Molecular Epidemiology of Pseudomonas aeruginosa in Brazil: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Material and Methods
2.1. Study Design
2.2. Eligibility Criteria and Definitions
2.3. Data Extraction
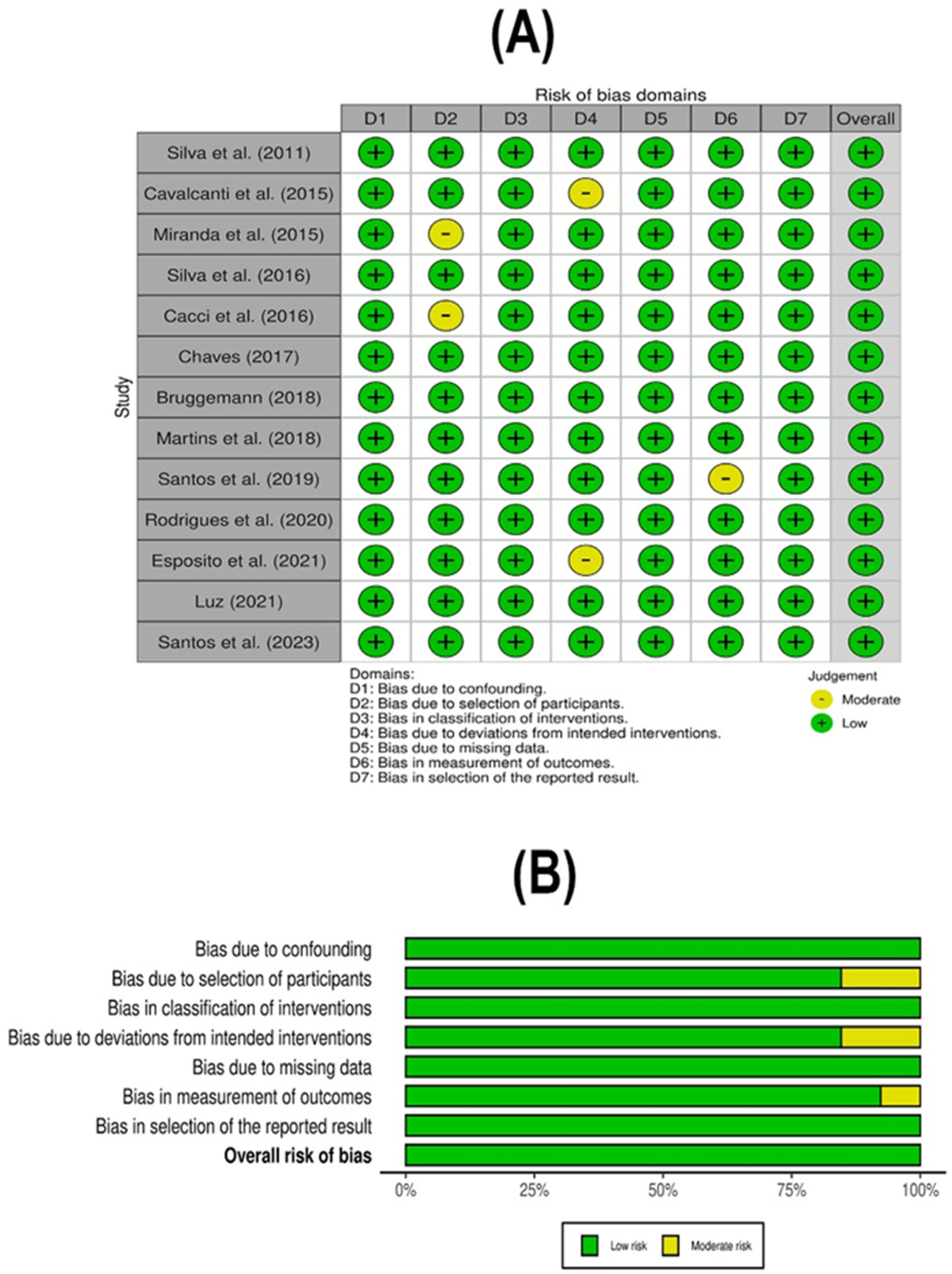
2.4. Methodological Quality and Risk of Bias Assessment
2.5. Statistical Analysis
3. Results
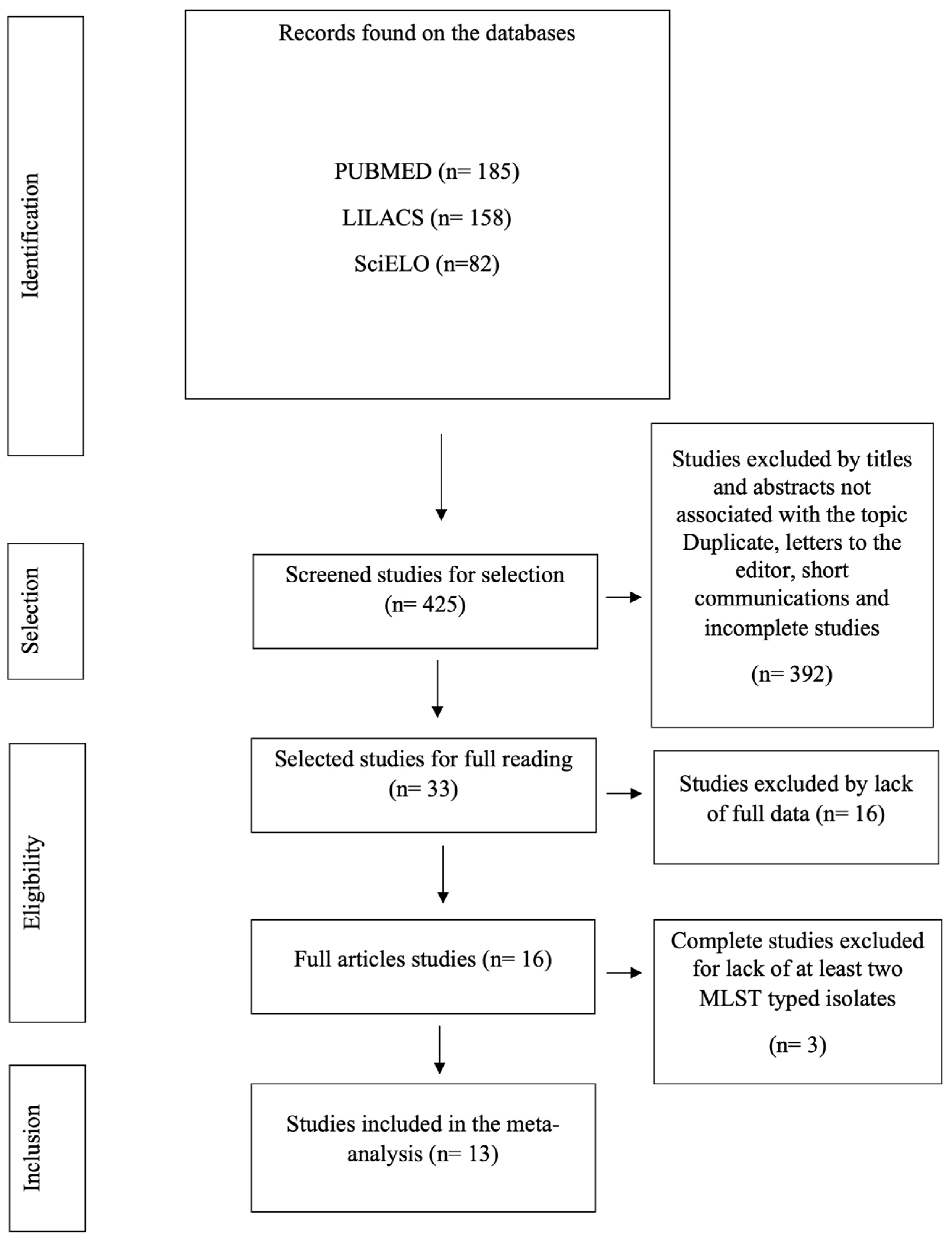
3.1. Literature Search
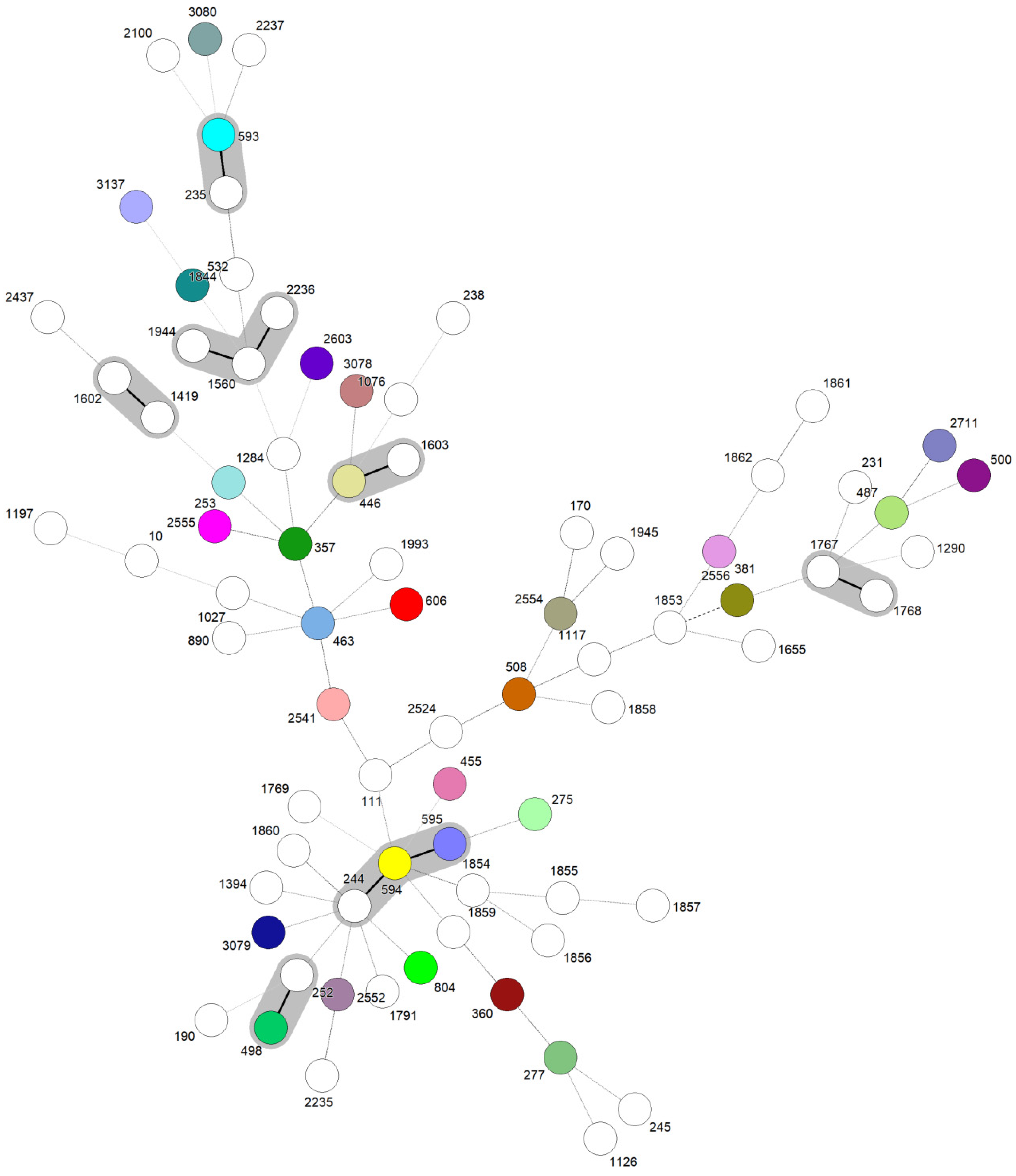
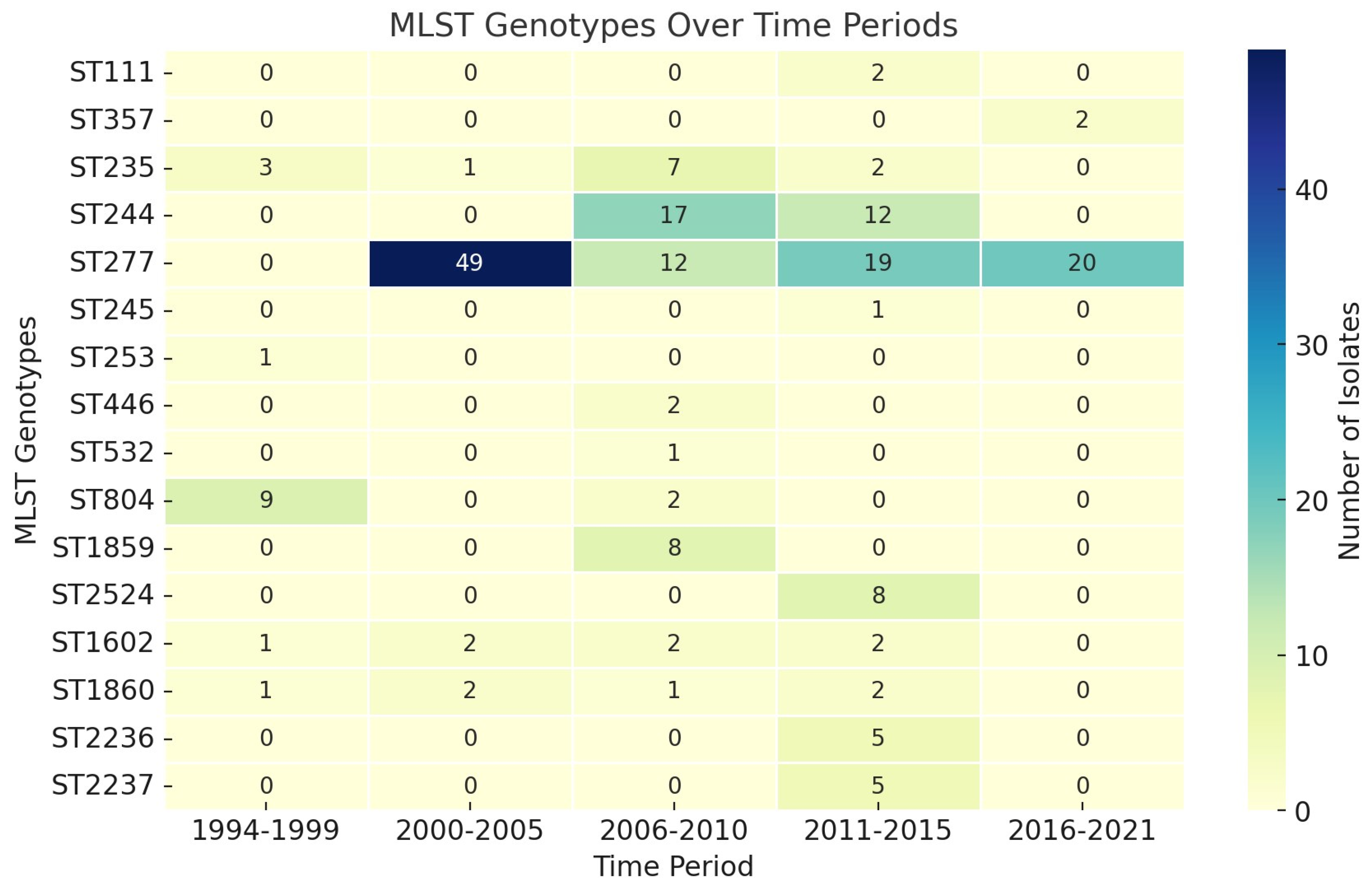
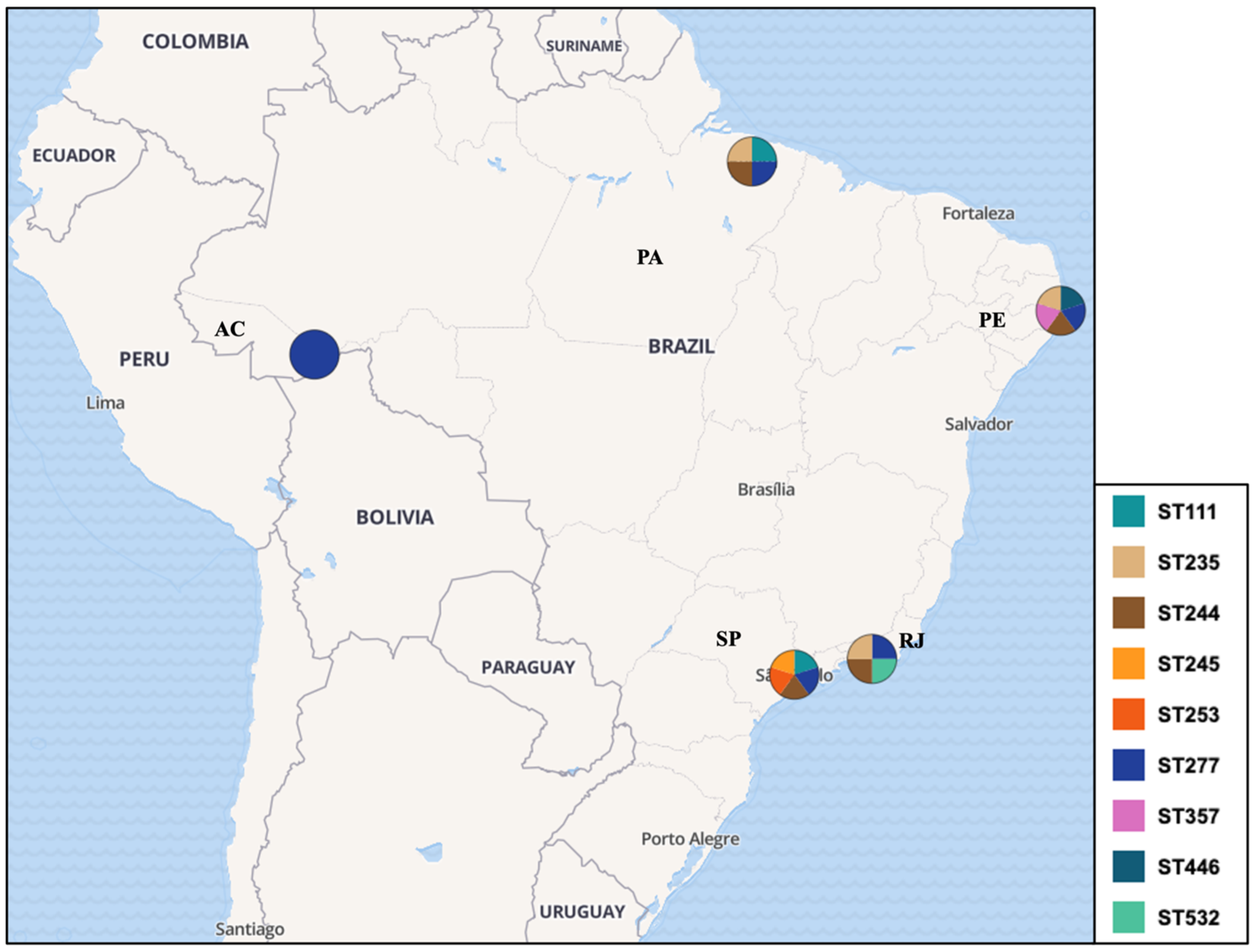
3.2. Characterization of Included Studies and Molecular Epidemiology Data
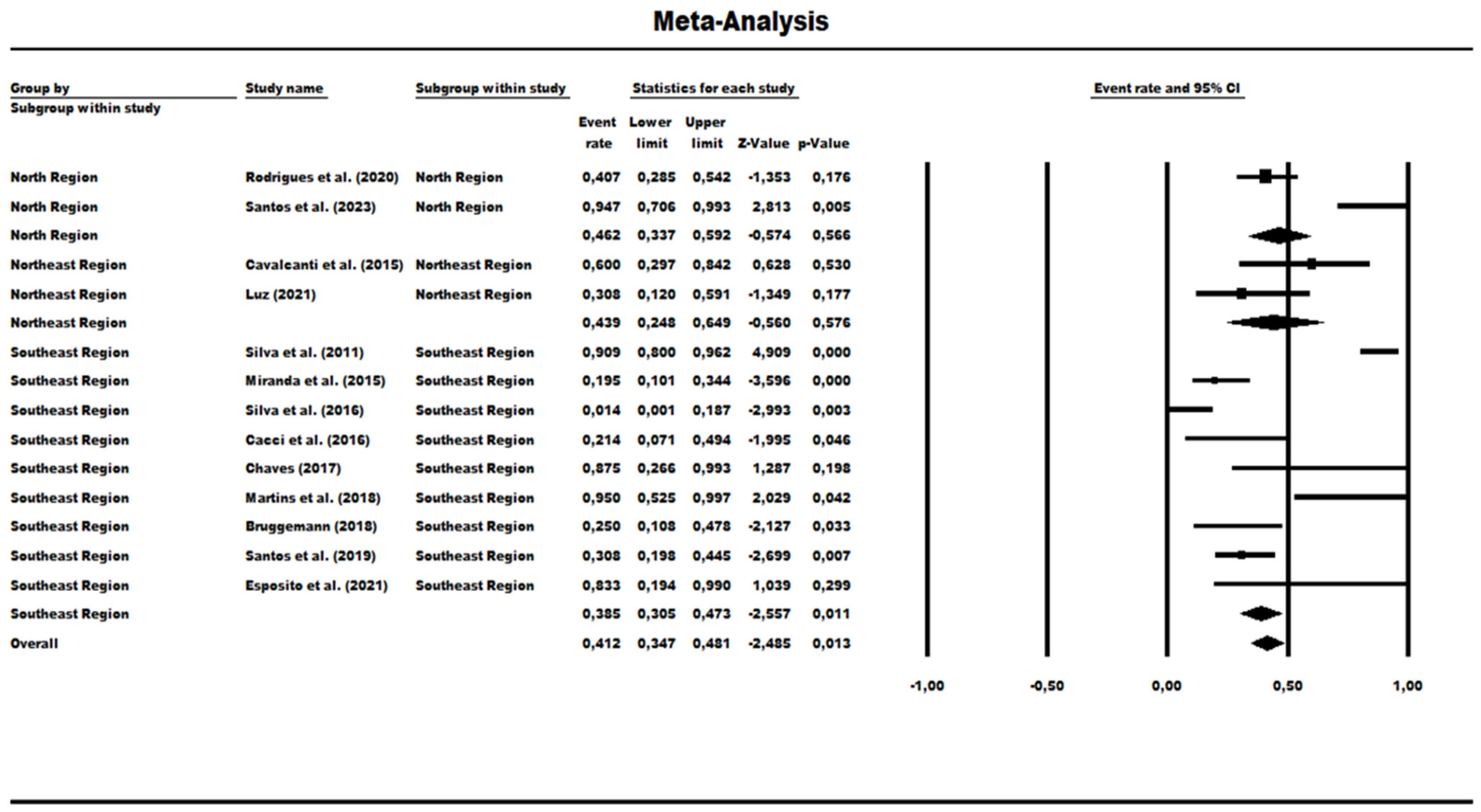
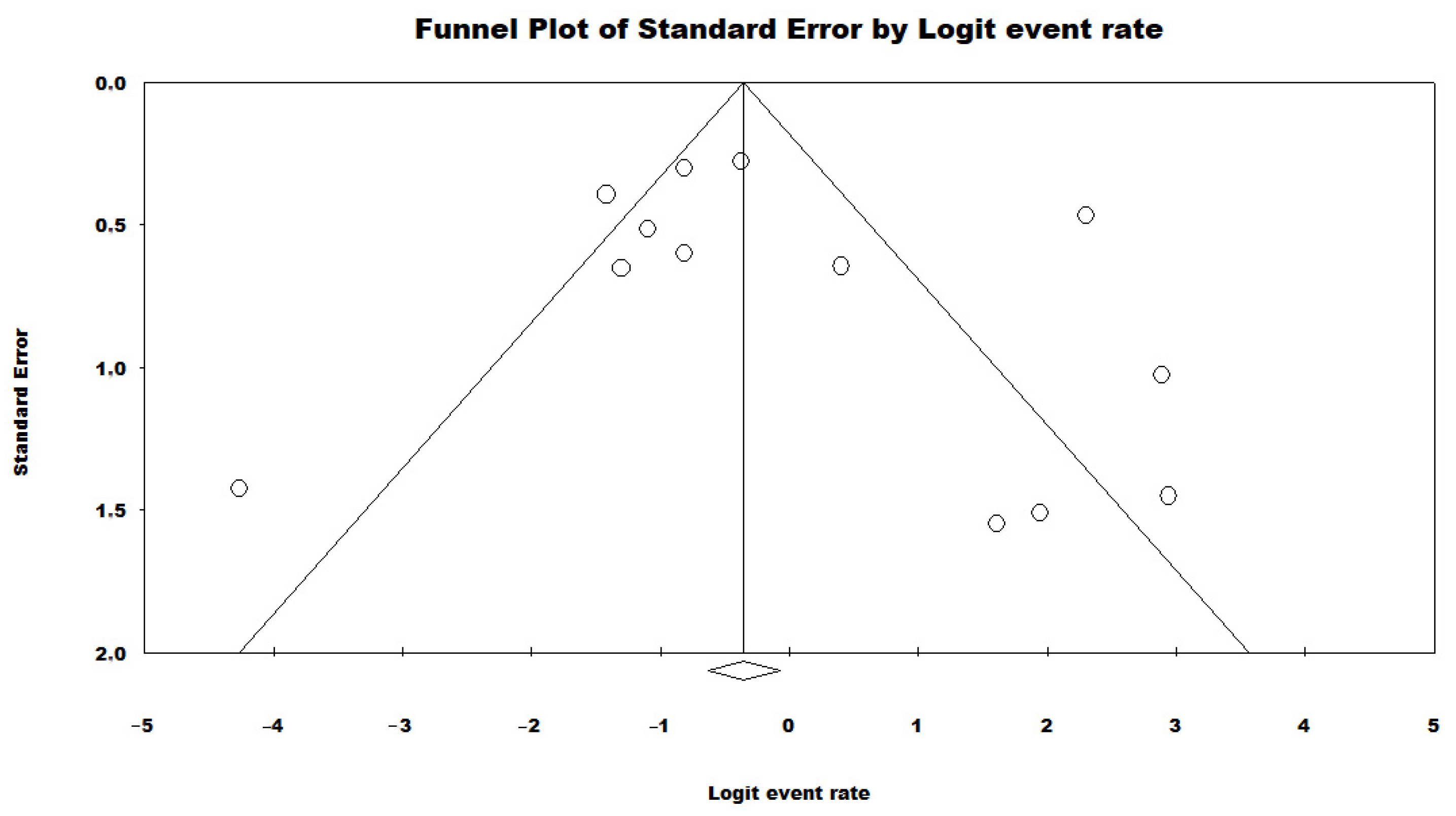
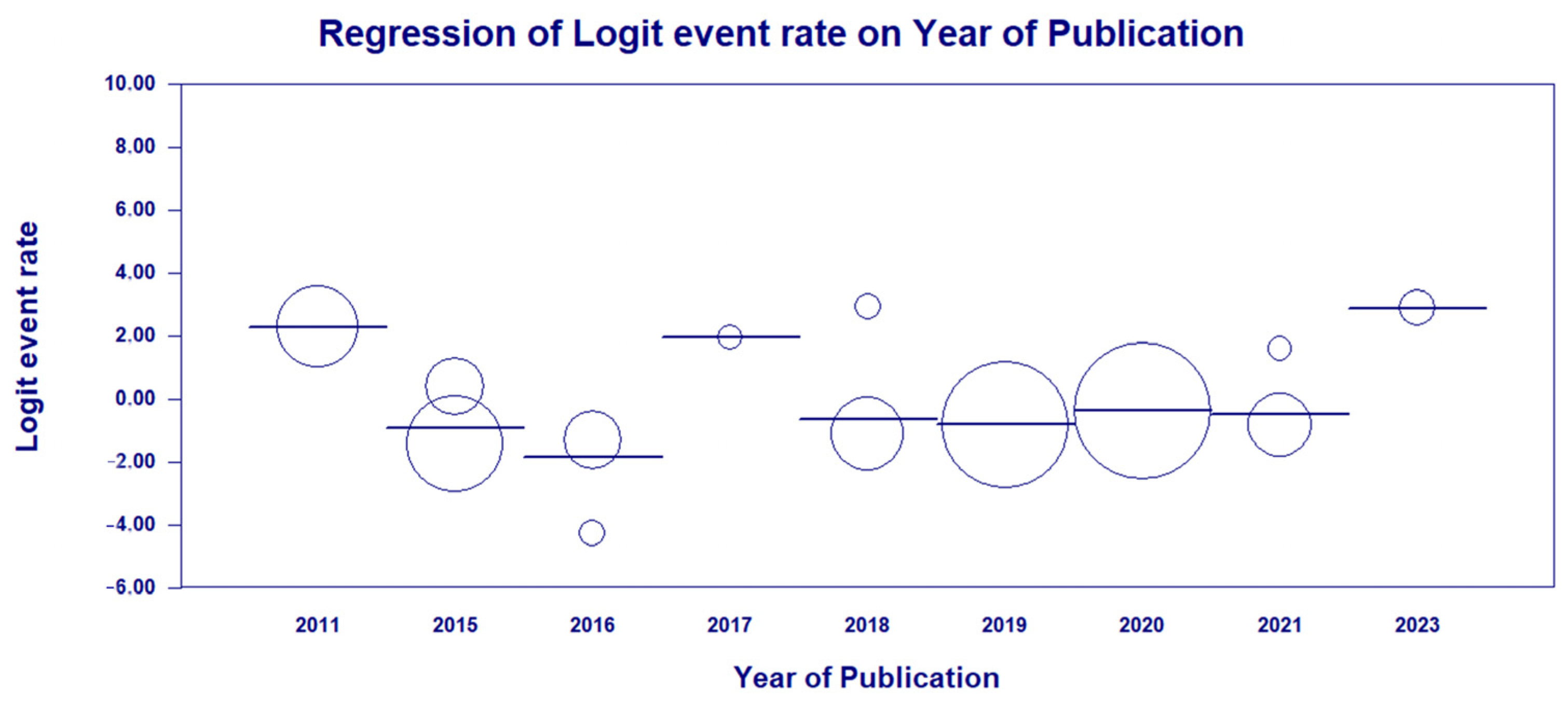
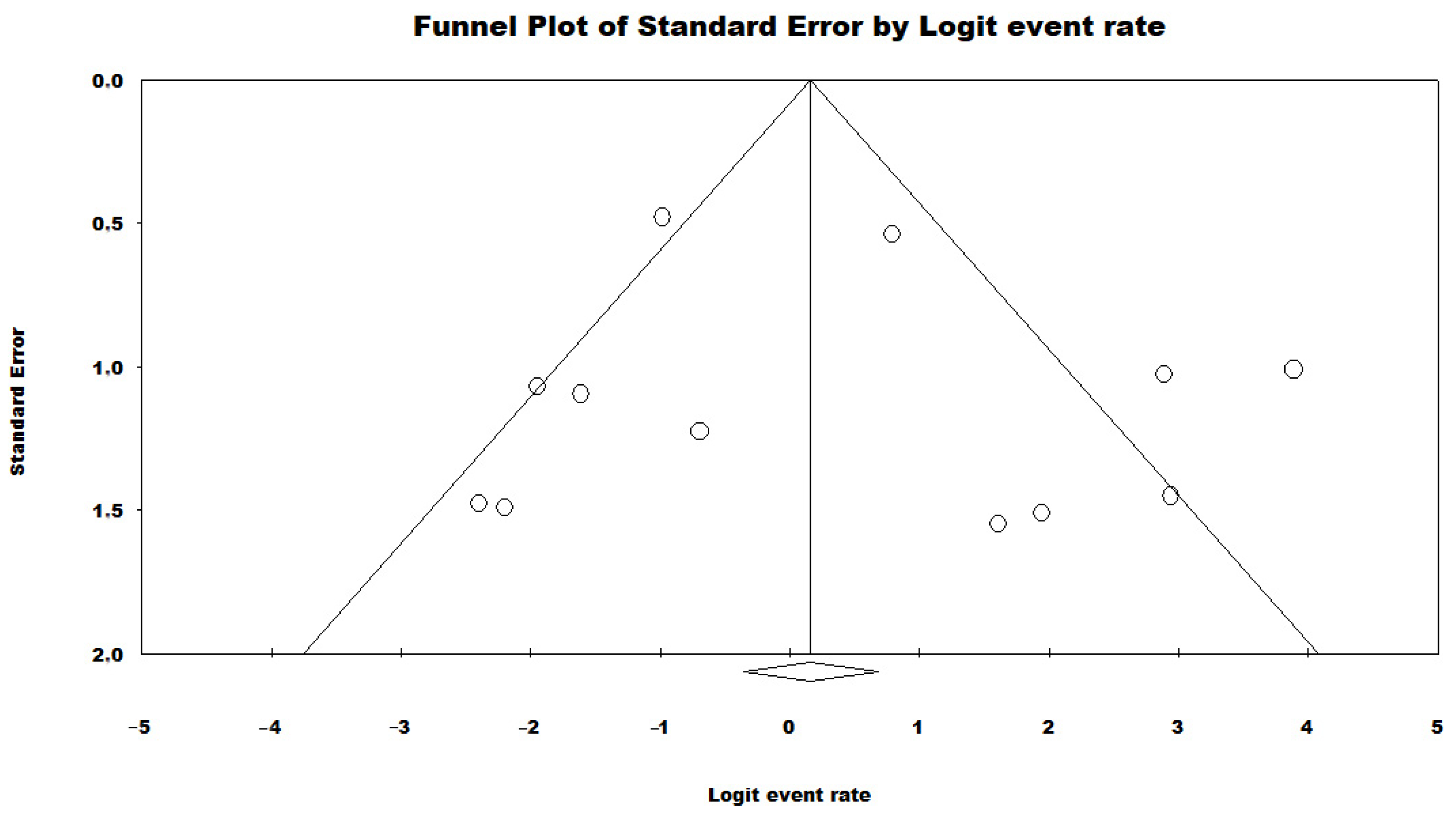
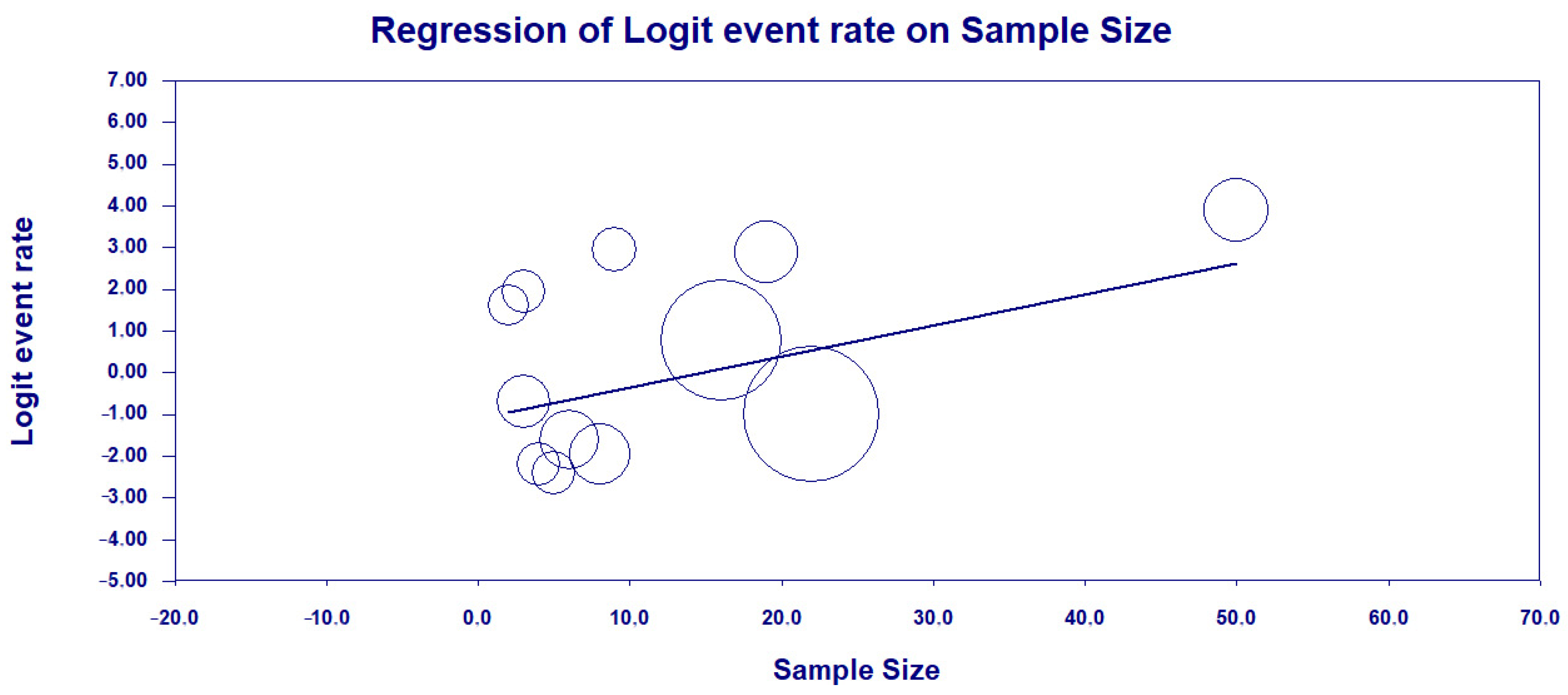
3.3. Results and Publication Bias of Meta-Analysis of Proportion of P. aeruginosa HRCs and Non-HRCs Strains
3.4. Results and Publication Bias of Meta-Analysis of ST277 within the HRCs Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Prevots, D.R.; Palmore, T.N.; Rhee, C.; Klompas, M.; Dekker, J.P. Difficult-to-Treat Resistance in Gram-Negative Bacteremia at 173 US Hospitals: Retrospective Cohort Analysis of Prevalence, Predictors, and Outcome of Resistance to All First-Line Agents. Clin. Infect. Dis. 2018, 67, 1803–1814. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Bertagnolio, S.; Dobreva, Z.; Centner, C.M.; Olaru, I.D.; Donà, D.; Burzo, S.; Huttner, B.D.; Chaillon, A.; Gebreselassie, N.; Wi, T. WHO Global Research Priorities for Antimicrobial Resistance in Human Health. Lancet Microbe 2024, 100902. [Google Scholar] [CrossRef]
- Coque, T.; Graham, D.W.; Pruden, A.; So, A.; Topp, E. Bracing for Superbugs: Strengthening Environmental Action in the One Health Response to Antimicrobial Resistance; United Nations Environment Programme: Geneva, Switzerland, 2023. [Google Scholar]
- Mendelson, M.; Lewnard, J.A.; Sharland, M.; Cook, A.; Pouwels, K.B.; Alimi, Y.; Mpundu, M.; Wesangula, E.; Weese, J.S.; Røttingen, J.-A. Ensuring Progress on Sustainable Access to Effective Antibiotics at the 2024 UN General Assembly: A Target-Based Approach. Lancet 2024, 403, 2551–2564. [Google Scholar] [CrossRef] [PubMed]
- Pillonetto, M.; Jordão, R.T.d.S.; Andraus, G.S.; Bergamo, R.; Rocha, F.B.; Onishi, M.C.; Almeida, B.M.M.d.; Nogueira, K.d.S.; Dal Lin, A.; Dias, V.M.d.C.H. The Experience of Implementing a National Antimicrobial Resistance Surveillance System in Brazil. Front. Public Health 2021, 8, 575536. [Google Scholar] [CrossRef]
- Pan American Health Organization Magnitude and Trends of Antimicrobial Resistance in Latin America. ReLAVRA 2014, 2015, 2016. Summary Report—PAHO/WHO | Pan American Health Organization; PAHO Antimicrobial Resistance Special Program: Washington, DC, USA, 2020. [Google Scholar]
- Wicky, P.-H.; Dupuis, C.; Cerf, C.; Siami, S.; Cohen, Y.; Laurent, V.; Mourvillier, B.; Reignier, J.; Goldgran-Toledano, D.; Schwebel, C. Ventilator-Associated Pneumonia in COVID-19 Patients Admitted in Intensive Care Units: Relapse, Therapeutic Failure and Attributable Mortality—A Multicentric Observational Study from the Outcomerea Network. J. Clin. Med. 2023, 12, 1298. [Google Scholar] [CrossRef]
- Velasquez-Garcia, L.; Mejia-Sanjuanelo, A.; Viasus, D.; Carratala, J. Causative Agents of Ventilator-Associated Pneumonia and Resistance to Antibiotics in COVID-19 Patients: A Systematic Review. Biomedicines 2022, 10, 1226. [Google Scholar] [CrossRef] [PubMed]
- ANVISA Boletim Segurança do Paciente e Qualidade em Serviços de Saúde no 30—Avaliação Nacional dos Indicadores de IRAS e RM—2022; Agência Nacional de Vigilância Sanitária: Brasília, Brazil, 2022; p. 11.
- Behzadi, P.; Baráth, Z.; Gajdács, M. It’s Not Easy Being Green: A Narrative Review on the Microbiology, Virulence and Therapeutic Prospects of Multidrug-Resistant Pseudomonas aeruginosa. Antibiotics 2021, 10, 42. [Google Scholar] [CrossRef]
- García-Betancur, J.C.; Appel, T.M.; Esparza, G.; Gales, A.C.; Levy-Hara, G.; Cornistein, W.; Vega, S.; Nuñez, D.; Cuellar, L.; Bavestrello, L. Update on the Epidemiology of Carbapenemases in Latin America and the Caribbean. Expert Rev. Anti Infect. Ther. 2021, 19, 197–213. [Google Scholar] [CrossRef]
- Halat, D.H.; Moubareck, C.A. The Intriguing Carbapenemases of Pseudomonas aeruginosa: Current Status, Genetic Profile, and Global Epidemiology. Yale J. Biol. Med. 2022, 95, 507–515. [Google Scholar]
- Sawa, T.; Shimizu, M.; Moriyama, K.; Wiener-Kronish, J.P. Association between Pseudomonas aeruginosa Type III Secretion, Antibiotic Resistance, and Clinical Outcome: A Review. Crit. Care 2014, 18, 668. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, Virulence Factors, Antibiotic Resistance, Interaction with Host, Technology Advances and Emerging Therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.H.; Kwon, K.C.; Kim, S.; Koo, S.H. Correlation between Virulence Genotype and Fluoroquinolone Resistance in Carbapenem-Resistant Pseudomonas aeruginosa. Ann. Lab. Med. 2014, 34, 286. [Google Scholar] [CrossRef]
- Peña, C.; Cabot, G.; Gómez-Zorrilla, S.; Zamorano, L.; Ocampo-Sosa, A.; Murillas, J.; Almirante, B.; Pomar, V.; Aguilar, M.; Granados, A. Influence of Virulence Genotype and Resistance Profile in the Mortality of Pseudomonas aeruginosa Bloodstream Infections. Clin. Infect. Dis. 2015, 60, 539–548. [Google Scholar] [CrossRef]
- Sánchez-Diener, I.; Zamorano, L.; López-Causapé, C.; Cabot, G.; Mulet, X.; Peña, C.; Del Campo, R.; Cantón, R.; Doménech-Sánchez, A.; Martínez-Martínez, L. Interplay among Resistance Profiles, High-Risk Clones, and Virulence in the Caenorhabditis Elegans Pseudomonas aeruginosa Infection Model. Antimicrob. Agents Chemother. 2017, 61, 5806–5816. [Google Scholar] [CrossRef]
- Recio, R.; Mancheño, M.; Viedma, E.; Villa, J.; Orellana, M.Á.; Lora-Tamayo, J.; Chaves, F. Predictors of Mortality in Bloodstream Infections Caused by Pseudomonas aeruginosa and Impact of Antimicrobial Resistance and Bacterial Virulence. Antimicrob. Agents Chemother. 2020, 64, e01759-19. [Google Scholar] [CrossRef] [PubMed]
- Horna, G.; Amaro, C.; Palacios, A.; Guerra, H.; Ruiz, J. High Frequency of the exoU+/exoS+ Genotype Associated with Multidrug-Resistant “High-Risk Clones” of Pseudomonas aeruginosa Clinical Isolates from Peruvian Hospitals. Sci. Rep. 2019, 9, 10874. [Google Scholar] [CrossRef]
- Sarges, E.d.S.N.F.; Rodrigues, Y.C.; Furlaneto, I.P.; de Melo, M.V.H.; Brabo, G.L.d.C.; Lopes, K.C.M.; Quaresma, A.J.P.G.; Lima, L.N.G.C.; Lima, K.V.B. Pseudomonas aeruginosa Type III Secretion System Virulotypes and Their Association with Clinical Features of Cystic Fibrosis Patients. Infect. Drug Resist. 2020, 13, 3771–3781. [Google Scholar] [CrossRef]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef]
- Achtman, M. Evolution, Population Structure, and Phylogeography of Genetically Monomorphic Bacterial Pathogens. Annu. Rev. Microbiol. 2008, 62, 53–70. [Google Scholar] [CrossRef]
- Woodford, N.; Turton, J.F.; Livermore, D.M. Multiresistant Gram-Negative Bacteria: The Role of High-Risk Clones in the Dissemination of Antibiotic Resistance. FEMS Microbiol. Rev. 2011, 35, 736–755. [Google Scholar] [CrossRef]
- Sawa, T.; Momiyama, K.; Mihara, T.; Kainuma, A.; Kinoshita, M.; Moriyama, K. Molecular Epidemiology of Clinically High-risk Pseudomonas aeruginosa Strains: Practical Overview. Microbiol. Immunol. 2020, 64, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Curran, B.; Jonas, D.; Grundmann, H.; Pitt, T.; Dowson, C.G. Development of a Multilocus Sequence Typing Scheme for the Opportunistic Pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 2004, 42, 5644–5649. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST. Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Mulet, X.; Cabot, G.; Ocampo-Sosa, A.A.; Domínguez, M.A.; Zamorano, L.; Juan, C.; Tubau, F.; Rodríguez, C.; Moyà, B.; Peña, C. Biological Markers of Pseudomonas aeruginosa Epidemic High-Risk Clones. Antimicrob. Agents Chemother. 2013, 57, 5527–5535. [Google Scholar] [CrossRef] [PubMed]
- Kocsis, B.; Gulyás, D.; Szabó, D. Diversity and Distribution of Resistance Markers in Pseudomonas aeruginosa International High-Risk Clones. Microorganisms 2021, 9, 359. [Google Scholar] [CrossRef]
- del Barrio-Tofiño, E.; López-Causapé, C.; Oliver, A. Pseudomonas aeruginosa Epidemic High-Risk Clones and Their Association with Horizontally-Acquired β-Lactamases: 2020 Update. Int. J. Antimicrob. Agents 2020, 56, 106196. [Google Scholar] [CrossRef]
- Oliver, A.; Rojo-Molinero, E.; Arca-Suarez, J.; Beşli, Y.; Bogaerts, P.; Cantón, R.; Cimen, C.; Croughs, P.D.; Denis, O.; Giske, C.G. Pseudomonas aeruginosa Antimicrobial Susceptibility Profiles, Resistance Mechanisms and International Clonal Lineages: Update from ESGARS-ESCMID/ISARPAE Group. Clin. Microbiol. Infect. 2024, 30, 469–480. [Google Scholar] [CrossRef]
- Oliver, A.; Mulet, X.; López-Causapé, C.; Juan, C. The Increasing Threat of Pseudomonas aeruginosa High-Risk Clones. Drug Resist. Updat. 2015, 21, 41–59. [Google Scholar] [CrossRef]
- Dos Santos, P.A.S.; Silva, M.J.A.; Gouveia, M.I.M.; Lima, L.N.G.C.; Quaresma, A.J.P.G.; De Lima, P.D.L.; Brasiliense, D.M.; Lima, K.V.B.; Rodrigues, Y.C. The Prevalence of Metallo-Beta-Lactamese-(MβL)-Producing Pseudomonas aeruginosa Isolates in Brazil: A Systematic Review and Meta-Analysis. Microorganisms 2023, 11, 2366. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. PHYLOViZ: Phylogenetic Inference and Data Visualization for Sequence Based Typing Methods. BMC Bioinform. 2012, 13, 87. [Google Scholar] [CrossRef] [PubMed]
- Peters, M.D.J.; Godfrey, C.M.; Khalil, H.; McInerney, P.; Parker, D.; Soares, C.B. Guidance for Conducting Systematic Scoping Reviews. Int. J. Evid. Based Healthc. 2015, 13, 141–146. [Google Scholar] [CrossRef]
- Munn, Z.; Aromataris, E.; Tufanaru, C.; Stern, C.; Porritt, K.; Farrow, J.; Lockwood, C.; Stephenson, M.; Moola, S.; Lizarondo, L.; et al. The Development of Software to Support Multiple Systematic Review Types: The Joanna Briggs Institute System for the Unified Management, Assessment and Review of Information (JBI SUMARI). Int. J. Evid. Based Healthc. 2019, 17, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.J.A.; Silva, C.S.; Da Silva Vieira, M.C.; Dos Santos, P.A.S.; Frota, C.C.; Lima, K.V.B.; Lima, L.N.G.C. The Relationship between TLR3 Rs3775291 Polymorphism and Infectious Diseases: A Meta-Analysis of Case-Control Studies. Genes 2023, 14, 1311. [Google Scholar] [CrossRef]
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A Tool for Assessing Risk of Bias in Non-Randomised Studies of Interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef]
- Silva, F.M.; Carmo, M.S.; Silbert, S.; Gales, A.C. SPM-1-Producing Pseudomonas aeruginosa: Analysis of the Ancestor Relationship Using Multilocus Sequence Typing, Pulsed-Field Gel Electrophoresis, and Automated Ribotyping. Microb. Drug Resist. 2011, 17, 215–220. [Google Scholar] [CrossRef]
- Cavalcanti, F.L.D.S.; Mirones, C.R.; Paucar, E.R.; Montes, L.Á.; Leal-Balbino, T.C.; Morais, M.M.C.D.; Martínez-Martínez, L.; Ocampo-Sosa, A.A. Mutational and Acquired Carbapenem Resistance Mechanisms in Multidrug Resistant Pseudomonas aeruginosa Clinical Isolates from Recife, Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 1003–1009. [Google Scholar] [CrossRef]
- Miranda, C.C.; De Filippis, I.; Pinto, L.H.; Coelho-Souza, T.; Bianco, K.; Cacci, L.C.; Picão, R.C.; Clementino, M.M. Genotypic Characteristics of Multidrug-Resistant Pseudomonas aeruginosa from Hospital Wastewater Treatment Plant in Rio de Janeiro, Brazil. J. Appl. Microbiol. 2015, 118, 1276–1286. [Google Scholar] [CrossRef]
- De Almeida Silva, K.D.C.F.; Calomino, M.A.; Deutsch, G.; De Castilho, S.R.; De Paula, G.R.; Esper, L.M.R.; Teixeira, L.A. Molecular Characterization of Multidrug-Resistant (MDR) Pseudomonas aeruginosa Isolated in a Burn Center. Burns 2017, 43, 137–143. [Google Scholar] [CrossRef]
- Cacci, L.C.; Chuster, S.G.; Martins, N.; Carmo, P.R.D.; Girão, V.B.D.C.; Nouér, S.A.; Freitas, W.V.D.; Matos, J.A.D.; Magalhães, A.C.D.G.; Ferreira, A.L.P.; et al. Mechanisms of Carbapenem Resistance in Endemic Pseudomonas aeruginosa Isolates after an SPM-1 Metallo-β-Lactamase Producing Strain Subsided in an Intensive Care Unit of a Teaching Hospital in Brazil. Mem. Inst. Oswaldo Cruz 2016, 111, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Chaves, L.; Tomich, L.M.; Salomão, M.; Leite, G.C.; Ramos, J.; Martins, R.R.; Rizek, C.; Neves, P.; Batista, M.V.; Amigo, U.; et al. High Mortality of Bloodstream Infection Outbreak Caused by Carbapenem-Resistant P. aeruginosa Producing SPM-1 in a Bone Marrow Transplant Unit. J. Med. Microbiol. 2017, 66, 1722–1729. [Google Scholar] [CrossRef] [PubMed]
- Martins, W.M.B.S.; Narciso, A.C.; Cayô, R.; Santos, S.V.; Fehlberg, L.C.C.; Ramos, P.L.; Da Cruz, J.B.; Gales, A.C. SPM-1-Producing Pseudomonas aeruginosa ST277 Clone Recovered from Microbiota of Migratory Birds. Diagn. Microbiol. Infect. Dis. 2018, 90, 221–227. [Google Scholar] [CrossRef]
- Brüggemann, H.; Migliorini, L.B.; Sales, R.O.D.; Koga, P.C.M.; Souza, A.V.D.; Jensen, A.; Poehlein, A.; Brzuszkiewicz, E.; Doi, A.M.; Pasternak, J.; et al. Comparative Genomics of Nonoutbreak Pseudomonas aeruginosa Strains Underlines Genome Plasticity and Geographic Relatedness of the Global Clone ST235. Genome Biol. Evol. 2018, 10, 1852–1857. [Google Scholar] [CrossRef]
- De Oliveira Santos, I.C.; Pereira De Andrade, N.F.; Da Conceição Neto, O.C.; Da Costa, B.S.; De Andrade Marques, E.; Rocha-de-Souza, C.M.; Asensi, M.D.; D’Alincourt Carvalho-Assef, A.P. Epidemiology and Antibiotic Resistance Trends in Clinical Isolates of Pseudomonas aeruginosa from Rio de Janeiro—Brazil: Importance of Mutational Mechanisms over the Years (1995–2015). Infect. Genet. Evol. 2019, 73, 411–415. [Google Scholar] [CrossRef]
- Rodrigues, Y.C.; Furlaneto, I.P.; Maciel, A.H.P.; Quaresma, A.J.P.G.; De Matos, E.C.O.; Conceição, M.L.; Vieira, M.C.D.S.; Brabo, G.L.D.C.; Sarges, E.D.S.N.F.; Lima, L.N.G.C.; et al. High Prevalence of Atypical Virulotype and Genetically Diverse Background among Pseudomonas aeruginosa Isolates from a Referral Hospital in the Brazilian Amazon. PLoS ONE 2020, 15, e0238741. [Google Scholar] [CrossRef]
- Esposito, F.; Cardoso, B.; Fontana, H.; Fuga, B.; Cardenas-Arias, A.; Moura, Q.; Fuentes-Castillo, D.; Lincopan, N. Genomic Analysis of Carbapenem-Resistant Pseudomonas aeruginosa Isolated From Urban Rivers Confirms Spread of Clone Sequence Type 277 Carrying Broad Resistome and Virulome Beyond the Hospital. Front. Microbiol. 2021, 12, 701921. [Google Scholar] [CrossRef]
- De Oliveira Luz, A.C.; Da Silva Junior, W.J.; Do Nascimento Junior, J.B.; Da Silva, J.M.A.; De Queiroz Balbino, V.; Leal-Balbino, T.C. Genetic Characteristics and Phylogenetic Analysis of Brazilian Clinical Strains of Pseudomonas aeruginosa Harboring CRISPR/Cas Systems. Curr. Genet. 2021, 67, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, P.A.S.; Rodrigues, Y.C.; Marcon, D.J.; Lobato, A.R.F.; Cazuza, T.B.; Gouveia, M.I.M.; Silva, M.J.A.; Souza, A.B.; Lima, L.N.G.C.; Quaresma, A.J.P.G.; et al. Endemic High-Risk Clone ST277 Is Related to the Spread of SPM-1-Producing Pseudomonas aeruginosa during the COVID-19 Pandemic Period in Northern Brazil. Microorganisms 2023, 11, 2069. [Google Scholar] [CrossRef]
- Mano, Y.; Saga, T.; Ishii, Y.; Yoshizumi, A.; Bonomo, R.A.; Yamaguchi, K.; Tateda, K. Molecular Analysis of the Integrons of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa Isolates Collected by Nationwide Surveillance Programs across Japan. BMC Microbiol. 2015, 15, 41. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, M.; Wang, M.; Lu, Y.; Yan, Z. Dissemination of IMP-6-Producing Pseudomonas aeruginosa ST244 in Multiple Cities in China. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1181–1187. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.L.; Meunier, D.; Findlay, J.; Mustafa, N.; Parsons, H.; Pike, R.; Wright, L.; Woodford, N. SPM-1 Metallo-β-Lactamase-Producing Pseudomonas aeruginosa ST277 in the UK. J. Med. Microbiol. 2016, 65, 696–697. [Google Scholar] [CrossRef] [PubMed]
- Galetti, R.; Andrade, L.N.; Varani, A.M.; Darini, A.L.C. SPM-1-Producing Pseudomonas aeruginosa ST277 Carries a Chromosomal Pack of Acquired Resistance Genes: An Example of High-Risk Clone Associated with ‘Intrinsic Resistome’. J. Glob. Antimicrob. Resist. 2019, 16, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Do Nascimento, A.P.B.; Medeiros Filho, F.; Pauer, H.; Antunes, L.C.M.; Sousa, H.; Senger, H.; Albano, R.M.; Trindade Dos Santos, M.; Carvalho-Assef, A.P.D.; Da Silva, F.A.B. Characterization of a SPM-1 Metallo-Beta-Lactamase-Producing Pseudomonas aeruginosa by Comparative Genomics and Phenotypic Analysis. Sci. Rep. 2020, 10, 13192. [Google Scholar] [CrossRef]
- Silveira, M.C.; Rocha-de-Souza, C.M.; Albano, R.M.; De Oliveira Santos, I.C.; Carvalho-Assef, A.P.D. Exploring the Success of Brazilian Endemic Clone Pseudomonas aeruginosa ST277 and Its Association with the CRISPR-Cas System Type I-C. BMC Genom. 2020, 21, 255. [Google Scholar] [CrossRef]
- Zhao, X.; Qin, J.; Chen, G.; Yang, C.; Wei, J.; Li, W.; Jia, W. Whole-Genome Sequencing, Multilocus Sequence Typing, and Resistance Mechanism of the Carbapenem-Resistant Pseudomonas aeruginosa in China. Microb. Pathog. 2024, 192, 106720. [Google Scholar] [CrossRef] [PubMed]
- Merradi, M.; Kassah-Laouar, A.; Ayachi, A.; Heleili, N.; Menasria, T.; Hocquet, D.; Cholley, P.; Sauget, M. Occurrence of VIM-4 Metallo-β-Lactamase-Producing Pseudomonas aeruginosa in an Algerian Hospital. J. Infect. Dev. Ctries. 2019, 13, 284–290. [Google Scholar] [CrossRef]
- Sadek, M.; Le Guern, R.; Kipnis, E.; Gosset, P.; Poirel, L.; Dessein, R.; Nordmann, P. Progressive in Vivo Development of Resistance to Cefiderocol in Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 61–66. [Google Scholar] [CrossRef]
- Fan, X.; Wu, Y.; Xiao, M.; Xu, Z.-P.; Kudinha, T.; Bazaj, A.; Kong, F.; Xu, Y.-C. Diverse Genetic Background of Multidrug-Resistant Pseudomonas aeruginosa from Mainland China and Emergence of an Extensively Drug-Resistant ST292 Clone in Kunming. Sci. Rep. 2016, 6, 26522. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, D.; Chen, K.; Xie, M.; Guo, J.; Chan, E.W.C.; Xie, L.; Wang, J.; Chen, E.; Chen, S.; et al. Epidemiological and Genetic Characteristics of Clinical Carbapenem-Resistant Pseudomonas aeruginosa Strains in Guangdong Province, China. Microbiol. Spectr. 2023, 11, e04261-22. [Google Scholar] [CrossRef]
- Fang, Y.; Baloch, Z.; Zhang, W.; Hu, Y.; Zheng, R.; Song, Y.; Tai, W.; Xia, X. Emergence of Carbapenem-Resistant ST244, ST292, and ST2446 Pseudomonas aeruginosa Clones in Burn Patients in Yunnan Province. Infect. Drug Resist. 2022, 15, 1103–1114. [Google Scholar] [CrossRef]
- Hu, Y.; Liu, C.; Wang, Q.; Zeng, Y.; Sun, Q.; Shu, L.; Lu, J.; Cai, J.; Wang, S.; Zhang, R.; et al. Emergence and Expansion of a Carbapenem-Resistant Pseudomonas aeruginosa Clone Are Associated with Plasmid-Borne Bla KPC-2 and Virulence-Related Genes. mSystems 2021, 6, e00154-21. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Vázquez, M.; Sola-Campoy, P.J.; Zurita, Á.M.; Ávila, A.; Gómez-Bertomeu, F.; Solís, S.; López-Urrutia, L.; Gónzalez-Barberá, E.M.; Cercenado, E.; Bautista, V.; et al. Carbapenemase-Producing Pseudomonas aeruginosa in Spain: Interregional Dissemination of the High-Risk Clones ST175 and ST244 Carrying blaVIM-2, blaVIM-1, blaIMP-8, blaVIM-20 and blaKPC-2. Int. J. Antimicrob. Agents 2020, 56, 106026. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Roldán, L.; Bellés, A.; Bueno, J.; Azcona-Gutiérrez, J.M.; Rojo-Bezares, B.; Torres, C.; Castillo, F.J.; Sáenz, Y.; Seral, C. Pseudomonas aeruginosa Isolates from Spanish Children: Occurrence in Faecal Samples, Antimicrobial Resistance, Virulence, and Molecular Typing. BioMed Res. Int. 2018, 2018, 8060178. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yang, L.; Wu, Y.; Li, H.; Shao, B. Spread of Multidrug-Resistant Pseudomonas aeruginosa in Animal-Derived Foods in Beijing, China. Int. J. Food Microbiol. 2023, 403, 110296. [Google Scholar] [CrossRef] [PubMed]
- Sastre-Femenia, M.À.; Fernández-Muñoz, A.; Gomis-Font, M.A.; Taltavull, B.; López-Causapé, C.; Arca-Suárez, J.; Martínez-Martínez, L.; Cantón, R.; Larrosa, N.; Oteo-Iglesias, J.; et al. Pseudomonas aeruginosa Antibiotic Susceptibility Profiles, Genomic Epidemiology and Resistance Mechanisms: A Nation-Wide Five-Year Time Lapse Analysis. Lancet Reg. Health Eur. 2023, 34, 100736. [Google Scholar] [CrossRef]
- Yano, H.; Hayashi, W.; Kawakami, S.; Aoki, S.; Anzai, E.; Zuo, H.; Kitamura, N.; Hirabayashi, A.; Kajihara, T.; Kayama, S.; et al. Nationwide Genome Surveillance of Carbapenem-Resistant Pseudomonas aeruginosa in Japan. Antimicrob. Agents Chemother. 2024, 68, e01669-23. [Google Scholar] [CrossRef]
- Zagui, G.S.; De Almeida, O.G.G.; Moreira, N.C.; Silva, N.G.A.; Meschede, M.S.C.; Darini, A.L.C.; Andrade, L.N.; Segura-Muñoz, S.I. Hospital Wastewater as Source of Human Pathogenic Bacteria: A Phenotypic and Genomic Analysis of International High-Risk Clone VIM-2-Producing Pseudomonas aeruginosa ST235/O11. Environ. Res. 2024, 255, 119166. [Google Scholar] [CrossRef]
- Talat, A.; Khan, F.; Khan, A.U. Genome Analyses of Colistin-Resistant High-Risk blaNDM-5 Producing Klebsiella Pneumoniae ST147 and Pseudomonas aeruginosa ST235 and ST357 in Clinical Settings. BMC Microbiol. 2024, 24, 174. [Google Scholar] [CrossRef]
- Antonelli, A.; Coppi, M.; Bonaiuto, C.; Giovacchini, N.; Vaggelli, G.; Farese, A.; Pollini, S.; Rossolini, G.M. Novel Resistance ICEs Carrying the Bla FIM-1 Metallo-β-Lactamase Gene from an ST235 Pseudomonas aeruginosa Sublineage. Antimicrob. Agents Chemother. 2024, 68, e01205-23. [Google Scholar] [CrossRef]
- Hayashi, W.; Izumi, K.; Yoshida, S.; Takizawa, S.; Sakaguchi, K.; Iyori, K.; Minoshima, K.; Takano, S.; Kitagawa, M.; Nagano, Y.; et al. Antimicrobial Resistance and Type III Secretion System Virulotypes of Pseudomonas aeruginosa Isolates from Dogs and Cats in Primary Veterinary Hospitals in Japan: Identification of the International High-Risk Clone Sequence Type 235. Microbiol. Spectr. 2021, 9, e00408-21. [Google Scholar] [CrossRef] [PubMed]
- Jangsangthong, A.; Lugsomya, K.; Apiratwarrasakul, S.; Phumthanakorn, N. Distribution of Sequence Types and Antimicrobial Resistance of Clinical Pseudomonas aeruginosa Isolates from Dogs and Cats Visiting a Veterinary Teaching Hospital in Thailand. BMC Vet. Res. 2024, 20, 234. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-G.; Liu, Z.-Y.; Liao, X.-P.; Sun, R.-Y.; Li, R.-B.; Liu, Y.; Fang, L.-X.; Sun, J.; Liu, Y.-H.; Zhang, R.-M. Retrospective Data Insight into the Global Distribution of Carbapenemase-Producing Pseudomonas aeruginosa. Antibiotics 2021, 10, 548. [Google Scholar] [CrossRef]
- Tickler, I.A.; Torre, J.C.G.D.L.; Alvarado, L.; Obradovich, A.E.; Tenover, F.C. Mechanisms of Carbapenemase-Mediated Resistance among High-Risk Pseudomonas aeruginosa Lineages in Peru. J. Glob. Antimicrob. Resist. 2022, 31, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.-J.; Jeong, S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef] [PubMed]
- Papagiannitsis, C.C.; Verra, A.; Galani, V.; Xitsas, S.; Bitar, I.; Hrabak, J.; Petinaki, E. Unravelling the Features of Success of VIM-Producing ST111 and ST235 Pseudomonas aeruginosa in a Greek Hospital. Microorganisms 2020, 8, 1884. [Google Scholar] [CrossRef]
- Papagiannitsis, C.C.; Medvecky, M.; Chudejova, K.; Skalova, A.; Rotova, V.; Spanelova, P.; Jakubu, V.; Zemlickova, H.; Hrabak, J. Molecular Characterization of Carbapenemase-Producing Pseudomonas aeruginosa of Czech Origin and Evidence for Clonal Spread of Extensively Resistant Sequence Type 357 Expressing IMP-7 Metallo-β-Lactamase. Antimicrob. Agents Chemother. 2017, 61, e01811-17. [Google Scholar] [CrossRef]
- Pirzadian, J.; Persoon, M.C.; Severin, J.A.; Klaassen, C.H.W.; De Greeff, S.C.; Mennen, M.G.; Schoffelen, A.F.; Wielders, C.C.H.; Witteveen, S.; Van Santen-Verheuvel, M.; et al. National Surveillance Pilot Study Unveils a Multicenter, Clonal Outbreak of VIM-2-Producing Pseudomonas aeruginosa ST111 in the Netherlands between 2015 and 2017. Sci. Rep. 2021, 11, 21015. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Costello, A.; Davies, T.A.; Jones, R.N. Epidemiology and Carbapenem Resistance Mechanisms of Carbapenem-Non-Susceptible Pseudomonas aeruginosa Collected during 2009–11 in 14 European and Mediterranean Countries. J. Antimicrob. Chemother. 2014, 69, 1804–1814. [Google Scholar] [CrossRef]
- Miyoshi-Akiyama, T.; Tada, T.; Ohmagari, N.; Viet Hung, N.; Tharavichitkul, P.; Pokhrel, B.M.; Gniadkowski, M.; Shimojima, M.; Kirikae, T. Emergence and Spread of Epidemic Multidrug-Resistant Pseudomonas aeruginosa. Genome Biol. Evol. 2017, 9, 3238–3245. [Google Scholar] [CrossRef]
- Pelegrin, A.C.; Saharman, Y.R.; Griffon, A.; Palmieri, M.; Mirande, C.; Karuniawati, A.; Sedono, R.; Aditianingsih, D.; Goessens, W.H.F.; Van Belkum, A.; et al. Erratum for Pelegrin et al. “High-Risk International Clones of Carbapenem-Nonsusceptible Pseudomonas aeruginosa Endemic to Indonesian Intensive Care Units: Impact of a Multifaceted Infection Control Intervention Analyzed at the Genomic Level”. mBio 2020, 11, e01372-20. [Google Scholar] [CrossRef] [PubMed]
- Çekin, Z.K.; Dabos, L.; Malkoçoğlu, G.; Fortineau, N.; Bayraktar, B.; Iorga, B.I.; Naas, T.; Aktaş, E. Carbapenemase -Producing Pseudomonas aeruginosa Isolates from Turkey: First Report of P. Aeruginosa High-Risk Clones with VIM-5– and IMP-7–Type Carbapenemases in a Tertiary Hospital. Diagn. Microbiol. Infect. Dis. 2021, 99, 115174. [Google Scholar] [CrossRef] [PubMed]
- Rossel, C.A.J.; Hendrickx, A.P.A.; Van Alphen, L.B.; Van Der Horst, R.P.J.; Janssen, A.H.J.W.; Kooyman, C.C.; Heddema, E.R. Tracing the Origin of NDM-1-Producing and Extensively Drug-Resistant Pseudomonas aeruginosa ST357 in the Netherlands. BMC Infect. Dis. 2024, 24, 817. [Google Scholar] [CrossRef]
- Mondol, S.M.; Islam, M.R.; Rakhi, N.N.; Shakil, S.K.; Islam, I.; Mustary, J.F.; Amiruzzaman; Shahjalal, H.M.; Gomes, D.J.; Rahaman, M.M. Unveiling a High-Risk Epidemic Clone (ST 357) of ‘Difficult to Treat Extensively Drug-Resistant’ (DT-XDR) Pseudomonas aeruginosa from a Burn Patient in Bangladesh: A Resilient Beast Revealing Coexistence of Four Classes of Beta Lactamases. J. Glob. Antimicrob. Resist. 2024, 36, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Orsi, T.D.; Perdigão Neto, L.V.; Martins, R.C.R.; Levin, A.S.; Costa, S.F. Polymyxin-Resistant Pseudomonas aeruginosa Assigned as ST245: First Report in an Intensive Care Unit in São Paulo, Brazil. J. Glob. Antimicrob. Resist. 2019, 16, 147–149. [Google Scholar] [CrossRef]
- Estepa, V.; Rojo-Bezares, B.; Torres, C.; Sáenz, Y. Faecal Carriage of Pseudomonas aeruginosa in Healthy Humans: Antimicrobial Susceptibility and Global Genetic Lineages. FEMS Microbiol. Ecol. 2014, 89, 15–19. [Google Scholar] [CrossRef]
- Fischer, S.; Dethlefsen, S.; Klockgether, J.; Tümmler, B. Phenotypic and Genomic Comparison of the Two Most Common ExoU-Positive Pseudomonas aeruginosa Clones, PA14 and ST235. mSystems 2020, 5, e01007-20. [Google Scholar] [CrossRef]
- Treepong, P.; Kos, V.N.; Guyeux, C.; Blanc, D.S.; Bertrand, X.; Valot, B.; Hocquet, D. Global Emergence of the Widespread Pseudomonas aeruginosa ST235 Clone. Clin. Microbiol. Infect. 2018, 24, 258–266. [Google Scholar] [CrossRef]
- Bean, D.C.; Wareham, D.W. Corrigendum to ‘Draft Genome Sequence of a Multidrug-Resistant Pseudomonas aeruginosa Producing blaSIM Metallo-β-Lactamase: London, UK’ [Journal of Global Antimicrobial Resistance 29 (2022) 222-224]. J. Glob. Antimicrob. Resist. 2022, 30, 490. [Google Scholar] [CrossRef]
- Rath, A.; Kieninger, B.; Fritsch, J.; Caplunik-Pratsch, A.; Blaas, S.; Ochmann, M.; Pfeifer, M.; Hartl, J.; Holzmann, T.; Schneider-Brachert, W. Whole-Genome Sequencing Reveals Two Prolonged Simultaneous Outbreaks Involving Pseudomonas aeruginosa High-Risk Strains ST111 and ST235 with Resistance to Quaternary Ammonium Compounds. J. Hosp. Infect. 2024, 145, 155–164. [Google Scholar] [CrossRef]
- Founou, R.C.; Founou, L.L.; Allam, M.; Ismail, A.; Essack, S.Y. First Report of a Clinical Multidrug-Resistant Pseudomonas aeruginosa ST532 Isolate Harbouring a Ciprofloxacin-Modifying Enzyme (CrpP) in South Africa. J. Glob. Antimicrob. Resist. 2020, 22, 145–146. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, R.; Fernández-Barat, L.; Vázquez, N.; Alcaraz-Serrano, V.; Bueno-Freire, L.; Amaro, R.; López-Aladid, R.; Oscanoa, P.; Muñoz, L.; Vila, J.; et al. Resistance Mechanisms and Molecular Epidemiology of Pseudomonas aeruginosa Strains from Patients with Bronchiectasis. J. Antimicrob. Chemother. 2022, 77, 1600–1610. [Google Scholar] [CrossRef]
- Kidd, T.J.; Grimwood, K.; Ramsay, K.A.; Rainey, P.B.; Bell, S.C. Comparison of Three Molecular Techniques for Typing Pseudomonas aeruginosa Isolates in Sputum Samples from Patients with Cystic Fibrosis. J. Clin. Microbiol. 2011, 49, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Kiffer, C.R.V.; Rezende, T.F.T.; Costa-Nobre, D.T.; Marinonio, A.S.S.; Shiguenaga, L.H.; Kulek, D.N.O.; Arend, L.N.V.S.; Santos, I.C.D.O.; Sued-Karam, B.R.; Rocha-de-Souza, C.M.; et al. A 7-Year Brazilian National Perspective on Plasmid-Mediated Carbapenem Resistance in Enterobacterales, Pseudomonas aeruginosa, and Acinetobacter Baumannii Complex and the Impact of the Coronavirus Disease 2019 Pandemic on Their Occurrence. Clin. Infect. Dis. 2023, 77, S29–S37. [Google Scholar] [CrossRef]
- Rocha, A.J.; Barsottini, M.R.D.O.; Rocha, R.R.; Laurindo, M.V.; Moraes, F.L.L.D.; Rocha, S.L.D. Pseudomonas aeruginosa: Virulence Factors and Antibiotic Resistance Genes. Braz. Arch. Biol. Technol. 2019, 62, e19180503. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence Factors of Pseudomonas aeruginosa and Antivirulence Strategies to Combat Its Drug Resistance. Front. Cell. Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E.W. Pseudomonas aeruginosa: New Insights into Pathogenesis and Host Defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Matos, E.C.O.D.; Andriolo, R.B.; Rodrigues, Y.C.; Lima, P.D.L.D.; Carneiro, I.C.D.R.S.; Lima, K.V.B. Mortality in Patients with Multidrug-Resistant Pseudomonas aeruginosa Infections: A Meta-Analysis. Rev. Soc. Bras. Med. Trop. 2018, 51, 415–420. [Google Scholar] [CrossRef]
- Rocha, M.F.G.; Diógenes, E.M.; Carvalho, V.L.; Marmontel, M.; Da Costa, M.O.; Da Silva, V.M.F.; De Souza Amaral, R.; Gravena, W.; Do Carmo, N.A.S.; Marigo, J.; et al. One Health Implications of Antimicrobial Resistance in Bacteria from Amazon River Dolphins. EcoHealth 2021, 18, 383–396. [Google Scholar] [CrossRef]
- Souza, C.O.; Cayô, R.; Lima, K.V.B.; Brasiliense, D.M.; Streling, A.P.; Siqueira, A.V.; Alberto-Lei, F.; Leal, J.T.; Nodari, C.S.; Pérez-Chaparro, P.J.; et al. Genetic and Biochemical Characterization of BIM-1, a Novel Acquired Subgroup B1 MBL Found in a Pseudomonas Sp. Strain from the Brazilian Amazon Region. J. Antimicrob. Chemother. 2023, 78, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
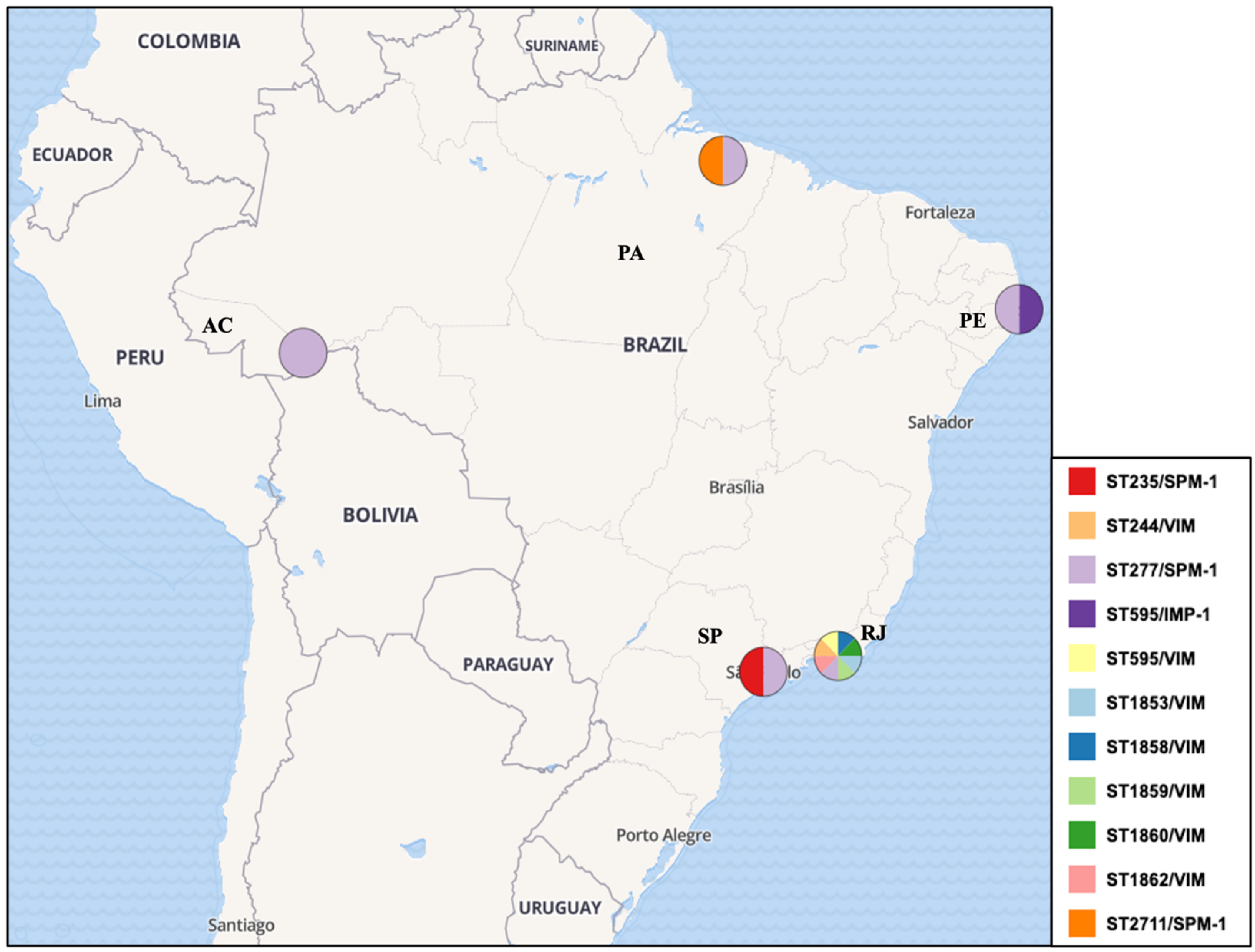

| Authors/Year | Type of Study | Isolates Period | Region/Location | Setting | N° of Samples (872)/ N° of MLST Typed (298) | MLST Genotypes | HRC | JBI Score |
|---|---|---|---|---|---|---|---|---|
| Silva et al. (2011) #,* [40] | Retrospective cross-sectional | 2000–2004 | Southeast region | Clinical | 55/55 | ST277 (49/55—89.09%) ST235 (1/55—1.82%) ST593 (3/55—5.45%) ST594 (1/55—1.82%) ST595 (1/55—1.82%) | Yes—50/55 (90.9%)—ST277, ST235 | (7/8) |
| Cavalcanti et al. (2015) #,* [41] | Retrospective cross-sectional | 2008–2010 | Northeast region | Clinical | 10/10 | ST235 (3/10—30.0%) ST446 (1/10—10.0%) ST1560 (1/10—10.0%) ST1419 (1/10—10.0%) ST1126 (1/10—10.0%) ST277 (1/10—10.0%) ST244 (2/10—20.0%) | Yes—6/10 (60.0%)—ST277, ST235, ST244 | (8/8) |
| Miranda et al. (2015) #,* [42] | Retrospective cross-sectional | 2008 and 2010 | Southeast region | Hospital wastewater treatment plant | 41/36 | ST244 (8/36—22.22%) ST1853 (2/36—5.56%) ST1854 (1/36—2.78%) ST1855 (2/36—5.56%) ST1856 (1/36—2.78%) ST1857 (1/36—2.78%) ST1858 (2/36—5.56%) ST1859 (8/36—22.22%) ST1860 (1/36—2.78%) ST1861 (1/36—2.78%) ST1862 (4/36—11.11%) ST238 (1/36—2.78%) ST381 (1/36—2.78%) ST595 (3/36—8.33%) | Yes—8/41 (19.51%)—ST244 | (6/8) |
| Silva et al. (2016) #,* [43] | Retrospective cross-sectional | 2012 | Southeast region | Burn center: Clinical and Clinical environment | 35/10 | ST 2236 (5/10—14.28%) ST 2237 (5/10—14.28%) | No (0.0%) | (8/8) |
| Cacci et al. (2016) #,* [44] | Retrospective cross-sectional | 2007–2008 | Southeast region | Clinical and Clinical environment | 482/14 | ST1027 (1/14—7.14%) ST1602 (2/14—14.29%) ST1603 (1/14—7.14%) ST1767 (1/14—7.14%) ST1768 (1/14—7.14%) ST1769 (1/14—7.14%) ST1844 (1/14—7.14%) ST235 (1/14—7.14%) ST244 (1/14—7.14%) ST277 (1/14—7.14%) ST446 (1/14—7.14%) ST532 (1/14—7.14%) ST890 (1/14—7.14%) | Yes—(3/14—21.42%)—ST235, ST244 and ST277 | (8/8) |
| Chaves et al. (2017) #,* [45] | Retrospective case-control study | 2011–2013 | Southeast region | Clinical | 29/3 | ST277 (3/3—100.0%) | Yes—ST277 (3/3—100.0%) | (10/10) |
| Martins et al. (2018) #,* [46] | Retrospective cross-sectional | 2012 | Southeast region | Environmental (Migratory birds) | 9/9 | ST277: 9/9 (100%) | Yes—9/9 (100.0%)—ST277 | (7/8) |
| Bruggemann et al. (2018) #,* [47] | Retrospective cross-sectional | 1994–2016 | Southeast region | Clinical | 20/20 | ST235 (3/20—15.0%) ST10 (1/20—5.0%) ST231 (1/20—5.0%) ST245 1/20—5.0%) ST252 (1/20—5.0%) ST446 (1/20—5.0%) ST455 (1/20—5.0%) ST498 (1/20—5.0%) ST606 (1/20—5.0%) ST1290 (1/20—5.0%) ST1993 (1/20—5.0%) ST2235 (1/20—5.0%) ST381 (1/20—5.0%) ST190 (1/20—5.0%) ST244 (2/20—10.0%) | Yes—5/20 (25.0%)—ST235, ST244 | (8/8) |
| Santos et al. (2019) #,* [48] | Retrospective cross-sectional | 1995–2015 | Southeast region | Clinical | 88/52 | ST1117 (3/52—5.77%) ST1560 (3/52—5.77%) ST1945 (3/52—5.77%) ST244 (5/52—9.62%) ST277 (11/52—21.15%) ST487 (4/52—7.69%) ST1791 (3/52—5.77%) ST1860 (4/52—7.69%) ST804 (11/52—21.15%) ST1602 (3/52—5.77%) ST1944 (2/52—3.85%) | Yes—16/52 (30.76%)—ST277, ST244 | (7/8) |
| Rodrigues et al. (2020) #,* [49] | Retrospective cross-sectional | 2010–2013 | North region | Referral hospital/Clinical | 54/54 | ST2524 (8/54—14.81%) ST2541 (2/54—3.70%) ST2552 (2/54—3.70%) ST2554 (1/54—1.85%) ST2555 (1/54—1.85%) ST2556 (2/54—3.70%) ST2603 (1/54—1.85%) ST111 (2/54—3.70%) ST170 (1/54—1.85%) ST235 (5/54—9.26%) ST244 (9/54—16.67%) ST277 (6/54—11.11%) ST360 (1/54—1.85%) ST463 (1/54—1.85%) ST500 (2/54—3.70%) ST508 (1/54—1.85%) ST1076 (1/54—1.85%) ST1197 (1/54—1.85%) ST1284 (4/54—7.41%) ST1655 (1/54—1.85%) ST2100 (1/54 —1.85%) ST2437 (1/54—1.85%) | Yes—22/54 (40.7%)—ST244, ST277, ST111, ST235 | (8/8) |
| Esposito et al. (2021) # [50] | Retrospective cross-sectional | 2016 | Southeast region | Impacted urban rivers (environmental) | 2/2 | ST277 (2/2—100.0%) | Yes—(2/2—100.0%) ST277 | (8/8) |
| De Oliveira Luz et al. (2021) #,* [51] | Retrospective cross-sectional | 2010, 2015, and 2016 | Northeast region | Clinical | 13/13 | ST244 (2/13—15.38%) ST1394 (1/13—7.69%) ST252 (1/13—7.69%) ST3079 (3/13—23.07%) ST357 (2/13—15.38%) ST3078 (1/13—7.69%) ST275 (1/13—7.69%) ST3080 (1/13—7.69%) ST3137 (1/13—7.69%) | Yes—(4/13—30.76%)—ST244, ST357 | (8/8) |
| Santos et al. (2023) # [52] | Retrospective cross-sectional | 2018–2021 | North region | Clinical | 34/19 | ST277 (18/19—94.74%) ST2711 (1/19—5.26%) | Yes—(18/19—94.74%)—ST277 | (8/8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, Y.C.; Silva, M.J.A.; dos Reis, H.S.; dos Santos, P.A.S.; Sardinha, D.M.; Gouveia, M.I.M.; dos Santos, C.S.; Marcon, D.J.; Aires, C.A.M.; Souza, C.d.O.; et al. Molecular Epidemiology of Pseudomonas aeruginosa in Brazil: A Systematic Review and Meta-Analysis. Antibiotics 2024, 13, 983. https://doi.org/10.3390/antibiotics13100983
Rodrigues YC, Silva MJA, dos Reis HS, dos Santos PAS, Sardinha DM, Gouveia MIM, dos Santos CS, Marcon DJ, Aires CAM, Souza CdO, et al. Molecular Epidemiology of Pseudomonas aeruginosa in Brazil: A Systematic Review and Meta-Analysis. Antibiotics. 2024; 13(10):983. https://doi.org/10.3390/antibiotics13100983
Chicago/Turabian StyleRodrigues, Yan Corrêa, Marcos Jessé Abrahão Silva, Herald Souza dos Reis, Pabllo Antonny Silva dos Santos, Daniele Melo Sardinha, Maria Isabel Montoril Gouveia, Carolynne Silva dos Santos, Davi Josué Marcon, Caio Augusto Martins Aires, Cintya de Oliveira Souza, and et al. 2024. "Molecular Epidemiology of Pseudomonas aeruginosa in Brazil: A Systematic Review and Meta-Analysis" Antibiotics 13, no. 10: 983. https://doi.org/10.3390/antibiotics13100983
APA StyleRodrigues, Y. C., Silva, M. J. A., dos Reis, H. S., dos Santos, P. A. S., Sardinha, D. M., Gouveia, M. I. M., dos Santos, C. S., Marcon, D. J., Aires, C. A. M., Souza, C. d. O., Quaresma, A. J. P. G., Lima, L. N. G. C., Brasiliense, D. M., & Lima, K. V. B. (2024). Molecular Epidemiology of Pseudomonas aeruginosa in Brazil: A Systematic Review and Meta-Analysis. Antibiotics, 13(10), 983. https://doi.org/10.3390/antibiotics13100983











