Phenotypic and Genotypic Characterization of Escherichia coli and Salmonella spp. Isolates from Pigs at Slaughterhouse and from Commercial Pork Meat in Portugal
Abstract
1. Introduction
2. Results
2.1. Microbiological Quality of Pork Meat
2.2. E. coli and Salmonella spp. Isolation and Typing
2.3. Virulence Determinants
2.4. Antimicrobial Resistance and Plasmids
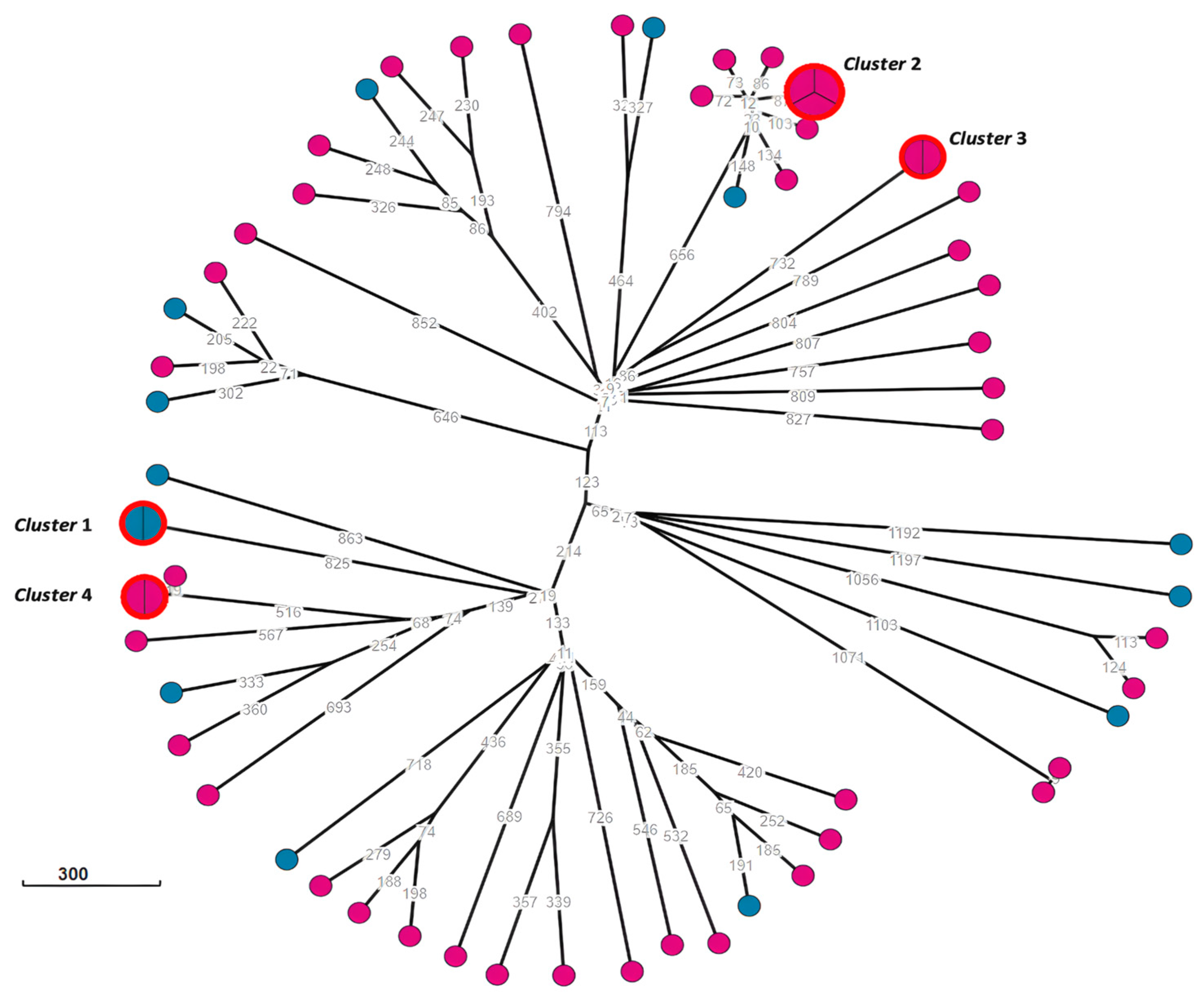
2.5. Phylogenetic Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Isolation Methodology from Faeces
4.3. Microbiological and Quality Analysis from Meat
4.4. Bacterial Typing and Serotyping
4.5. Antimicrobial Susceptibility Testing
4.6. Detection of mcr 1–9 Genes
4.7. Whole-Genome Sequencing and Genomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akbar, A.; Anal, A.K. Food Safety Concerns and Food-borne Pathogens, Salmonella, Escherichia coli and Campylobacter. FUUAST J. Biol. 2011, 1, 5–17. [Google Scholar]
- Hemalata, V.B.; Virupakshaiah, D.B.M. Isolation and Identification of food borne pathogens from Spoiled food samples. Int. J. Curr. Microbiol. App. Sci. 2016, 20165, 1017–1025. [Google Scholar] [CrossRef]
- Abebe, E.; Gugsa, G.; Ahmed, M. Review on Major Food-Borne Zoonotic Bacterial Pathogens. J. Trop. Med. 2020, 2020, 4674235. [Google Scholar] [CrossRef] [PubMed]
- Karimi, F. Safety of Food throughout the Supply Chain. In Logistics Engineering; Chelladurai, S.J.S., Mayilswamy, S., Gnanasekaran, S., Thirumalaisamy, R., Eds.; IntechOpen: London, UK, 2022. [Google Scholar] [CrossRef]
- World Health Organization. Estimating the Burden of Foodborne Diseases. 2015. Available online: https://www.who.int/activities/estimating-the-burden-of-foodborne-diseases (accessed on 1 August 2023).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [CrossRef]
- OECD. Food and Agriculture Organization of the United Nations. Meat. In OECD-FAO Agricultural Outlook 2023–2032; OECD Publishing: Paris, France, 2023. [Google Scholar] [CrossRef]
- Costa, R.D.; Silva, V.; Leite, A.; Saraiva, M.; Lopes, T.T.; Themudo, P.; Campos, J.; Vieira-Pinto, M. Salmonella spp., Escherichia coli and Enterobacteriaceae Control at a Pig Abattoir: Are We Missing Lairage Time Effect, Pig Skin, and Internal Carcass Surface Contamination? Foods 2023, 12, 2910. [Google Scholar] [CrossRef]
- Gabinete de Planeamento. Políticas e Administração Geral. Atividade Suinícola—Diagnóstico Do Setor E Propostas de Medidas de Promoção Da Sustentabilidade Da Atividade Suinícola. 2022. Available online: https://www.gpp.pt/images/Observatorio_Precos/Relatorios_analise_setores/Relatorio_GTSuinicultura_Diagnostico.pdf (accessed on 1 August 2023).
- Rajaei, M.; Moosavy, M.H.; Gharajalar, S.N.; Khatibi, S.A. Antibiotic resistance in the pathogenic foodborne bacteria isolated from raw kebab and hamburger: Phenotypic and genotypic study. BMC Microbiol. 2021, 21, 272. [Google Scholar] [CrossRef]
- Gomes-Neves, E.; Antunes, P.; Manageiro, V.; Gärtner, F.; Caniça, M.; da Costa, J.M.C.; Peixe, L. Clinically relevant multidrug resistant Salmonella enterica in swine and meat handlers at the abattoir. Vet. Microbiol. 2014, 168, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Lei, C.; Kong, L.; Jiang, W.; Liu, B.; Men, S.; Yang, Y.; Cheng, G.; Chen, Y.; Wang, H. Prevalence, Antimicrobial Resistance, and Relatedness of Salmonella Isolated from Chickens and Pigs on Farms, Abattoirs, and Markets in Sichuan Province, China. Foodborne Pathog. Dis. 2017, 14, 667–677. [Google Scholar] [CrossRef]
- Campos, J.; Mourão, J.; Peixe, L.; Antunes, P. Non-typhoidal Salmonella in the Pig Production Chain: A Comprehensive Analysis of Its Impact on Human Health. Pathogens 2019, 8, 19. [Google Scholar] [CrossRef]
- Martelli, F.; Oastler, C.; Barker, A.; Jackson, G.; Smith, R.P.; Davies, R. Abattoir-based study of Salmonella prevalence in pigs at slaughter in Great Britain. Epidemiol. Infect. 2021, 149, e218. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Lauteri, C.; Festino, A.R.; Conter, M.; Vergara, A. Prevalence and antimicrobial resistance profile in Salmonella spp. isolates from swine food chain. Ital. J. Food Saf. 2022, 11, 9980. [Google Scholar] [CrossRef] [PubMed]
- Roasto, M.; Bonardi, S.; Mäesaar, M.; Alban, L.; Gomes-Neves, E.; Vieira-Pinto, M.; Vågsholm, I.; Elias, T.; Lindegaard, L.L.; Blagojevic, B. Salmonella enterica prevalence, serotype diversity, antimicrobial resistance and control in the European pork production chain. Trends Food Sci. Technol. 2023, 131, 210–219. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. Available online: https://iris.who.int/bitstream/handle/10665/112642/9789241564748_eng.pdf (accessed on 1 August 2023).
- World Health Organization. Antimicrobial Resistance. 2023. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 1 August 2023).
- European Centre for Disease Prevention and Control. Assessing the Health Burden of Infections with Antibiotic-Resistant Bacteria in the EU/EEA, 2016–2020; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/health-burden-infections-antibiotic-resistant-bacteria-2016-2020 (accessed on 10 November 2023).
- O’Neill, J.; Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance. 2016. Available online: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (accessed on 21 March 2023).
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Organisation des Nations Unies Pour L’Alimentation et L’Agriculture. Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production. 2016. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/95efae06-4da2-4689-b12a-1b33fffc2244/content (accessed on 30 March 2023).
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A. Antibiotic Resistance in Bacteria—A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Costa, M.; Cardo, M.; Cara d’Anjo, M.; Leite, A. Assessing antimicrobial resistance occurrence in the Portuguese food system: Poultry, pigs and derived food, 2014–2018. Zoonoses Public Health 2022, 69, 312–324. [Google Scholar] [CrossRef]
- O’Neill, L.; Manzanilla, E.G.; Ekhlas, D.; Leonard, F.C. Antimicrobial Resistance in Commensal Escherichia coli of the Porcine Gastrointestinal Tract. Antibiotics 2023, 12, 1616. [Google Scholar] [CrossRef]
- European Medicines Agency. European Medicines Agency. European Surveillance of Veterinary Antimicrobial Consumption, 2022. In Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2022; European Medicines Agency: Amsterdam, The Netherlands, 2023; EMA/299538/2023. [Google Scholar]
- European Commission. Regulation (EC) No 1441/2007 of 5 December 2007. Amending Regulation (EC) No 2073/2005 on Microbiological Criteria for Foodstuffs; L322/12; Official Journal of the European Communities: Luxembourg, 2007. Available online: https://eur-lex.europa.eu/eli/reg/2007/1441/oj (accessed on 30 March 2023).
- Kanokudom, S.; Assawakongkarat, T.; Akeda, Y.; Ratthawongjirakul, P.; Chuanchuen, R.; Chaichanawongsaroj, N. Rapid detection of extended spectrum β-lactamase producing Escherichia coli isolated from fresh pork meat and pig cecum samples using multiplex recombinase polymerase amplification and lateral flow strip analysis. PLoS ONE 2021, 16, e0248536. [Google Scholar] [CrossRef]
- Chonsin, K.; Changkwanyeun, R.; Siriphap, A.; Intarapuk, A.; Prapasawat, W.; Changkaew, K.; Pulsrikarn, C.; Isoda, N.; Nakajima, C.; Suzuki, Y.; et al. Prevalence and Multidrug Resistance of Salmonella in Swine Production Chain in a Central Province, Thailand. J. Food Prot. 2021, 84, 2174–2184. [Google Scholar] [CrossRef]
- Marín, C.; Chinillac, M.C.; Cerdà-Cuéllar, M.; Montoro-Dasi, L.; Sevilla-Navarro, S.; Ayats, T. Contamination of pig carcass with Salmonella enterica serovar Typhimurium monophasic variant 1,4[5],12:i: Originates mainly in live animals. Sci. Total Environ. 2020, 703, 134609. [Google Scholar] [CrossRef]
- Moura-Alves, M.; Carvalho, M.; Baggio-Ribeiro, D.H.; Barbosa, J.; Silveira, L.; Pista, Â.; Pinto, H.P.; Saraiva, C.; Teixeira, P.; Esteves, A. Hygiene Indicators and Salmonella on Surfaces of Swine Carcasses from Two Slaughterhouses in Northern Portugal. J. Food Prot. 2022, 85, 1566–1575. [Google Scholar] [CrossRef] [PubMed]
- Deane, A.; Murphy, D.; Leonard, F.C.; Byrne, W.; Clegg, T.; Madigan, G.; Griffin, M.; Egan, J.; Prendergast, D.M. Prevalence of Salmonella spp. in slaughter pigs and carcasses in Irish abattoirs and their antimicrobial resistance. Ir. Vet. J. 2022, 75, 4. [Google Scholar] [CrossRef]
- Bantawa, K.; Rai, K.; Subba Limbu, D.; Khanal, H. Food-borne bacterial pathogens in marketed raw meat of Dharan, eastern Nepal. BMC Res. Notes 2018, 11, 618. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Vázquez, A.V.; Rivera-Sánchez, G.; Lira-Méndez, K.; Reyes-López, M.Á.; Bocanegra-García, V. Prevalence, antimicrobial resistance and virulence genes of Escherichia coli isolated from retail meat in Tamaulipas, Mexico. J. Glob. Antimicrob. Resist. 2018, 14, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Gomes, V.T.M.; Moreno, L.Z.; Silva, A.P.S.; Thakur, S.; La Ragione, R.M.; Mather, A.E.; Moreno, A.M. Characterization of Salmonella enterica Contamination in Pork and Poultry Meat from São Paulo/Brazil: Serotypes, Genotypes and Antimicrobial Resistance Profiles. Pathogens 2022, 11, 358. [Google Scholar] [CrossRef]
- Gambino, D.; Gargano, V.; Butera, G.; Sciortino, S.; Pizzo, M.; Oliveri, G.; Cardamone, C.; Piraino, C.; Cassata, G.; Vicari, D.; et al. Food Is Reservoir of MDR Salmonella: Prevalence of ESBLs Profiles and Resistance Genes in Strains Isolated from Food. Microorganisms 2022, 10, 780. [Google Scholar] [CrossRef]
- Zhang, L.; Fu, Y.; Xiong, Z.; Ma, Y.; Wei, Y.; Qu, X.; Zhang, H.; Zhang, J.; Liao, M. Highly Prevalent Multidrug-Resistant Salmonella From Chicken and Pork Meat at Retail Markets in Guangdong, China. Front. Microbiol. 2018, 9, 2140. [Google Scholar] [CrossRef]
- Tîrziu, E.; Barbalan, G.; Morar, A.; Herman, V.; Cristina, R.T.; Imre, K. Occurrence and Antimicrobial Susceptibility Profile of Salmonella spp. in Raw and Ready-To-Eat Foods and Campylobacter spp. in Retail Raw Chicken Meat in Transylvania, Romania. Foodborne Pathog. Dis. 2020, 17, 479–484. [Google Scholar] [CrossRef]
- Maio, R.; García-Díez, J.; Saraiva, C. Microbiological Quality of Foodstuffs Sold on Expiry Date at Retail in Portugal: A Preliminary Study. Foods 2020, 9, 919. [Google Scholar] [CrossRef]
- Schill, F.; Abdulmawjood, A.; Klein, G.; Reich, F. Prevalence and characterization of extended-spectrum β-lactamase (ESBL) and AmpC β-lactamase producing Enterobacteriaceae in fresh pork meat at processing level in Germany. Int. J. Food Microbiol. 2017, 257, 58–66. [Google Scholar] [CrossRef]
- Manyi-Loh, C.E.; Lues, R. A South African Perspective on the Microbiological and Chemical Quality of Meat: Plausible Public Health Implications. Microorganisms 2023, 11, 2484. [Google Scholar] [CrossRef] [PubMed]
- Dang-Xuan, S.; Nguyen-Viet, H.; Pham-Duc, P.; Grace, D.; Unger, F.; Nguyen-Hai, N.; Nguyen-Tien, T.; Makita, K. Simulating Cross-Contamination of Cooked Pork with Salmonella enterica from Raw Pork through Home Kitchen Preparation in Vietnam. Int. J. Environ. Res. Public Health 2018, 15, 2324. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. STEC Infection—Annual Epidemiological Report for 2022; ECDC: Stockholm, Sweden, 2024. Available online: https://www.ecdc.europa.eu/en/publications-data/stec-infection-annual-epidemiological-report-2022 (accessed on 10 February 2024).
- Hu, J.; Li, J.; Huang, X.; Xia, J.; Cui, M.; Huang, Y.; Wen, Y.; Xie, Y.; Zhao, Q.; Cao, S.; et al. Genomic traits of multidrug resistant enterotoxigenic Escherichia coli isolates from diarrheic pigs. Front. Microbiol. 2023, 14, 1244026. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, Y.; Liu, Q.; Sun, H.; Luo, M.; Xiong, Y.; Matussek, A.; Hu, B.; Bai, X. Genomic Characteristics of Stx2e-Producing Escherichia coli Strains Derived from Humans, Animals, and Meats. Pathogens 2021, 10, 1551. [Google Scholar] [CrossRef]
- Serrano, N.S.; Zweifel, C.; Corti, S.; Stephan, R. Microbiological quality and presence of foodborne pathogens in raw milk cheeses and raw meat products marketed at farm level in Switzerland. Ital. J. Food Saf. 2018, 7, 7337. [Google Scholar] [CrossRef]
- Shen, J.; Zhi, S.; Guo, D.; Jiang, Y.; Xu, X.; Zhao, L.; Lv, J. Prevalence, Antimicrobial Resistance, and Whole Genome Sequencing Analysis of Shiga Toxin-Producing Escherichia coli (STEC) and Enteropathogenic Escherichia coli (EPEC) from Imported Foods in China during 2015–2021. Toxins 2022, 14, 68. [Google Scholar] [CrossRef] [PubMed]
- von Mentzer, A.; Blackwell, G.A.; Pickard, D.; Boinett, C.J.; Joffré, E.; Page, A.J.; Svennerholm, A.-M.; Dougan, G.; Sjöling, Å. Long-read-sequenced reference genomes of the seven major lineages of enterotoxigenic Escherichia coli (ETEC) circulating in modern time. Sci. Rep. 2021, 11, 9256. [Google Scholar] [CrossRef]
- Reid, C.J.; DeMaere, M.Z.; Djordjevic, S.P. Australian porcine clonal complex 10 (CC10) Escherichia coli belong to multiple sublineages of a highly diverse global CC10 phylogeny. Microb. Genom. 2019, 5, e000225. [Google Scholar] [CrossRef]
- Bernreiter-Hofer, T.; Schwarz, L.; Müller, E.; Cabal-Rosel, A.; Korus, M.; Misic, D.; Frankenfeld, K.; Abraham, K.; Grünzweil, O.; Weiss, A.; et al. The Pheno- and Genotypic Characterization of Porcine Escherichia coli Isolates. Microorganisms 2021, 9, 1676. [Google Scholar] [CrossRef]
- Massella, E.; Giacometti, F.; Bonilauri, P.; Reid, C.J.; Djordjevic, S.P.; Merialdi, G.; Bacci, C.; Fiorentini, L.; Massi, P.; Bardasi, L.; et al. Antimicrobial Resistance Profile and ExPEC Virulence Potential in Commensal Escherichia coli of Multiple Sources. Antibiotics 2021, 10, 351. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; BuglaPloskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence, and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Sora, V.M.; Meroni, G.; Martino, P.A.; Soggiu, A.; Bonizzi, L.; Zecconi, A. Extraintestinal Pathogenic Escherichia coli: Virulence Factors and Antibiotic Resistance. Pathogens 2021, 10, 1355. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microb. 2013, 21, 3107–3117. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
- Shen, W.; Chen, H.; Geng, J.; Wu, R.A.; Wang, X.; Ding, T. Prevalence, serovar distribution, and antibiotic resistance of Salmonella spp. isolated from pork in China: A systematic review and meta-analysis. Int. J. Food Microb. 2022, 361, 109473. [Google Scholar] [CrossRef]
- Diab, M.S.; Thabet, A.S.; Elsalam, M.A.; Ewida, R.M.; Sotohy, S.A. Detection of Virulence and β-lactamase resistance genes of non-typhoidal Salmonella isolates from human and animal origin in Egypt “one health concern”. Gut Pathog. 2023, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Sabry, M.A.; Abdel-Moein, K.A.; Abdel-Kader, F.; Hamza, E. Extended-spectrum β-lactamase-producing Salmonella serovars among healthy and diseased chickens and their public health implication. J. Glob. Antim. Resist. 2020, 22, 742–748. [Google Scholar] [CrossRef]
- Nikiema, M.E.M.; Kakou-Ngazoa, S.; Ky-Ba, A.; Sylla, A.; Bako, E.; Addablah, A.Y.A.; Ouoba, J.B.; Sampo, E.; Gnada, K.; Zongo, O.; et al. Characterization of virulence factors of Salmonella isolated from human stools and street food in urban areas of Burkina Faso. BMC Microb. 2021, 21, 338. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.L.E.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef]
- Day, M.J.; Rodríguez, I.; van Essen-Zandbergen, A.; Dierikx, C.; Kadlec, K.; Schink, A.-K.; Wu, G.; Chattaway, M.A.; DoNascimento, V.; Wain, J.; et al. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J. Antim. Chemoth. 2016, 71, 1178–1182. [Google Scholar] [CrossRef]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2021–2022. EFSA J. 2024, 22, e8583. [Google Scholar] [CrossRef]
- Clemente, L.; Leão, C.; Moura, L.; Albuquerque, T.; Amaro, A. Prevalence and Characterization of ESBL/AmpC Producing Escherichia coli from Fresh Meat in Portugal. Antibiotics 2021, 10, 1333. [Google Scholar] [CrossRef] [PubMed]
- Amaro, A.; Leão, C.; Guerra, V.; Albuquerque, T.; Clemente, L. Plasmid-Mediated Colistin Resistance Genes mcr-1 and mcr-4 in Multidrug-Resistant Escherichia coli Strains Isolated from a Healthy Pig in Portugal. Microb. Drug Resist. 2023, 29, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- Rebuffat, S. Microcins in action: Amazing defence strategies of Enterobacteria. Biochem. Soc. Trans. 2012, 40, 1456–1462. [Google Scholar] [CrossRef]
- Massip, C.; Oswald, E. Siderophore-Microcins in Escherichia coli: Determinants of Digestive Colonization, the First Step Toward Virulence. Front. Cell. Infect. Microbiol. 2020, 10, 381. [Google Scholar] [CrossRef]
- Mey, A.R.; Gómez-Garzón, C.; Payne, S.M. Iron Transport and Metabolism in Escherichia, Shigella, and Salmonella. EcoSal Plus 2021, 9. [Google Scholar] [CrossRef]
- Pista, A.; Silveira, L.; Ribeiro, S.; Fontes, M.; Castro, R.; Coelho, A.; Furtado, R.; Lopes, T.; Maia, C.; Mixão, V.; et al. Pathogenic Escherichia coli, Salmonella spp. and Campylobacter spp. in Two Natural Conservation Centers of Wildlife in Portugal: Genotypic and Phenotypic Characterization. Microorganisms 2022, 10, 2132. [Google Scholar] [CrossRef]
- Sabat, G.; Rose, P.; Hickey, W.J.; Harkin, J.M. Selective and Sensitive Method for PCR Amplification of Escherichia coli 16S rRNA Genes in Soil. Appl. Environ. Microbiol. 2000, 66, 844–849. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food Chain-Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. ISO: Geneva, Switzerland, 2017.
- ISO 7218:2007; Microbiology of Food and Animal Feeding Stuffs—General Requirements and Guidance for Microbiological Examinations. ISO: Geneva, Switzerland, 2007.
- Persson, S.; Olsen, K.E.; Scheutz, F.; Krogfelt, K.A.; Gerner-Smidt, P. A method for fast and simple detection of major diarrhoeagenic Escherichia coli in the routine diagnostic laboratory. Clin. Microbiol. Infect. 2007, 13, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Boisen, N.; Scheutz, F.; Rasko, D.A.; Redman, J.C.; Persson, S.; Simon, J.; Kotloff, K.L.; Levine, M.M.; Sow, S.; Tamboura, B.; et al. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J. Infect. Dis. 2012, 205, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, M.; Otomo, Y.; Ahsan, C.R. A novel single-step multiplex polymerase chain reaction assay for the detection of diarrheagenic Escherichia coli. J. Microbiol. Methods 2013, 92, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Piérard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef]
- Grimont, P.A.D.; Weill, F.-X. WHO Collaborating Centre for Reference and Research on Salmonella. In Antigenic Formulae of the Salmonella Serovars, 9th ed.; Grimont, P.A.D., Weill, F.X., Eds.; Institut Pasteur: Paris, France, 2007; pp. 1–166. [Google Scholar]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 13.1. 2023. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.1_Breakpoint_Tables.pdf (accessed on 30 March 2023).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for detection of plasmid mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018, 23, 17–00672. [Google Scholar] [CrossRef]
- Borowiak, M.; Baumann, B.; Fischer, J.; Thomas, K.; Deneke, C.; Hammerl, J.A.; Szabo, I.; Malorny, B. Development of a Novel mcr-6 to mcr-9 Multiplex PCR and Assessment of mcr-1 to mcr-9 Occurrence in Colistin-Resistant Salmonella enterica Isolates From Environment, Feed, Animals and Food (2011–2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef]
- Bertelloni, F.; Cagnoli, G.; Turchi, B.; Ebani, V.V. Low Level of Colistin Resistance and mcr Genes Presence in Salmonella spp.: Evaluation of Isolates Collected between 2000 and 2020 from Animals and Environment. Antibiotics 2022, 11, 272. [Google Scholar] [CrossRef]
- Achtman, M.; Zhou, Z.; Alikhan, N.F.; Tyne, W.; Parkhill, J.; Cormican, M.; Chiou, C.S.; Torpdahl, M.; Litrup, E.; Prendergast, D.M.; et al. Genomic diversity of Salmonella enterica—The UoWUCC 10K genomes project. Wellcome Open Res. 2020, 5, 223. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Agama Study Group; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carrico, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed]

| Microbiological Quality | Satisfactory (%) | Borderline (%) | Unsatisfactory (%) |
|---|---|---|---|
| E. coli count (cfu/g) | 48 (92.3) | 0 | 4 (7.7) |
| Presence of Salmonella spp. | 49 (94.2) | N/A | 3 (5.8) |
| No. of Tested Samples | Pork Meat | Pig Faeces | Total | |
|---|---|---|---|---|
| 52 | 100 | 152 | ||
| E. coli isolates | Total (% +ve) | 36 (69.2) | 100 (100) | 136 (89.5) |
| Pathogenic (% +ve) | 14 (26.9) | 47 (47.0 a) | 61 (40.1) | |
| STEC (% +ve) | 0 | 2 (2.0 b) | 2 (1.3) | |
| ETEC (% +ve) | 0 | 4 (4.0 c) | 4 (2.6) | |
| ExPEC (% +ve) | 14 (26.9) | 41 (41.0 b,c) | 55 (36.2) | |
| Salmonella isolates | Total (% +ve) | 3 (5.8) | 0 | 3 (2.0) |
| S. London (% +ve) | 1 (1.9) | 0 | 1 (0.7) | |
| S. Typhimurium (% +ve) | 1 (1.9) | 0 | 1 (0.7) | |
| S. monophasic Typhimurium (% +ve) | 1 (1.9) | 0 | 1 (0.7) | |
| E. coli | |||
| Isolate ID | Phenotypic profile | Resistance genes and point mutations related to phenotype profile | Other resistance genes |
| Ec_SM1-1 | AMP, AMC, TET, CHL, TMP, SMX | blaTEM-1C, tet(A), floR, dfrA14, sul2 | aph(3″)-Ib, aph(6)-Id |
| Ec_SM3-1 | AMP, TET, CHL, TMP | blaTEM-1B, tet(A), cmlA1, dfrA12 | aadA1, aadA2 |
| Ec_SM5-1 | AMP, TET, TMP, SMX | blaTEM-1B, tet(B), dfrA1, sul3, | aadA1 |
| Ec_SM6-1 | AMP, TET, TMP, SMX | blaTEM-1B, tet(A), dfrA1, sul3 | qnrS1, aadA1, aph(6)-Id |
| Ec_SM8-1 | Susceptible | No | No |
| Ec_SM8-3 | TET, CHL, TMP, SMX | tet(A), cmlA1, dfrA12, sul3 | aadA1, aadA2, qacL |
| Ec_SM10-1 | TET | tet(A) | No |
| Ec_SM11-2 | AMP, TET, TMP, SMX | blaTEM-1B, tet(A), dfrA1, sul3 | aadA1 |
| Ec_SM12-2 | TET, CHL, TMP, SMX | tet(A), cmlA1, dfrA12, sul3 | mef(B), aadA1, aadA2 |
| Ec_SM13-3 | AMP, TET, SMX | blaTEM-1B, tet(B), sul3 | No |
| Ec_SM15-3 | AMP, TET, CHL | blaTEM-1B, tet(B), floR | sul2, aph(3″)-Ib, aph(6)-Id |
| Ec_SM16-2 | AMP, TET, CHL, TMP | blaTEM-1B, tet(A), cmlA1, dfrA12 | aadA2 |
| Ec_SM17-4 | AMP, AMC, TET, CHL | blaTEM-1B, tet(B), floR | aph(6)-Id |
| Ec_SM19-4 | AMP, AMC, TET, CHL, TMP, SMX | blaTEM-1B, tet(A), cmlA1, dfrA12, sul3 | aadA1, aadA2 |
| Ec_SM24-1 | AMP, TET, SMX, NAL, CIP | blaTEM-1B, tet(B), sul3, qnrS1, gyrA (p.S83A) | aph(3″)-Ib, aph(6)-Id |
| Ec_SM28-3 | AMP, AMC, TET, TMP, SMX | blaTEM-1A, tet(A), dfrA1, sul3 | No |
| Ec_SM30-2 | TET | tet(B) | No |
| Ec_SM31-3 | AMP, TET, TMP, SMX, NAL | blaTEM-1B, tet(A), dfrA14, sul2, gyrA (p.D87Y) | mph(G), aph(6)-Id |
| Ec_SM33-4 | AMP, CHL, SMX | blaTEM-1B, floR, sul2 | aph(3″)-Ib, aph(6)-Id |
| Ec_SM34-2 | TET | tet(A) | No |
| Ec_SM39-3 | Susceptible | No | No |
| Ec_SM43-2 | AMP, AMC, TET, TMP, SMX | blaTEM-1B, tet(B), dfrA1, sul1 | aadA1 |
| Ec_SM43-5 | TET | tet(A) | No |
| Ec_SM44-3 | AMP, TET, SMX, NAL | blaTEM-1B, tet(A), sul3, gyrA (p.S83A) | mef(B), aph(6)-Id |
| Ec_SM45-4 | AMP, TET, CHL, TMP, SMX | blaTEM-1B, tet(B), cmlA1, dfrA12, sul3 | aadA1, aadA2, aph(6)-Id |
| Ec_SM73-1 | AMP, TET, CHL, SMX | blaTEM-1B, tet(A), cmlA1, sul2, sul3 | qnrS1, dfrA12, aadA1 |
| Ec_SM74-1 | AMP, TET, CHL, SMX | blaTEM-1B, tet(A), floR, sul2 | qnrS1 |
| Ec_SM77-1 | AMP, TET, TMP, SMX, NAL, CIP | blaTEM-1B, tet(A), dfrA17, sul2, gyrA (p.S83L), gyrA (p.D87N), parC (p.S80I) | mph(A), aadA5, aph(3″)-Ib, aph(6)-Id |
| Ec_SM78-1 | AMP, TET, TMP, SMX | blaTEM-1A, tet(B), dfrA1, sul3 | aadA1 |
| Ec_SM79-1 | AMP, TET, SMX, NAL | blaTEM-1B, tet(B), sul3, gyrA (p.S83A) | mef(B), aph(3″)-Ib, aph(6)-Id |
| Ec_SM114-1 | AMP, TET, SMX | blaTEM-1B, tet(A), sul1 | aadA12 |
| Ec_SM141-1 | AMP, TET, CHL, TMP, SMX | blaTEM-1B, tet(A), cmlA1, dfrA12, sul3 | mef(B), aadA1, aadA2 |
| Ec_SM142-1 | AMP, TET, CHL, TMP, SMX, CIP | blaTEM-1B, tet(A), cmlA1, dfrA12, sul3, qnrS1 | No |
| Ec_SM145-1 | AMP, AZM, TMP, SMX, NAL | blaTEM-1B, mph(G), dfrA5, sul2, gyrA (p.S83L) | aph(3″)-Ib, aph(6)-Id |
| Ec_SM146-1 | AMP, AZM, TMP, SMX, NAL | blaTEM-1B, mph(G), dfrA5, sul2, gyrA (p.S83L) | aph(3″)-Ib, aph(6)-Id |
| Ec_SM147-1 | AMP, TET, TMP | blaTEM-1A, tet(B), dfrA1, dfrA5 | aadA1 |
| Ec_SM148-1 | AMP, AZM, TMP, SMX, NAL | blaTEM-1B, mph(G), dfrA5, sul2, gyrA (p.S83L) | aph(3″)-Ib, aph(6)-Id |
| Ec_SM150-1 | AMP, FEP, COX, CRO, TMP, SMX | blaCTX-M-1, dfrA17, sul2 | aadA5 |
| Ec_SM152-1 | AMP, TMP, SMX | blaTEM-1B, dfrA5, sul2 | aph(3″)-Ib, aph(6)-Id |
| Ec_SM153-1 | AMP, TET, TMP, SMX | blaTEM-1A, tet(A), dfrA8, sul2 | aph(6)-Id |
| Ec_SM154-3 | AMP, TET, TMP, SMX | blaTEM-1B, tet(A), dfrA14, sul2 | aph(6)-Id |
| Ec_SM164-1 | AMP, TET, CHL | blaTEM-1B, tet(A), floR | qnrS1 |
| Ec_SM164-3 | AMP, TET, SMX | blaTEM-1B, tet(B), sul3 | aph(6)-Id |
| Ec_SM165-2 | CHL, SMX, CIP, NAL | floR, sul2, qnrS1, gyrA (p.S83L) | No |
| Ec_SM166-3 | CHL, SMX | floR, sul2 | qnrS1 |
| Ec_SM196-1 | AMP, TET, CHL, SMX | blaTEM-1C, tet(A), floR, sul2 | aph(3″)-Ib |
| Ec_SM206-1 | TMP, SMX, NAL | dfrA1, sul3, gyrA (p.S83L) | aadA1, aph(3″)-Ib, aph(6)-Id |
| Isolate ID | Phenotypic profile | Resistance genes and point mutations related to phenotype profile | Other resistance genes |
| Ec_CC8-1 | SMX, CIP, NAL | sul3, gyrA (p.S83L), gyrA (p.D87N), parC (p.S80I) | No |
| Ec_CC9-5 | AMC, AMP, TET, CHL, TMP, SMX | blaTEM-1B, tet(A), cmlA1, dfrA8, sul3 | qnrS1, aadA1, aph(6)-Id |
| Ec_CC10-1 | AMC, AMP, TET, CHL, TMP, SMX | blaTEM-1B, tet(A), dfrA1, sul2 | mph(B), aadA1, aph(3″)-Ib, aph(6)-Id |
| Ec_CC13-4 | AMP, TET, SMX | blaTEM-1B, tet(A), sul3 | No |
| Ec_CC15-4 | AMC, AMP, TET | blaTEM-1C, tet(A) | No |
| Ec_CC22-3 | AMP, TET, SMX | blaTEM-1B, tet(A), sul1 | aadA12 |
| Ec_CC24-1 | TET, CHL, SMX, NAL | tet(A), cmlA1, sul3, gyrA (p.S83L) | dfrA12, aadA2 |
| Ec_CC27-4 | AMP, TET, SMX | blaTEM-1B, tet(A), sul1 | No |
| Ec_CC28-2 | AMP, TET, TMP, SMX | blaTEM-1B, tet(A), dfrA1, sul1 | aadA1 |
| Ec_CC34-1 | AMP, TET, TMP, SMX, CIP, NAL | blaTEM-1B, tet(A), dfrA14, sul2, gyrA (p.S83L), gyrA (p.D87N), parC (p.S80I) | aph(6)-Id |
| Ec_CC44-5 | TET, TMP, SMX | tet(A), dfrA14, sul2 | qnrS1, aph(3″)-Ib, aph(6)-Id |
| Ec_CC46-1 | AMP, TET, SMX | blaTEM-1B, tet(A), sul2 | aph(3″)-Ib, aph(6)-Id |
| Ec_CC47-2 | AMP, TET, CHL, TMP, SMX, NAL | blaTEM-1C, tet(A), cmlA1, dfrA12, sul3, gyrA (p.S83L), parE (p.I529L) | aadA1, aadA2 |
| Ec_CC48-2 | AMP, TET, CHL, TMP, SMX | blaTEM-1A, tet(A), cmlA1, dfrA1, sul2, sul3 | aph(6)-Id |
| Salmonella spp. | |||
| Isolate ID | Phenotypic profile | Resistance genes and point mutations related to phenotype profile | Other resistance genes |
| Se_CC10 | PEF, NAL | qnrB19, qnrB82 | aac(6′)-Iaa |
| Se_CC26 | AMP, TET, SMX | blaTEM-1B, tet(B), sul2 | aac(6′)-Iaa, aph(3″)-Ib, aph(6)-Id |
| Se_CC47 | AMP, CHL, TMP, SMX | blaTEM-1B, cmlA1, dfrA12, sul3 | aac(6′)-Iaa, aadA1, aadA2 |
| Microbiological Quality Parameters | Interpretation | ||
|---|---|---|---|
| Satisfactory | Borderline | Unsatisfactory | |
| E. coli count (cfu/g) | ≤500 | >500 and ≤5000 | >5000 |
| Presence of Salmonella spp. (in 25 g) | Not detected | N/A | Detected |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, C.; Silveira, L.; Rodrigues, J.; Furtado, R.; Ramos, S.; Nunes, A.; Pista, Â. Phenotypic and Genotypic Characterization of Escherichia coli and Salmonella spp. Isolates from Pigs at Slaughterhouse and from Commercial Pork Meat in Portugal. Antibiotics 2024, 13, 957. https://doi.org/10.3390/antibiotics13100957
Gonçalves C, Silveira L, Rodrigues J, Furtado R, Ramos S, Nunes A, Pista Â. Phenotypic and Genotypic Characterization of Escherichia coli and Salmonella spp. Isolates from Pigs at Slaughterhouse and from Commercial Pork Meat in Portugal. Antibiotics. 2024; 13(10):957. https://doi.org/10.3390/antibiotics13100957
Chicago/Turabian StyleGonçalves, Carlota, Leonor Silveira, João Rodrigues, Rosália Furtado, Sónia Ramos, Alexandra Nunes, and Ângela Pista. 2024. "Phenotypic and Genotypic Characterization of Escherichia coli and Salmonella spp. Isolates from Pigs at Slaughterhouse and from Commercial Pork Meat in Portugal" Antibiotics 13, no. 10: 957. https://doi.org/10.3390/antibiotics13100957
APA StyleGonçalves, C., Silveira, L., Rodrigues, J., Furtado, R., Ramos, S., Nunes, A., & Pista, Â. (2024). Phenotypic and Genotypic Characterization of Escherichia coli and Salmonella spp. Isolates from Pigs at Slaughterhouse and from Commercial Pork Meat in Portugal. Antibiotics, 13(10), 957. https://doi.org/10.3390/antibiotics13100957






