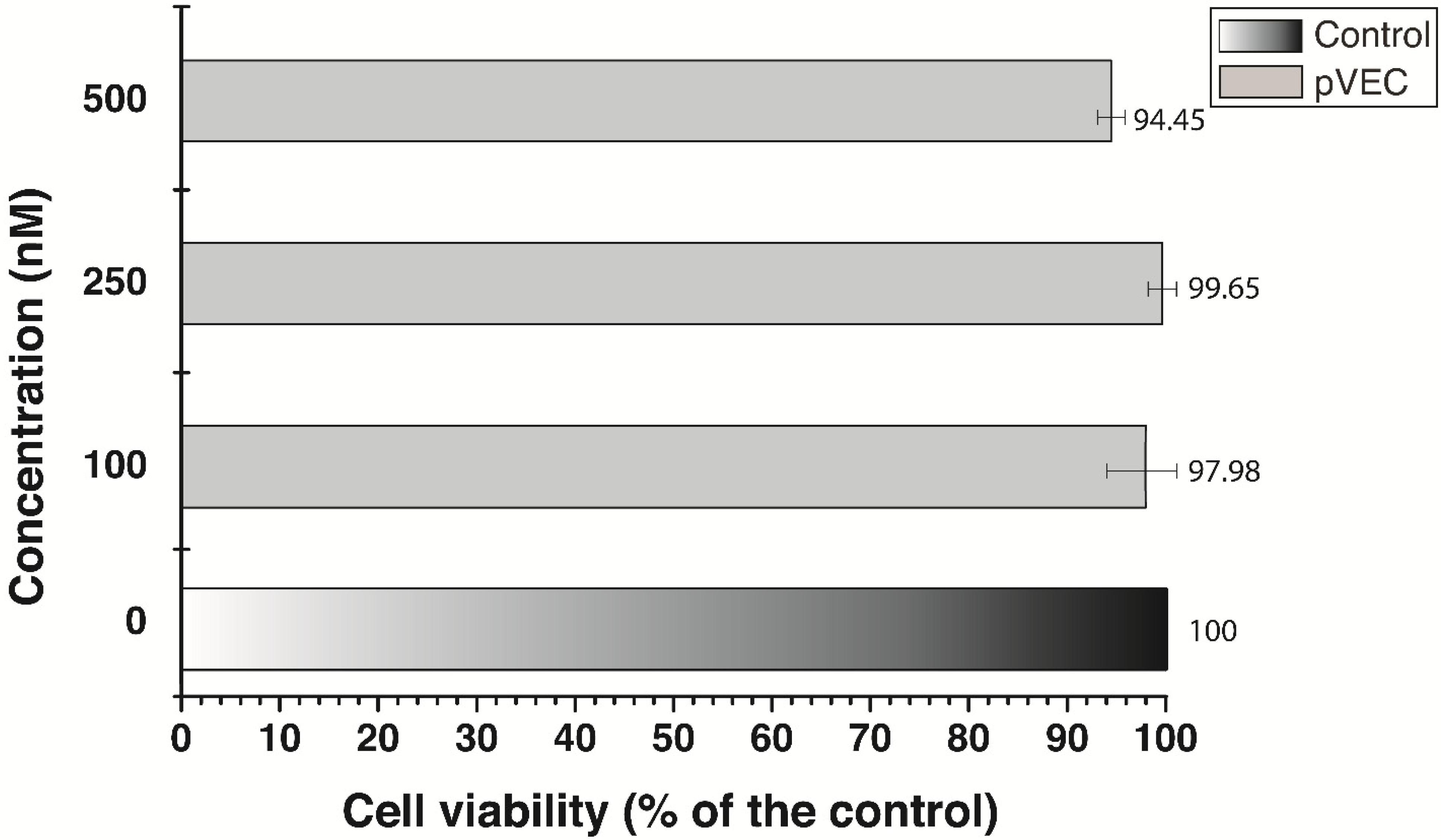
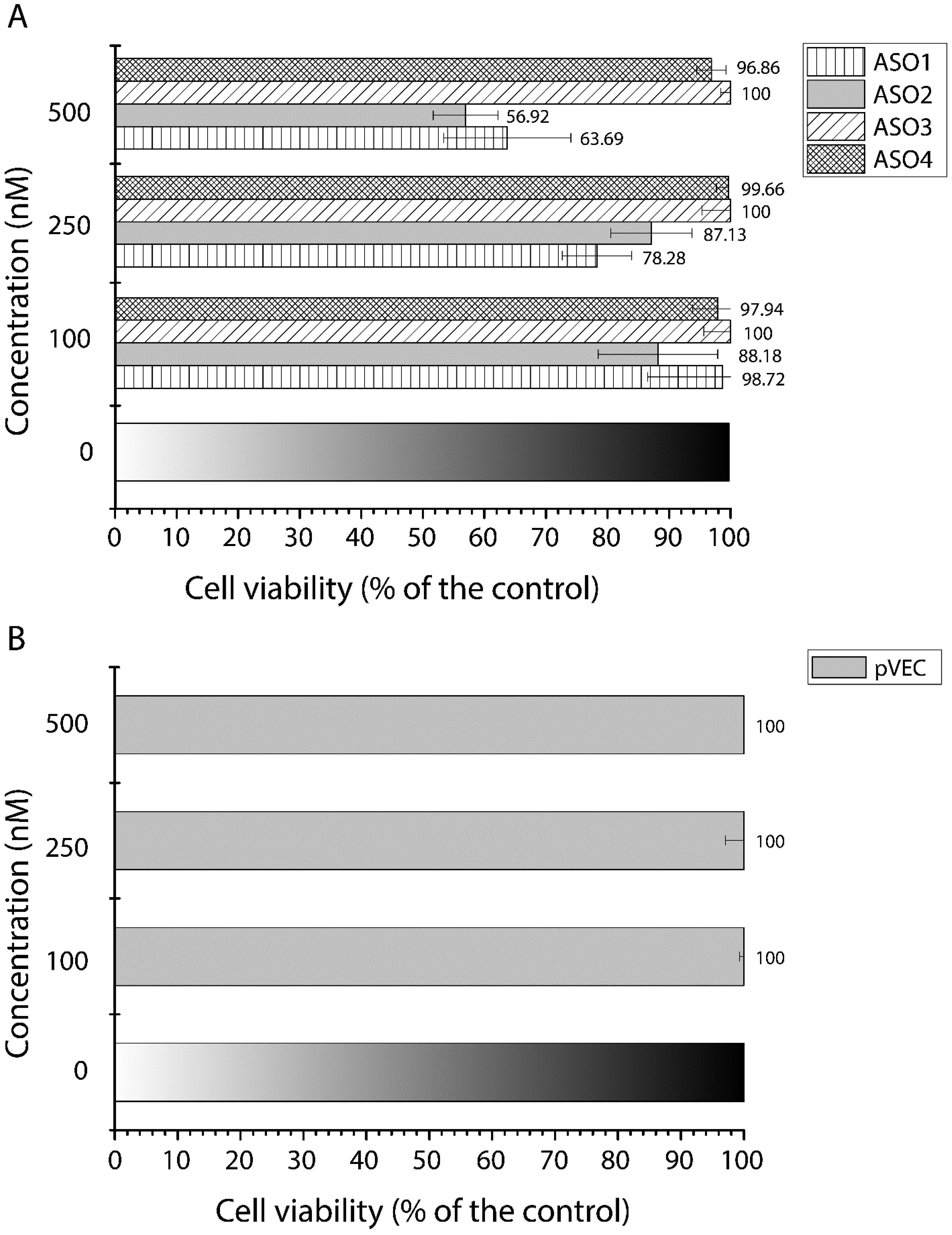
3. Results
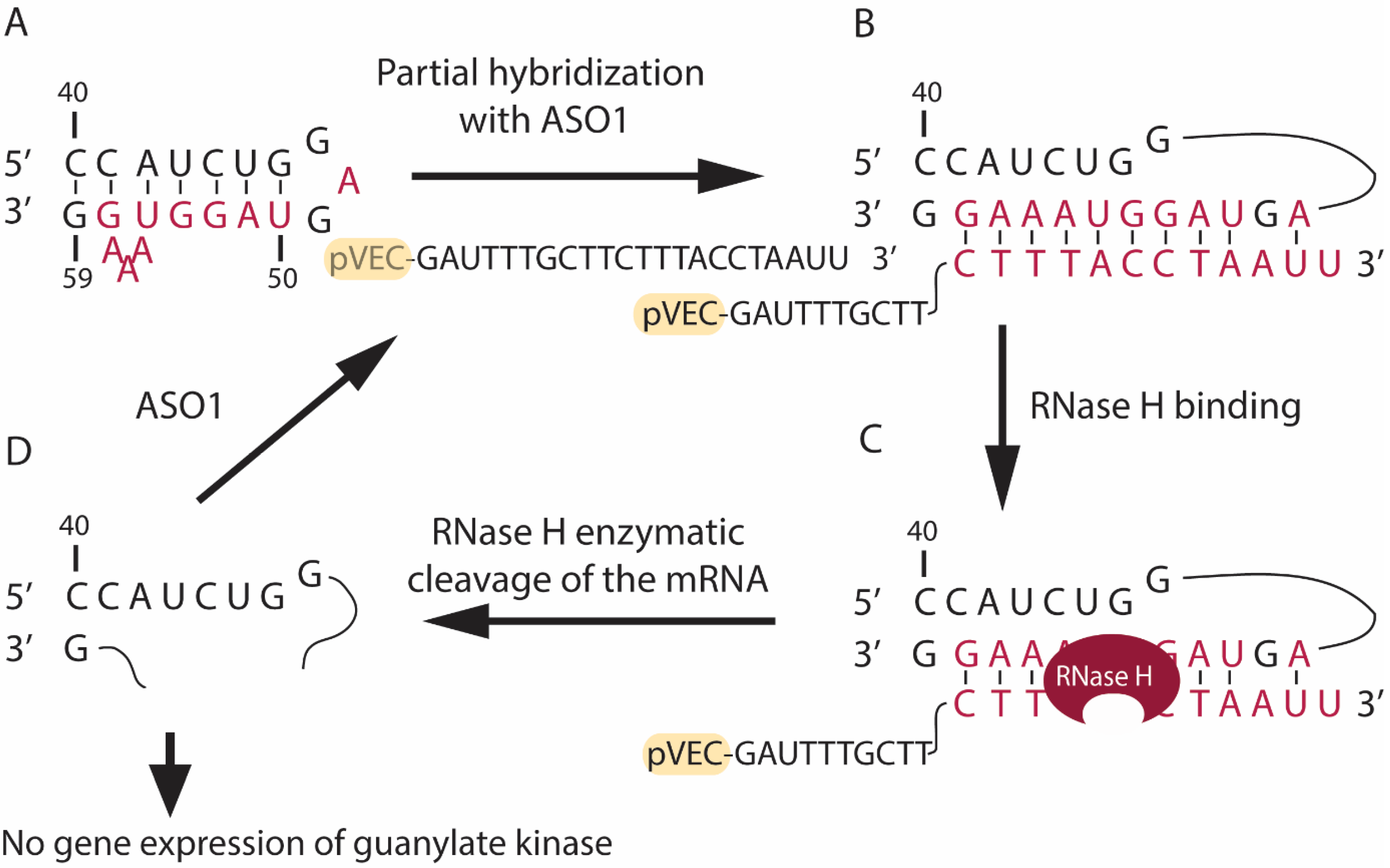
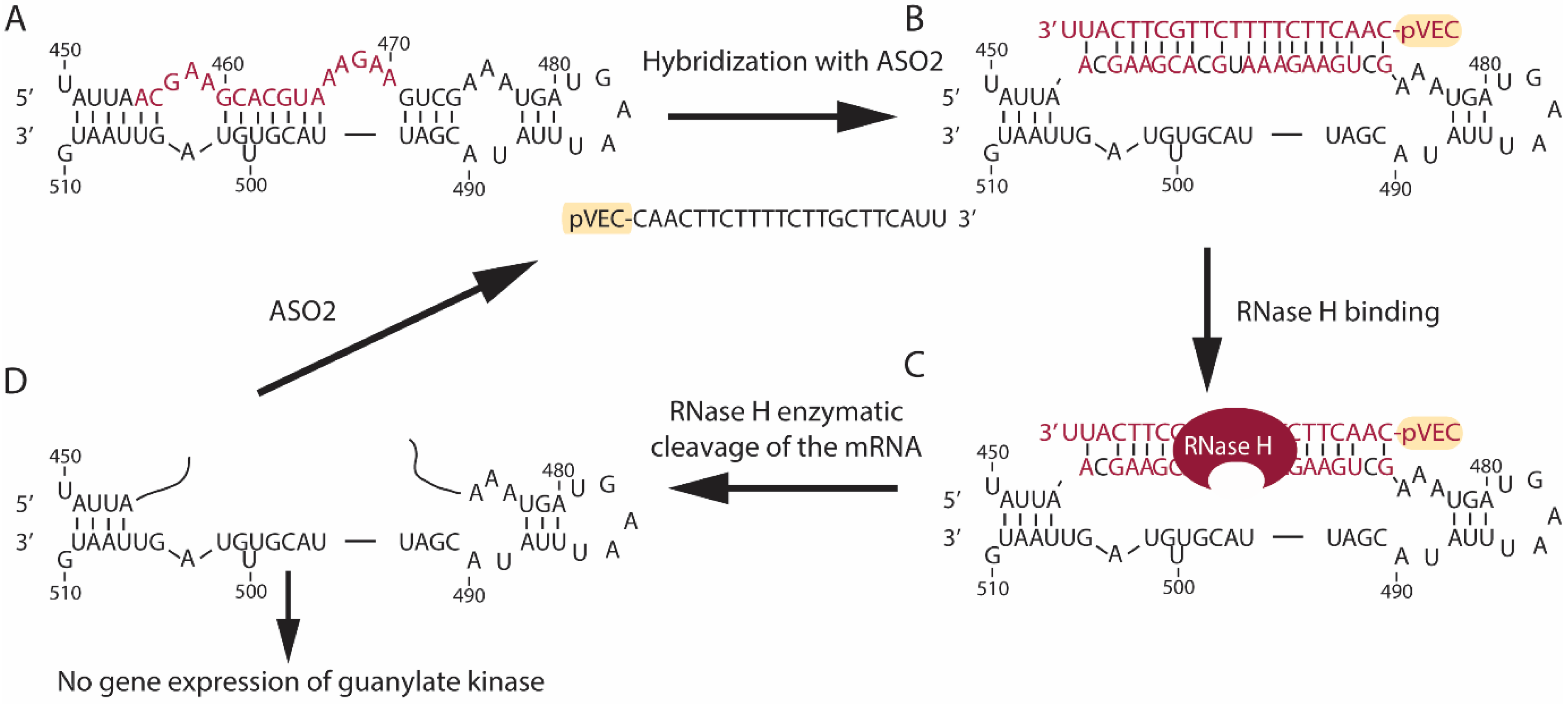
3.1. Design of Antisense Oligonucleotides
3.2. Bioinformatics Analyses
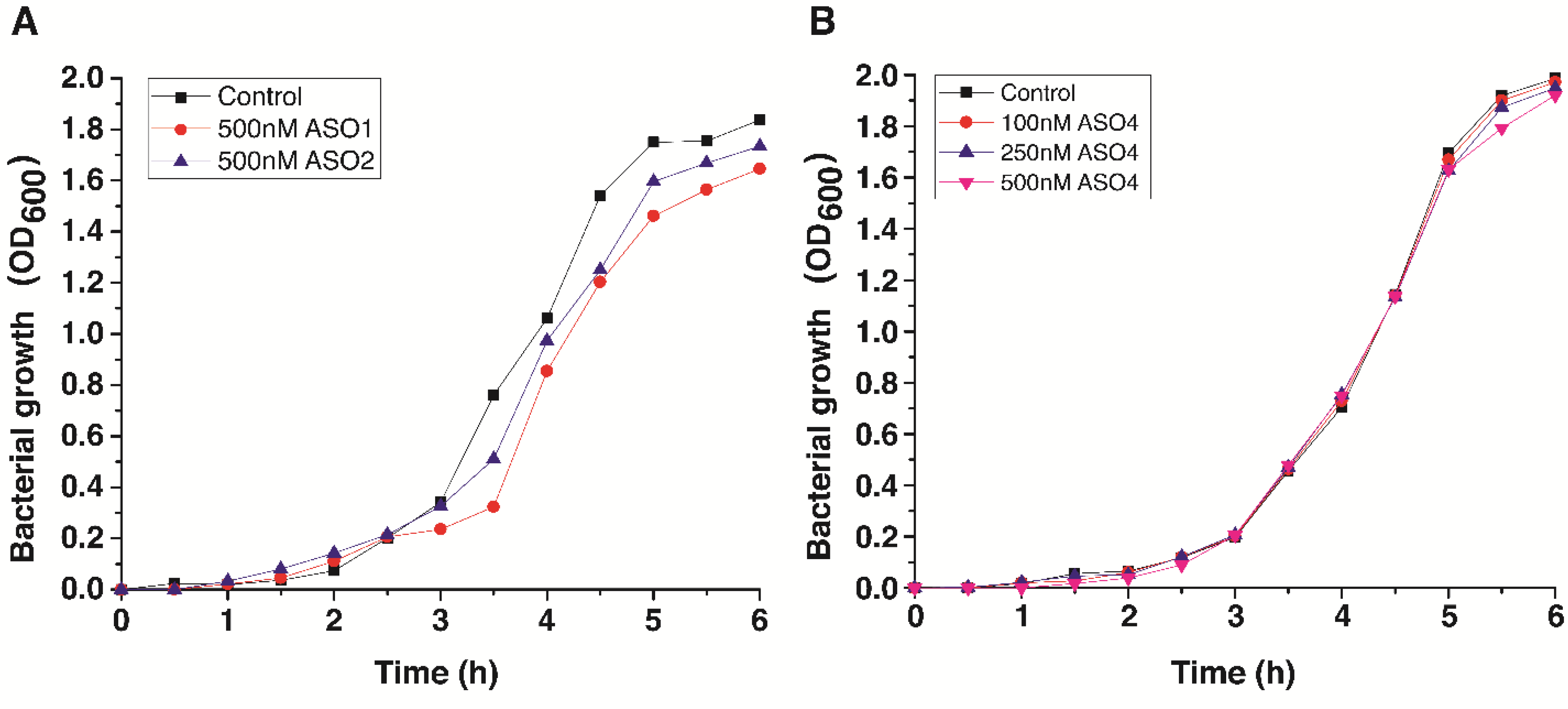
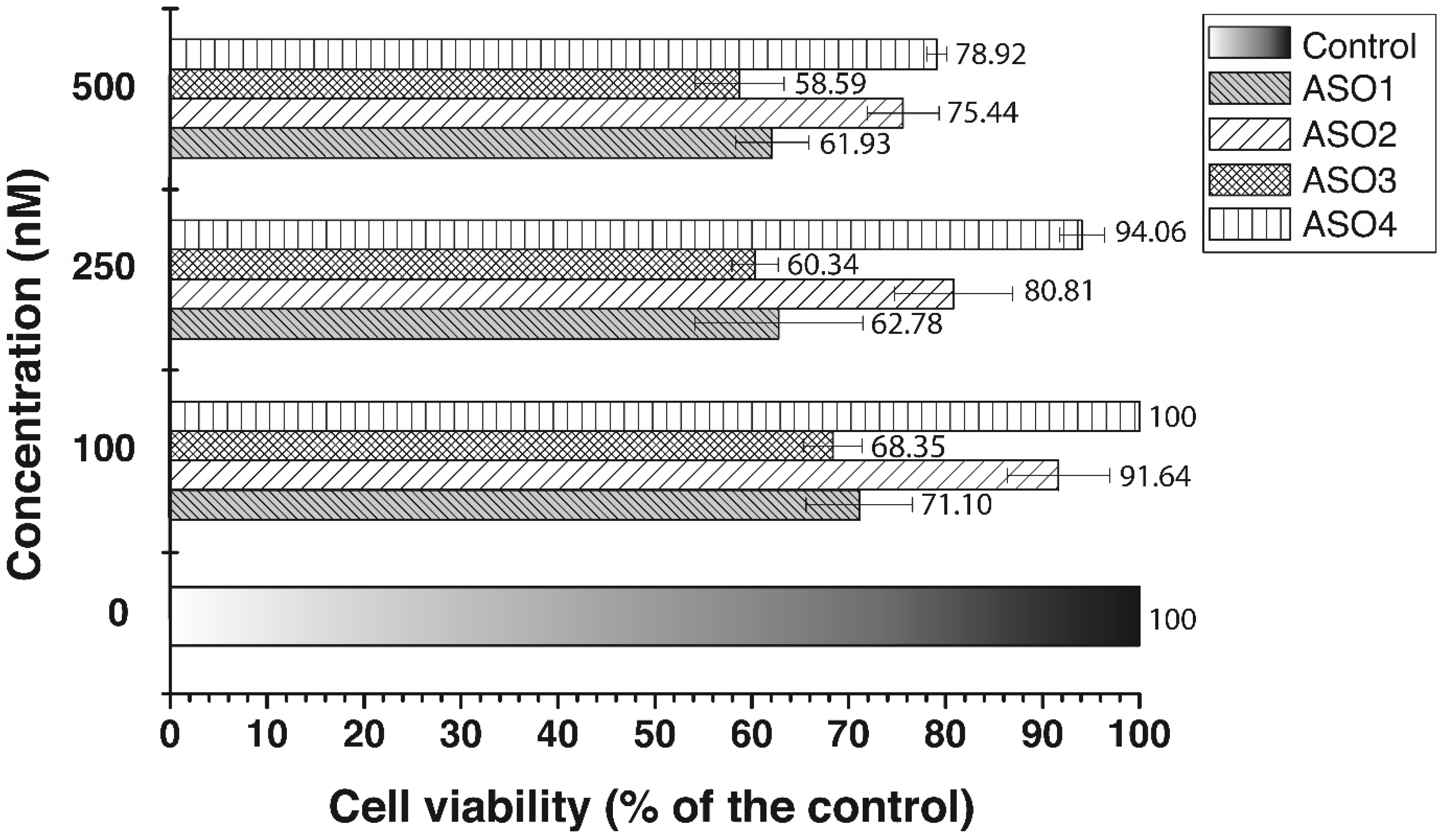
3.3. Cell Growth Inhibition by Antisense Oligonucleotides
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Aboul-Fadl, T. Antisense oligonucleotide technologies in drug discovery. Expert Opin. Drug Discov. 2006, 1, 285–288. [Google Scholar] [CrossRef]
- Chan, J.H.; Lim, S.; Wong, W.S. Antisense oligonucleotides: From design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 533–540. [Google Scholar] [CrossRef]
- Mansoor, M.; Melendez, A.J. Advances in antisense oligonucleotide development for target identification, validation, and as novel therapeutics. Gene Regul. Syst. Biol. 2008, 2, 275–295. [Google Scholar] [CrossRef]
- Pavlova, N.; Traykovska, M.; Penchovsky, R. Targeting FMN, TPP, SAM-I, and glmS Riboswitches with Chimeric Antisense Oligonucleotides for Completely Rational Antibacterial Drug Development. Antibiotics 2023, 12, 1607. [Google Scholar] [CrossRef]
- Traykovska, M.; Penchovsky, R. Engineering Antisense Oligonucleotides as Antibacterial Agents That Target FMN Riboswitches and Inhibit the Growth of Staphylococcus aureus, Listeria monocytogenes, and Escherichia coli. ACS Synth. Biol. 2022, 11, 1845–1855. [Google Scholar] [CrossRef]
- Traykovska, M.; Popova, K.B.; Penchovsky, R. Targeting glmS Ribozyme with Chimeric Antisense Oligonucleotides for Antibacterial Drug Development. ACS Synth. Biol. 2021, 10, 3167–3176. [Google Scholar] [CrossRef]
- Traykovska, M.; Penchovsky, R. Targeting SAM-I Riboswitch Using Antisense Oligonucleotide Technology for Inhibiting the Growth of Staphylococcus aureus and Listeria monocytogenes. Antibiotics 2022, 11, 1662. [Google Scholar] [CrossRef]
- Traykovska, M.; Otcheva, L.A.; Penchovsky, R. Targeting TPP Riboswitches Using Chimeric Antisense Oligonucleotide Technology for Antibacterial Drug Development. ACS Appl. Bio Mater. 2022, 5, 4896–4902. [Google Scholar] [CrossRef]
- Marafini, I.; Monteleone, G. Inflammatory bowel disease: New therapies from antisense oligonucleotides. Ann. Med. 2018, 50, 361–370. [Google Scholar] [CrossRef]
- Di Fusco, D.; Dinallo, V.; Marafini, I.; Figliuzzi, M.M.; Romano, B.; Monteleone, G. Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease. Front. Pharmacol. 2019, 10, 305. [Google Scholar] [CrossRef]
- Lane, R.M.; Smith, A.; Baumann, T.; Gleichmann, M.; Norris, D.; Bennett, C.F.; Kordasiewicz, H. Translating Antisense Technology into a Treatment for Huntington’s Disease. Methods Mol. Biol. 2018, 1780, 497–523. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Leavitt, B.R.; Landwehrmeyer, G.B.; Wild, E.J.; Saft, C.; Barker, R.A.; Blair, N.F.; Craufurd, D.; Priller, J.; Rickards, H.; et al. Targeting Huntingtin Expression in Patients with Huntington’s Disease. N. Engl. J. Med. 2019, 380, 2307–2316. [Google Scholar] [CrossRef]
- Fischbeck, K.H.; Wexler, N.S. Oligonucleotide Treatment for Huntington’s Disease. N. Engl. J. Med. 2019, 380, 2373–2374. [Google Scholar] [CrossRef]
- Wood, M.J.A.; Talbot, K.; Bowerman, M. Spinal muscular atrophy: Antisense oligonucleotide therapy opens the door to an integrated therapeutic landscape. Hum. Mol. Genet. 2017, 26, R151–R159. [Google Scholar] [CrossRef]
- Corey, D.R. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat. Neurosci. 2017, 20, 497–499. [Google Scholar] [CrossRef]
- Klim, J.R.; Vance, C.; Scotter, E.L. Antisense oligonucleotide therapies for Amyotrophic Lateral Sclerosis: Existing and emerging targets. Int. J. Biochem. Cell Biol. 2019, 110, 149–153. [Google Scholar] [CrossRef]
- Ly, C.V.; Miller, T.M. Emerging antisense oligonucleotide and viral therapies for amyotrophic lateral sclerosis. Curr. Opin. Neurol. 2018, 31, 648–654. [Google Scholar] [CrossRef]
- Laikova, K.V.; Oberemok, V.V.; Krasnodubets, A.M.; Gal’chinsky, N.V.; Useinov, R.Z.; Novikov, I.A.; Temirova, Z.Z.; Gorlov, M.V.; Shved, N.A.; Kumeiko, V.V.; et al. Advances in the Understanding of Skin Cancer: Ultraviolet Radiation, Mutations, and Antisense Oligonucleotides as Anticancer Drugs. Molecules 2019, 24, 1516. [Google Scholar] [CrossRef]
- Moreno, P.M.; Pego, A.P. Therapeutic antisense oligonucleotides against cancer: Hurdling to the clinic. Front. Chem. 2014, 2, 87. [Google Scholar] [CrossRef]
- Le, B.T.; Raguraman, P.; Kosbar, T.R.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Antisense Oligonucleotides Targeting Angiogenic Factors as Potential Cancer Therapeutics. Mol. Ther. Nucleic Acids 2019, 14, 142–157. [Google Scholar] [CrossRef]
- Cooper, T.A.; Wan, L.; Dreyfuss, G. RNA and disease. Cell 2009, 136, 777–793. [Google Scholar] [CrossRef]
- Toonen, L.J.A.; Casaca-Carreira, J.; Pellise-Tintore, M.; Mei, H.; Temel, Y.; Jahanshahi, A.; van Roon-Mom, W.M.C. Intracerebroventricular Administration of a 2′-O-Methyl Phosphorothioate Antisense Oligonucleotide Results in Activation of the Innate Immune System in Mouse Brain. Nucleic Acid Ther. 2018, 28, 63–73. [Google Scholar] [CrossRef]
- Imbert, M.; Blandel, F.; Leumann, C.; Garcia, L.; Goyenvalle, A. Lowering Mutant Huntingtin Using Tricyclo-DNA Antisense Oligonucleotides As a Therapeutic Approach for Huntington’s Disease. Nucleic Acid Ther. 2019, 29, 256–265. [Google Scholar] [CrossRef]
- Huang, L.; Aghajan, M.; Quesenberry, T.; Low, A.; Murray, S.F.; Monia, B.P.; Guo, S. Targeting Translation Termination Machinery with Antisense Oligonucleotides for Diseases Caused by Nonsense Mutations. Nucleic Acid Ther. 2019, 29, 175–186. [Google Scholar] [CrossRef]
- Penchovsky, R.; Kostova, G.T. Computational selection and experimental validation of allosteric ribozymes that sense a specific sequence of human telomerase reverse transcriptase mRNAs as universal anticancer therapy agents. Nucleic Acid Ther. 2013, 23, 408–417. [Google Scholar] [CrossRef]
- Penchovsky, R. Clinical Trials of Functional Nucleic Acids: Antisense Oligonucleotides and Aptamers. Int. J. Med. Eng. Inform. 2018, 7, 46–60. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.A.; Castanotto, D. FDA-Approved Oligonucleotide Therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Post, N.; Yu, R.; Greenlee, S.; Gaus, H.; Hurh, E.; Matson, J.; Wang, Y. Metabolism and Disposition of Volanesorsen, a 2′-O-(2 methoxyethyl) Antisense Oligonucleotide, Across Species. Drug Metab. Dispos. 2019, 47, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hu, C.; Moufawad El Achkar, C.; Black, L.E.; Douville, J.; Larson, A.; Pendergast, M.K.; Goldkind, S.F.; Lee, E.A.; Kuniholm, A.; et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N. Engl. J. Med. 2019, 381, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Wareham, D.W.; on behalf of the UK Antibacterial Antisense Study Group. Tackling antibiotic resistance: A dose of common antisense? J. Antimicrob. Chemother. 2009, 63, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Hegarty, J.P.; Stewart, D.B., Sr. Advances in therapeutic bacterial antisense biotechnology. Appl. Microbiol. Biotechnol. 2018, 102, 1055–1065. [Google Scholar] [CrossRef]
- Sully, E.K.; Geller, B.L. Antisense antimicrobial therapeutics. Curr. Opin. Microbiol. 2016, 33, 47–55. [Google Scholar] [CrossRef]
- Bai, H.; Sang, G.; You, Y.; Xue, X.; Zhou, Y.; Hou, Z.; Meng, J.; Luo, X. Targeting RNA polymerase primary sigma70 as a therapeutic strategy against methicillin-resistant Staphylococcus aureus by antisense peptide nucleic acid. PLoS ONE 2012, 7, e29886. [Google Scholar] [CrossRef]
- Meng, J.; Da, F.; Ma, X.; Wang, N.; Wang, Y.; Zhang, H.; Li, M.; Zhou, Y.; Xue, X.; Hou, Z.; et al. Antisense growth inhibition of methicillin-resistant Staphylococcus aureus by locked nucleic acid conjugated with cell-penetrating peptide as a novel FtsZ inhibitor. Antimicrob. Agents Chemother. 2015, 59, 914–922. [Google Scholar] [CrossRef]
- Wagner, A.; Bock, C.T.; Fechner, H.; Kurreck, J. Application of modified antisense oligonucleotides and siRNAs as antiviral drugs. Future Med. Chem. 2015, 7, 1637–1642. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.; Penchovsky, R. Genome-wide bioinformatics analysis of FMN, SAM-I, glmS, TPP, lysine, purine, cobalamin, and SAH riboswitches for their applications as allosteric antibacterial drug targets in human pathogenic bacteria. Expert Opin. Ther. Targets 2019, 23, 631–643. [Google Scholar] [CrossRef]
- Pavlova, N.; Kaloudas, D.; Penchovsky, R. Riboswitch distribution, structure, and function in bacteria. Gene 2019, 708, 38–48. [Google Scholar] [CrossRef]
- Penchovsky, R.; Traykovska, M. Designing drugs that overcome antibacterial resistance: Where do we stand and what should we do? Expert Opin. Drug Discov. 2015, 10, 631–650. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Griffiths, A.J.F.; Miller, J.H.; Suzuki, D.T.; Lewontin, R.C.; Gelbart, W.M. An Introduction to Genetic Analysis, 7th ed.; W. H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Hodi, F.S.; Bastian, B.C. Mutation-driven drug development in melanoma. Curr. Opin. Oncol. 2010, 22, 178–183. [Google Scholar] [CrossRef]
- Ribas, A.; Flaherty, K.T. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat. Rev. Clin. Oncol. 2011, 8, 426–433. [Google Scholar] [CrossRef]
- Secq, V.; Villeret, J.; Fina, F.; Carmassi, M.; Carcopino, X.; Garcia, S.; Metellus, I.; Boubli, L.; Iovanna, J.; Charpin, C. Triple negative breast carcinoma EGFR amplification is not associated with EGFR, Kras or ALK mutations. Br. J. Cancer 2014, 110, 1045–1052. [Google Scholar] [CrossRef]
- Soffietti, R.; Ahluwalia, M.; Lin, N.; Rudà, R. Management of brain metastases according to molecular subtypes. Nat. Rev. Neurol. 2020, 16, 557–574. [Google Scholar] [CrossRef]
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Sabo, A.; Guccione, E. Targeting MYC in cancer therapy: RNA processing offers new opportunities. Bioessays 2016, 38, 266–275. [Google Scholar] [CrossRef] [PubMed]


| No | Name | ASO Sequence (5′-3′) | Size (nt) | Molecular Mass (Da) |
|---|---|---|---|---|
| 1 | pVEC-ASO1 | pVEC-G1A1U1T2T2T2G2C2T2T2C2T2T2T2A2C2C2T2A2A1U1U1 | 22 | 7906 |
| 2 | pVEC-ASO2 | pVEC-C1A1A1C2T2T2C2T2T2T2T2C2T2T2G2C2T2T2C2A1U1U1 | 22 | 7856 |
| 3 | pVEC-ASO3 | pVEC-G1A1U1C1A2A2T2C2A2G2G2T2C2G2A2T2G2T2G2G1G1A1G1 | 23 | 8490 |
| 4 | pVEC-ASO4 | pVEC-U1A2C2G2C2T2C2G2G2A2C1 | 11 | 4362 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popova, K.B.; Penchovsky, R. General and Specific Cytotoxicity of Chimeric Antisense Oligonucleotides in Bacterial Cells and Human Cell Lines. Antibiotics 2024, 13, 122. https://doi.org/10.3390/antibiotics13020122
Popova KB, Penchovsky R. General and Specific Cytotoxicity of Chimeric Antisense Oligonucleotides in Bacterial Cells and Human Cell Lines. Antibiotics. 2024; 13(2):122. https://doi.org/10.3390/antibiotics13020122
Chicago/Turabian StylePopova, Katya B., and Robert Penchovsky. 2024. "General and Specific Cytotoxicity of Chimeric Antisense Oligonucleotides in Bacterial Cells and Human Cell Lines" Antibiotics 13, no. 2: 122. https://doi.org/10.3390/antibiotics13020122
APA StylePopova, K. B., & Penchovsky, R. (2024). General and Specific Cytotoxicity of Chimeric Antisense Oligonucleotides in Bacterial Cells and Human Cell Lines. Antibiotics, 13(2), 122. https://doi.org/10.3390/antibiotics13020122






