Short Antimicrobial Peptide Derived from the Venom Gland Transcriptome of Pamphobeteus verdolaga Increases Gentamicin Susceptibility of Multidrug-Resistant Klebsiella pneumoniae
Abstract
:1. Introduction
2. Results
2.1. Prediction and Synthesis of Short Cationic Hypothetical AMPs in P. verdolaga’s Transcriptome
2.2. Screening the Potential Antimicrobial Activity of P. verdolaga’s Hypothetical AMPs
2.3. MIC, MBC, and IC50 Values of P. verdolaga’s AMPs against ESKAPE Group Bacteria
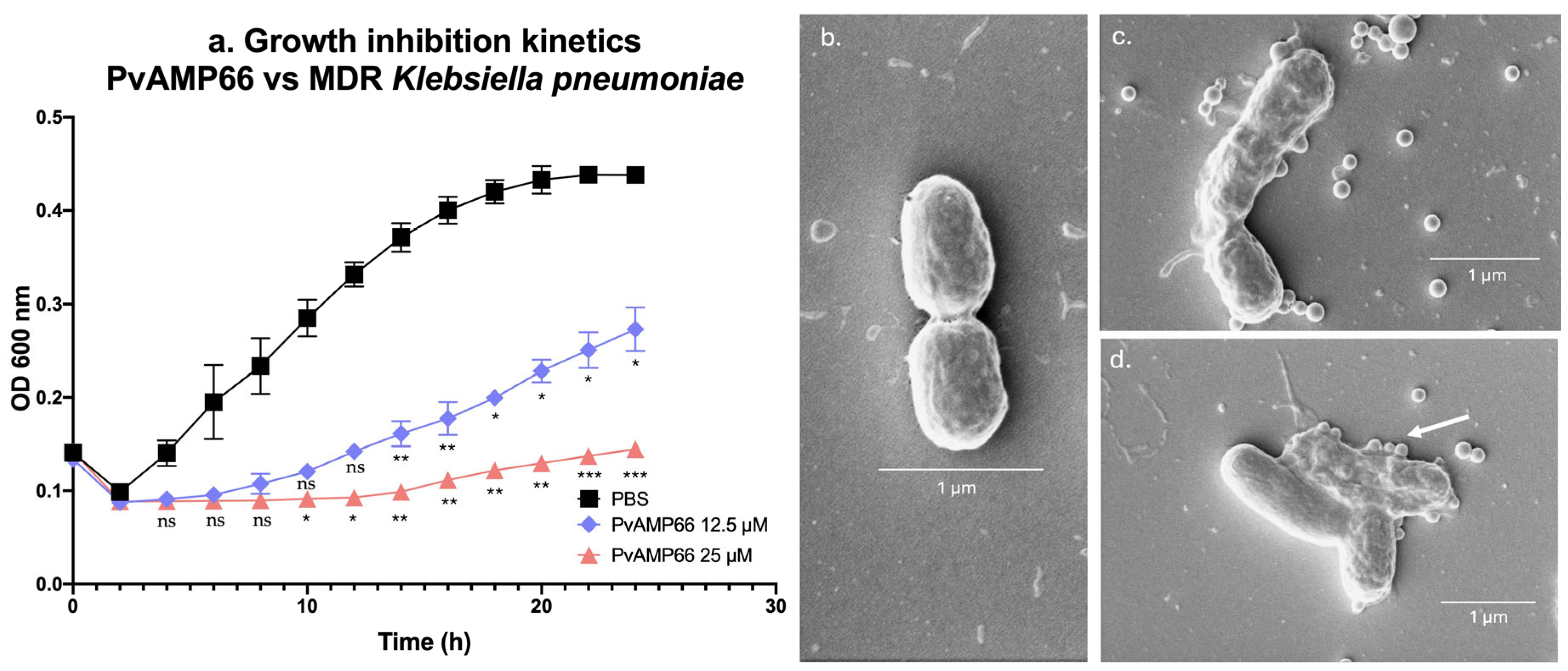
2.4. Assessing the Susceptibility of a Multidrug-Resistant K. pneumoniae Strain to PvAMP66, and Its Interaction with Gentamicin
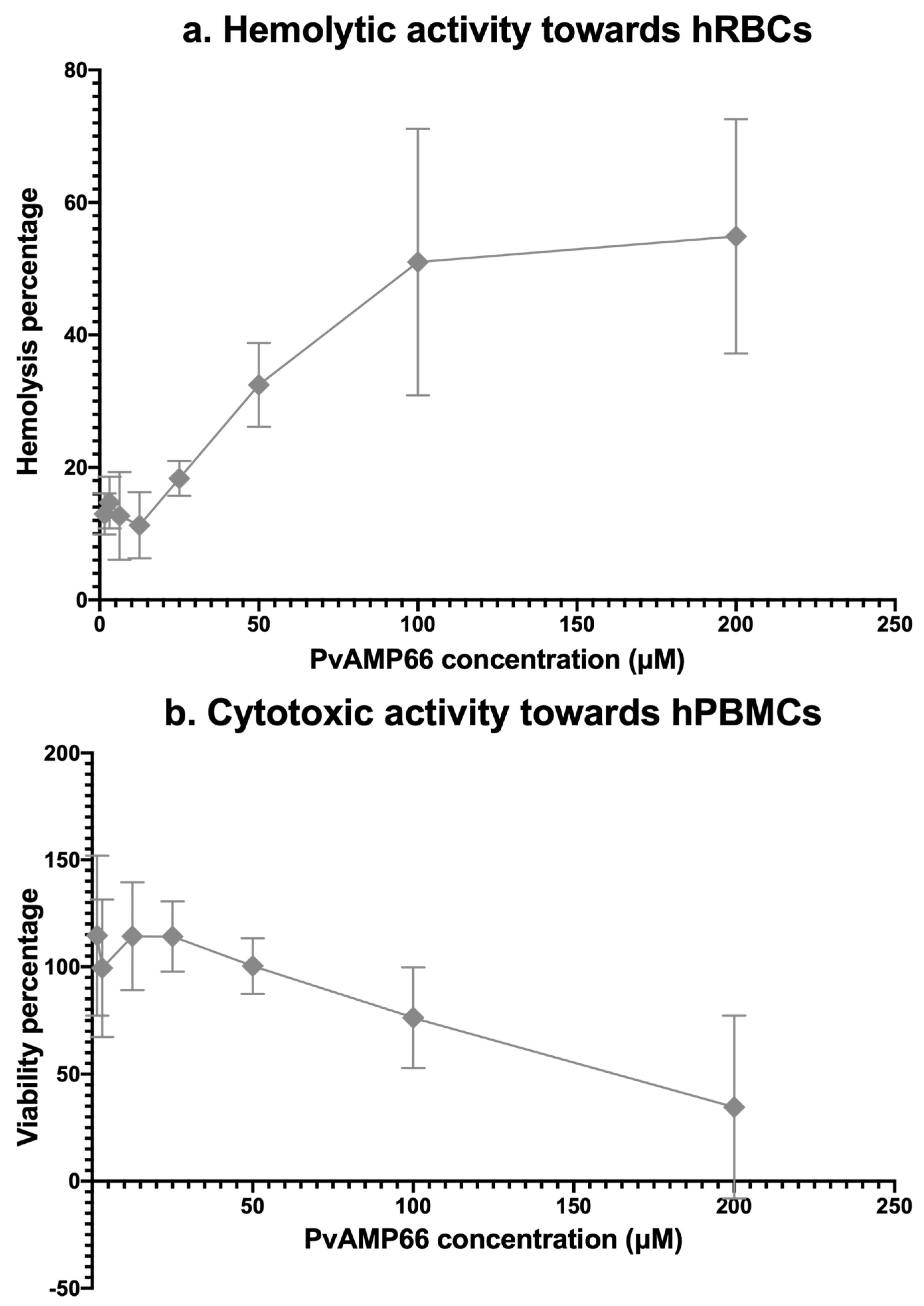
2.5. Assessing the Cytotoxicity of PvAMP66
3. Discussion
3.1. Prospecting Hypothetical AMPs from the Venom Gland Transcriptome of P. verdolaga
3.2. Activity of the Hypothetical AMPs from P. verdolaga
3.3. PvAMP66 Enhances Gentamicin Inhibition of MDR K. pneumoniae Growth
3.4. PvAMP66 Is P. verdolaga’s Most Active AMP
4. Materials and Methods
4.1. Reagents & Bacterial Strains
4.2. Creation of the Custom AMP Database and Prediction of Antimicrobial Peptides from the Venom Gland Transcriptome of P. verdolaga
4.3. Synthesis, Purification, Disulfide Bridge Formation, and Quality Control of Candidate AMPs
4.4. Evaluation of the Antimicrobial Activity of P. verdolaga’s AMPs
4.5. Scanning Electron Microscopy Analysis
4.6. Evaluation of the Hemolytic Activity and Cytotoxicity of P. verdolaga’s AMPs
4.7. Tables, Graphs, and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). The Top 10 Causes of Death. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 30 April 2021).
- Woolhouse, M.; Waugh, C.; Perry, M.R.; Nair, H. Global disease burden due to antibiotic resistance—State of the evidence. J. Glob. Health 2016, 6, 010306. [Google Scholar] [CrossRef]
- World Health Organization (WHO), WHO, Department of Health Systems, WHO, Financing Health Systems and Services. Who Guide to Identifying the Economic Consequences of Disease and Injury. 2009. Available online: https://www.who.int/publications/i/item/9789241598293 (accessed on 31 May 2023).
- Bloom, D.E.; Kuhn, M.; Prettner, K. Modern Infectious Diseases: Macroeconomic Impacts and Policy Responses; IZA—Institute of Labor Economics: Bonn, Germany, 2020; Available online: www.iza.org (accessed on 31 May 2023).
- World Health Organization (WHO). Worldwide Country Situation Analysis: Response to Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; pp. 1–50. ISBN 9789241564946. Available online: https://iris.who.int/bitstream/handle/10665/163468/9789241564946_eng.pdf?sequence=1&isAllowed=y (accessed on 31 March 2018).
- World Health Organization (WHO). Antimicrobial resistance. Glob. Rep. Surveill. 2014, 61, 12–28. [Google Scholar]
- World Health Organization (WHO). 2021 Antibacterial Agents in Clinical and Preclinical Development: An Overview and Analysis. 2022. Available online: https://www.who.int/publications/i/item/9789240047655 (accessed on 31 March 2022).
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Her Majesty Government: London, UK, 2014; pp. 1–18. [Google Scholar]
- Yewale, V.N. Antimicrobial Resistance—A Ticking Bomb! Cadman, H., Martinez, L., Eds.; World Health Organization: Geneva, Switzerland, 2014; Volume 51, pp. 171–172. [Google Scholar]
- Centers for Disease Control (CDC). Antibiotic Resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 31 March 2023).
- El-Mahallawy, H.A.; Hassan, S.S.; El-Wakil, M.; Moneer, M.M. Bacteremia due to ESKAPE pathogens: An emerging problem in cancer patients. J. Egypt. Natl. Cancer Inst. 2016, 28, 157–162. [Google Scholar] [CrossRef]
- Ye, Q.F.; Zhao, J.; Wan, Q.Q.; Qiao, B.B.; Zhou, J.D. Frequency and clinical outcomes of ESKAPE bacteremia in solid organ transplantation and the risk factors for mortality. Transpl. Infect Dis. Off. J. Transpl. Soc. 2014, 16, 767–774. [Google Scholar] [CrossRef]
- Faye, I.; Lindberg, B.G. Towards a paradigm shift in innate immunity—Seminal work by Hans G. Boman and co-workers. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 1695. [Google Scholar] [CrossRef]
- Kim, H.S.; Yoon, H.; Minn, I.; Park, C.B.; Lee, W.T.; Zasloff, M.; Kim, S.C. Pepsin-Mediated Processing of the Cytoplasmic Histone H2A to Strong Antimicrobial Peptide Bufori. J. Immunol. 2000, 165, 3268–3274. [Google Scholar] [CrossRef]
- Wada, Y.; Lönnerdal, B. Bioactive peptides derived from human milk proteins: An update. Curr. Opin. Clin. Nutr. Metab. Care 2020, 23, 217–222. [Google Scholar] [CrossRef]
- Melo, M.N.; Ferre, R.; Castanho, M.A.R.B. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef]
- Makurvet, F.D. Biologics vs. small molecules: Drug costs and patient access. Med. Drug Discov. 2020, 9, 100075. [Google Scholar] [CrossRef]
- Jochumsen, N.; Marvig, R.L.; Damkiær, S.; Jensen, R.L.; Paulander, W.; Molin, S.; Jelsbak, L.; Folkesson, A. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 2016, 7, 13002. [Google Scholar] [CrossRef]
- Kintses, B.; Jangir, P.K.; Fekete, G.; Számel, M.; Méhi, O.; Spohn, R.; Daruka, L.; Martins, A.; Hosseinnia, A.; Gagarinova, A.; et al. Chemical-genetic profiling reveals limited cross-resistance between antimicrobial peptides with different modes of action. Nat. Commun. 2019, 10, 5731. [Google Scholar] [CrossRef]
- Spohn, R.; Daruka, L.; Lázár, V.; Martins, A.; Vidovics, F.; Grézal, G.; Méhi, O.; Kintses, B.; Számel, M.; Jangir, P.K.; et al. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019, 10, 4538. [Google Scholar] [CrossRef]
- Maron, B.; Rolff, J.; Friedman, J.; Hayouka, Z. Antimicrobial Peptide Combination Can Hinder Resistance Evolution. Microbiol. Spectr. 2022, 10, e0097322. [Google Scholar] [CrossRef]
- Peschel, A.; Sahl, H.G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529–536. [Google Scholar] [CrossRef]
- Cifuentes, Y.; Estrada-Gomez, S.; Vargas-Muñoz, L.J.; Perafán, C. Description and molecular characterization of a new species of tarantula, pamphobeteus verdolaga, from colombia (Araneae: Mygalomorphae: Theraphosidae). Zoologia 2016, 33, 6–11. [Google Scholar] [CrossRef]
- Estrada-Gomez, S.; Cardoso, F.C.; Vargas-Muñoz, L.J.; Quintana-Castillo, J.C.; Gómez, C.M.A.; Pineda, S.S.; Saldarriaga-Cordoba, M.M. Venomic, transcriptomic, and bioactivity analyses of pamphobeteus verdolaga venom reveal complex disulfide-rich peptides that modulate calcium channels. Toxins 2019, 11, 21. [Google Scholar] [CrossRef]
- Salinas-Restrepo, C.; Misas, E.; Estrada-Gómez, S.; Quintana-Castillo, J.C.; Guzman, F.; Calderón, J.C.; Giraldo, M.A.; Segura, C. Improving the Annotation of the Venom Gland Transcriptome of Pamphobeteus verdolaga, Prospecting Novel Bioactive Peptides. Toxins 2022, 14, 408. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Wayne, P. Clinical and Laboratory Standards Institute. CLSI. Performance Standards for Antimicrobial Susceptability Testing, 30th ed.; CLSI Suplement M100; Clinical and Laboratory Standards Institute: Wayne, IL, USA, 2020. [Google Scholar]
- Huang, R.-Y.; Pei, L.; Liu, Q.; Chen, S.; Dou, H.; Shu, G.; Yuan, Z.-X.; Lin, J.; Peng, G.; Zhang, W.; et al. Isobologram analysis: A comprehensive review of methodology and current research. Front. Pharmacol. 2019, 10, 1222. [Google Scholar] [CrossRef]
- Ebou, A.; Koua, D.; Addablah, A.; Kakou-Ngazoa, S.; Dutertre, S. Combined Proteotranscriptomic-Based Strategy to Discover Novel Antimicrobial Peptides from Cone Snails. Biomedicines 2021, 9, 344. [Google Scholar] [CrossRef]
- Habet, S. Narrow Therapeutic Index drugs: Clinical pharmacology perspective. J. Pharm. Pharmacol. 2021, 73, 1285–1291. [Google Scholar] [CrossRef]
- Tamargo, J.; Le Heuzey, J.-Y.; Mabo, P. Narrow therapeutic index drugs: A clinical pharmacological consideration to flecainide. Eur. J. Clin. Pharmacol. 2015, 71, 549–567. [Google Scholar] [CrossRef]
- Yu, L.X.; Jiang, W.; Zhang, X.; Lionberger, R.; Makhlouf, F.; Schuirmann, D.J.; Muldowney, L.; Chen, M.-L.; Davit, B.; Conner, D.; et al. Novel bioequivalence approach for narrow therapeutic index drugs. Clin. Pharmacol. Ther. 2015, 97, 286–291. [Google Scholar] [CrossRef]
- Platnick, N.I.; NMBE—World Spider Catalog. Natural History Museum Bern. 2015, pp. 1–4. Available online: https://wsc.nmbe.ch/ (accessed on 30 June 2023).
- Macedo, K.W.R.; Costa, L.J.; de Souza, J.O.; de Vasconcelos, I.A.; de Castro, J.S.; de Santana, C.J.C.; Magalhães, A.C.M.; Castro, M.S.; Pires, O.R. Brazilian Theraphosidae: A toxicological point of view. J. Venom. Anim. Toxins Trop. Dis. 2021, 27, e20210004. [Google Scholar] [CrossRef]
- Cooper, A.M.; Nelsen, D.R.; Hayes, W.K. The Strategic Use of Venom by Spiders. In Evolution of Venomous Animals and Their Toxins; Gopalakrishnakone, P., Malhotra, A., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 1–18. [Google Scholar]
- Saez, N.J.; Herzig, V. Versatile spider venom peptides and their medical and agricultural applications. Toxicon 2018, 158, 109–126. [Google Scholar] [CrossRef]
- Harrison, P.L.; Abdel-Rahman, M.A.; Miller, K.; Strong, P.N. Antimicrobial peptides from scorpion venoms. Toxicon 2014, 88, 115–137. [Google Scholar] [CrossRef]
- Estrada-Gomez, S.; Vargas Muñoz, L.J.; Quintana Castillo, J.C. Extraction and partial characterization of venom from the Colombian spider Pamphobeteus aff. nigricolor (Aranae: Theraphosidae). Toxicon 2013, 76, 301–309. [Google Scholar] [CrossRef]
- NIH. NCBI Refseq. Available online: https://www.ncbi.nlm.nih.gov/refseq/ (accessed on 30 June 2020).
- Pineda, S.S.; Chaumeil, P.-A.; Kunert, A.; Kaas, Q.; Thang, M.W.C.; Le, L.; Nuhn, M.; Herzig, V.; Saez, N.J.; Cristofori-Armstrong, B.; et al. ArachnoServer 3.0: An online resource for automated discovery, analysis and annotation of spider toxins. Bioinformatics 2018, 34, 1074–1076. [Google Scholar] [CrossRef]
- Pearson, W.R. An introduction to sequence similarity (“homology”) searching. Curr. Protoc. Bioinform. 2013, 42, 3.1.1–3.1.8. [Google Scholar] [CrossRef]
- Mount, D.W.; Comparison of the PAM and BLOSUM Amino Acid Substitution Matrices. CSH Protoc. 2008. Available online: https://pubmed.ncbi.nlm.nih.gov/21356840/ (accessed on 31 May 2023).
- Bystroff, C.; Krogh, A. Hidden Markov Models for prediction of protein features. Methods Mol. Biol. 2008, 413, 173–198. [Google Scholar]
- Yu, H.M.; Chen, S.T.; Wang, K.T. Enhanced Coupling Efficiency in Solid-Phase Peptide Synthesis by Microwave Irradiation. J. Org. Chem. 1992, 57, 4781–4784. [Google Scholar] [CrossRef]
- Domingues, T.M.; Riske, K.A.; Miranda, A. Revealing the lytic mechanism of the antimicrobial peptide gomesin by observing giant unilamellar vesicles. Langmuir 2010, 26, 11077–11084. [Google Scholar] [CrossRef]
- Lipkin, R.B.; Lazaridis, T. Implicit Membrane Investigation of the Stability of Antimicrobial Peptide β-Barrels and Arcs. J. Membr. Biol. 2015, 248, 469–486. [Google Scholar] [CrossRef]
- Barbosa, F.M.; Daffre, S.; Maldonado, R.A.; Miranda, A.; Nimrichter, L.; Rodrigues, M.L. Gomesin, a peptide produced by the spider Acanthoscurria gomesiana, is a potent anticryptococcal agent that acts in synergism with fluconazole. FEMS Microbiol. Lett. 2007, 274, 279–286. [Google Scholar] [CrossRef]
- Kozlov, S.A.; Vassilevski, A.A.; Feofanov, A.V.; Surovoy, A.Y.; Karpunin, D.V.; Grishin, E.V. Latarcins, antimicrobial and cytolytic peptides from the venom of the spider Lachesana tarabaevi (Zodariidae) that exemplify biomolecular diversity. J. Biol. Chem. 2006, 281, 20983–20992. [Google Scholar] [CrossRef]
- Lee, H.; Hwang, J.-S.; Lee, J.; Kim, J.I.; Lee, D.G. Scolopendin 2, a cationic antimicrobial peptide from centipede, and its membrane-active mechanism. Biochim. Biophys. Acta Biomembr. 2015, 1848, 634–642. [Google Scholar] [CrossRef]
- Lee, W.; Hwang, J.S.; Lee, D.G. A novel antimicrobial peptide, scolopendin, from Scolopendra subspinipes mutilans and its microbicidal mechanism. Biochimie 2015, 118, 176–184. [Google Scholar] [CrossRef]
- Choi, H.; Hwang, J.S.; Lee, D.G. Identification of a novel antimicrobial peptide, scolopendin 1, derived from centipede Scolopendra subspinipes mutilans and its antifungal mechanism. Insect. Mol. Biol. 2014, 23, 788–799. [Google Scholar] [CrossRef]
- Menk, J.J.; Matuhara, Y.E.; Sebestyen-França, H.; Henrique-Silva, F.; Ferro, M.; Rodrigues, R.S.; Santos-Júnior, C.D. Antimicrobial Peptide Arsenal Predicted from the Venom Gland Transcriptome of the Tropical Trap-Jaw Ant Odontomachus chelifer. Toxins 2023, 15, 345. [Google Scholar] [CrossRef]
- Shin, M.K.; Lee, B.; Kim, S.T.; Yoo, J.S.; Sung, J.S. Designing a Novel Functional Peptide with Dual Antimicrobial and Anti-inflammatory Activities via in Silico Methods. Front. Immunol. 2022, 13, 821070. [Google Scholar] [CrossRef]
- Brinckerhoff, L.H.; Kalashnikov, V.V.; Thompson, L.W.; Yamshchikov, G.V.; Pierce, R.A.; Galavotti, H.S.; Engelhard, V.H.; Slingluff, C.L., Jr. Terminal modifications inhibit proteolytic degradation of an immunogenic mart-1 27-35 peptide: Implications for peptide vaccines. J. Cancer 1999, 83, 326–334. [Google Scholar]
- USP-NF. Chapter <51> Antimicrobial Effectiveness Testing. In U.S. Pharmacopeia General; United States Pharmacopeia: Rockville, MD, USA, 2023. [Google Scholar]
- Giacometti, A.; Cirioni, O.; Kamysz, W.; D’amato, G.; Silvestri, C.; Licci, A.; Nadolski, P.; Riva, A.; Lukasiak, J.; Scalise, G. In Vitro activity of MSI-78 alone and in combination with antibiotics against bacteria responsible for bloodstream infections in neutropenic patients. Int. J. Antimicrob. Agents 2005, 26, 235–240. [Google Scholar] [CrossRef]
- Oguiura, N.; Boni-Mitake, M.; Affonso, R.; Zhang, G. In Vitro antibacterial and hemolytic activities of crotamine, a small basic myotoxin from rattlesnake Crotalus durissus. J. Antibiot. 2011, 64, 327–331. [Google Scholar] [CrossRef]
- Miyazawa, H.; Ueda, C.; Yahata, K.; Su, Z.-H. Molecular phylogeny of Myriapoda provides insights into evolutionary patterns of the mode in post-embryonic development. Sci. Rep. 2014, 4, 4127. [Google Scholar] [CrossRef]
- Aloke, C.; Achilonu, I. Coping with the ESKAPE Pathogens: Evolving Strategies, Challenges and Future Prospects. Microb. Pathog. 2023, 175, 105963. [Google Scholar] [CrossRef]
- Marturano, J.E.; Lowery, T.J. ESKAPE Pathogens in Bloodstream Infections Are Associated with Higher Cost and Mortality but Can Be Predicted Using Diagnoses Upon Admission. Open Forum Infect. Dis. 2019, 6, ofz503. [Google Scholar] [CrossRef]
- Alves, E.; Santos, N.; Melo, T.; Maciel, E.; Dória, M.L.; Faustino, M.A.F.; Tomé, J.P.C.; Neves, M.G.P.M.S.; Cavaleiro, J.A.S.; Cunha, A.; et al. Photodynamic oxidation of Escherichia coli membrane phospholipids: New insights based on lipidomics. Rapid Commun. Mass Spectrom. 2013, 27, 2717–2728. [Google Scholar] [CrossRef]
- Furse, S.; Wienk, H.; Boelens, R.; de Kroon, A.I.; Killian, J.A.E. coli MG1655 modulates its phospholipid composition through the cell cycle. FEBS Lett. 2015, 589, 2726–2730. [Google Scholar] [CrossRef]
- Swain, J.; El Khoury, M.; Kempf, J.; Briée, F.; Van Der Smissen, P.; Décout, J.-L.; Mingeot-Leclercq, M.-P. Effect of cardiolipin on the antimicrobial activity of a new amphiphilic aminoglycoside derivative on Pseudomonas aeruginosa. PLoS ONE 2018, 13, e0201752. [Google Scholar] [CrossRef]
- Hagiya, H.; Aoki, K.; Akeda, Y.; Yamamoto, N.; Shanmugakani, R.K.; Ishii, Y.; Tomono, K. In Vitro Effectiveness of Meropenem and Cefmetazole Combination Treatment Against KPC-2-Producing Enterobacteriaceae. Microb. Drug Resist. 2019, 25, 839–845. [Google Scholar] [CrossRef]
- Campos, M.A.; Vargas, M.A.; Regueiro, V.; Llompart, C.M.; Albertí, S.; Bengoechea, J.A. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 2004, 72, 7107–7114. [Google Scholar] [CrossRef]
- Khan, A.; Davlieva, M.; Panesso, D.; Rincon, S.; Miller, W.R.; Diaz, L.; Reyes, J.; Cruz, M.R.; Pemberton, O.; Nguyen, A.H.; et al. Antimicrobial sensing coupled with cell membrane remodeling mediates antibiotic resistance and virulence in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 2019, 26, 26925–26932. [Google Scholar] [CrossRef]
- Reyes, J.; Panesso, D.; Tran, T.T.; Mishra, N.N.; Cruz, M.R.; Munita, J.M.; Singh, K.V.; Yeaman, M.R.; Murray, B.E.; Shamoo, Y.; et al. A liaR deletion restores susceptibility to daptomycin and antimicrobial peptides in multidrug-resistant Enterococcus faecalis. J. Infect. Dis. 2014, 211, 1317–1325. [Google Scholar] [CrossRef]
- Harp, J.R.; Saito, H.E.; Bourdon, A.K.; Reyes, J.; Arias, C.A.; Campagna, S.R.; Fozo, E.M. Exogenous Fatty Acids Protect Enterococcus faecalis from Daptomycin-Induced Membrane Stress Independently of the Response Regulator LiaR. Appl. Environ. Microbiol. 2016, 82, 4410–4420. [Google Scholar] [CrossRef]
- Fernandes, P.B.; Reed, P.; Monteiro, J.M.; Pinho, M.G. Revisiting the Role of VraTSR in Staphylococcus aureus Response to Cell Wall-Targeting Antibiotics. J. Bacteriol. 2022, 204, e0016222. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. Cad Pesqui. 2017, 43, 348–365. [Google Scholar]
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.A.; Albertí, S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef]
- Al-Farsi, H.M.; Al-Adwani, S.; Ahmed, S.; Vogt, C.; Ambikan, A.T.; Leber, A.; Al-Jardani, A.; Al-Azri, S.; Al-Muharmi, Z.; Toprak, M.S.; et al. Effects of the Antimicrobial Peptide LL-37 and Innate Effector Mechanisms in Colistin-Resistant Klebsiella pneumoniae with mgrB Insertions. Front. Microbiol. 2019, 10, 2632. [Google Scholar] [CrossRef]
- Kidd, T.J.; Mills, G.; Sá-Pessoa, J.; Dumigan, A.; Frank, C.G.; Insua, J.L.; Ingram, R.; Hobley, L.; Bengoechea, J.A. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol. Med. 2017, 9, 430–447. [Google Scholar] [CrossRef]
- Ainavarapu, S.R.K.; Brujić, J.; Huang, H.H.; Wiita, A.P.; Lu, H.; Li, L.; Walther, K.A.; Carrion-Vazquez, M.; Li, H.; Fernandez, J.M. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys. J. 2007, 92, 225–233. [Google Scholar] [CrossRef]
- Tripathi, J.K.; Kathuria, M.; Kumar, A.; Mitra, K.; Ghosh, J.K. An unprecedented alteration in mode of action of IsCT resulting its translocation into bacterial cytoplasm and inhibition of macromolecular syntheses. Sci. Rep. 2015, 5, 9127. [Google Scholar] [CrossRef]
- de Almeida, N.R.; Catazaro, J.; Krishnaiah, M.; Chhonker, Y.S.; Murry, D.J.; Powers, R.; Conda-Sheridan, M. Understanding interactions of Citropin 1.1 analogues with model membranes and their influence on biological activity. Peptides 2019, 119, 170119. [Google Scholar] [CrossRef]
- Devanga Ragupathi, N.K.; Muthuirulandi Sethuvel, D.P.; Triplicane Dwarakanathan, H.; Murugan, D.; Umashankar, Y.; Monk, P.N.; Karunakaran, E.; Veeraraghavan, B. The Influence of Biofilms on Carbapenem Susceptibility and Patient Outcome in Device Associated, K. pneumoniae Infections: Insights into Phenotype vs Genome-Wide Analysis and Correlation. Front. Microbiol. 2020, 12, 11. [Google Scholar] [CrossRef]
- Di Bella, S.; Giacobbe, D.R.; Maraolo, A.E.; Viaggi, V.; Luzzati, R.; Bassetti, M.; Luzzaro, F.; Principe, L. Resistance to ceftazidime/avibactam in infections and colonisations by KPC-producing Enterobacterales: A systematic review of observational clinical studies. J. Glob. Antimicrob. Resist. 2021, 25, 268–281. [Google Scholar] [CrossRef]
- Muraleedharan, C.; Talreja, D.; Kanwar, M.; Kumar, A.; Walia, S.K. Occurrence of extended-spectrum β-lactamase-producing bacteria in urban Clinton River habitat. J. Glob. Antimicrob. Resist. 2019, 16, 225–235. [Google Scholar] [CrossRef]
- Hancock, R.E.; Raffle, V.J.; Nicas, T.I. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1981, 19, 777–785. [Google Scholar] [CrossRef]
- Taber, H.W.; Mueller, J.P.; Miller, P.F.; Arrow, A.S. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 1987, 51, 439–457. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Lewis, L.A.; Radulović, M.; Kim, T.K.; Porter, L.M.; Mulenga, A. Identification of 24h Ixodes scapularis immunogenic tick saliva proteins. Ticks Tick-Borne Dis. 2015, 6, 424–434. [Google Scholar] [CrossRef]
- De la Salud Bea, R.; Petraglia, A.F.; Ascuitto, M.R.; Buck, Q.M. Antibacterial activity and toxicity of analogs of scorpion venom IsCT peptides. Antibiotics 2017, 6, 13. [Google Scholar] [CrossRef]
- Acevedo, I.C.C.; Silva, P.I., Jr.; Silva, F.D.; Araujo, I.; Alves, F.L.; Oliveira, C.S.; Oliveira, V.X., Jr. IsCT-based analogs intending better biological activity. J. Pept. Sci. 2019, 25, e3219. [Google Scholar] [CrossRef]
- Morita, S.Y.; Shirakawa, S.; Kobayashi, Y.; Nakamura, K.; Teraoka, R.; Kitagawa, S.; Terada, T. Enzymatic measurement of phosphatidylserine in cultured cells. J. Lipid. Res. 2012, 53, 325. [Google Scholar] [CrossRef]
- Kay, J.G.; Fairn, G.D. Distribution, dynamics and functional roles of phosphatidylserine within the cell. Cell Commun. Signal. 2019, 17, 1–8. [Google Scholar] [CrossRef]
- Virtanen, J.A.; Cheng, K.H.; Somerharju, P. Phospholipid composition of the mammalian red cell membrane can be rationalized by a superlattice model. Proc. Natl. Acad. Sci. USA 1998, 95, 4964–4969. [Google Scholar] [CrossRef]
- Ridyard, K.E.; Overhage, J. The Potential of Human Peptide LL-37 as an Antimicrobial and Anti-Biofilm Agent. Antibiotics 2021, 10, 650. [Google Scholar] [CrossRef]
- Saporito, P.; Vang Mouritzen, M.; Løbner-Olesen, A.; Jenssen, H. LL-37 fragments have antimicrobial activity against Staphylococcus epidermidis biofilms and wound healing potential in HaCaT cell line. J. Pept. Sci. Off. Publ. Eur. Pept. Soc. 2018, 24, e3080. [Google Scholar] [CrossRef]
- Quemé-Peña, M.; Ricci, M.; Juhász, T.; Horváti, K.; Bösze, S.; Biri-Kovács, B.; Szeder, B.; Zsila, F.; Beke-Somfai, T. Old Polyanionic Drug Suramin Suppresses Detrimental Cytotoxicity of the Host Defense Peptide LL-37. ACS Pharmacol. Transl. Sci. 2021, 4, 155–167. [Google Scholar] [CrossRef]
- Chernov, A.N.; Tsapieva, A.; Alaverdian, D.A.; Filatenkova, T.A.; Galimova, E.S.; Suvorova, M.; Shamova, O.V.; Suvorov, A.N. In Vitro Evaluation of the Cytotoxic Effect of Streptococcus pyogenes Strains, Protegrin PG-1, Cathelicidin LL-37, Nerve Growth Factor and Chemotherapy on the C6 Glioma Cell Line. Molecules 2022, 27, 569. [Google Scholar] [CrossRef]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2006, 6, 29–40. [Google Scholar] [CrossRef]
- Tommasi, R.; Brown, D.G.; Walkup, G.K.; Manchester, J.I.; Miller, A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015, 14, 529–542. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2010, 38, D774–D780. [Google Scholar] [CrossRef]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2020, 49, D288–D297. [Google Scholar] [CrossRef]
- Ye, G.; Wu, H.; Huang, J.; Wang, W.; Ge, K.; Li, G.; Zhong, J.; Huang, Q. LAMP2: A major update of the database linking antimicrobial peptides. Database 2020, 2020, 61. [Google Scholar] [CrossRef]
- Piotto, S.P.; Sessa, L.; Concilio, S.; Iannelli, P. YADAMP: Yet another database of antimicrobial peptides. Int. J. Antimicrob. Agents 2012, 39, 346–351. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Guzmán, F.; Gauna, A.; Roman, T.; Luna, O.; Álvarez, C.; Pareja-Barrueto, C.; Mercado, L.; Albericio, F.; Cárdenas, C. Tea Bags for Fmoc Solid-Phase Peptide Synthesis: An Example of Circular Economy. Molecules 2021, 26, 5035. [Google Scholar] [CrossRef]
- Zhang, J.; Diamond, S.; Arvedson, T.; Sasu, B.J.; Miranda, L.P. Oxidative folding of hepcidin at acidic pH. Biopolymers 2010, 94, 257–264. [Google Scholar] [CrossRef]

| Stimulus | E. coli ATCC® 8739 (Inhibition %) | E. coli ATCC® 25922 (Inhibition %) | S. aureus ATCC® 6538 (Inhibition %) | S. aureus ATCC® 29213 (Inhibition %) |
|---|---|---|---|---|
| PBS (C−) | 0.00 ± 3.90 | 0.00 ± 5.72 | 0.07 ± 6.61 | 0.00 ± 4.34 |
| BTM-P1 (C+) | 85.52 ± 2.57 **** | 96.89 ± 0.51 **** | 71.10 ± 1.84 **** | 94.12 ± 0.66 **** |
| PvAMP7 | 26.74 ± 0.68 **** | 18.21 ± 2.54 **** | 37.89 ± 1.50 **** | 23.66 ± 3.34 ** |
| PvAMP8 | NA | NA | 47.95 ± 3.62 **** | 11.90 ± 8.93 ns |
| PvAMP15 | NA | NA | 43.62 ± 10.56 **** | 27.48 ± 29.69 ns |
| PvAMP16 | NA | NA | 42.06 ± 4.29 **** | 13.78 ± 6.48 ns |
| PvAMP30 | NA | NA | 35.52 ± 7.29 *** | 7.34 ± 10.33 ns |
| PvAMP32 | 30.73 ± 5.21 **** | 21.19 ± 3.82 *** | 43.62 ± 4.90 * | 33.17 ± 8.46 * |
| PvAMP66 | 88.81 ± 18.15 **** | 73.59 ± 21.83 **** | 139.72 ± 1.90 **** | 98.25 ± 0.39 **** |
| PvAMP69 | 1.75 ± 9.78 ns | 19.08 ± 0.12 ** | 32.32 ± 7.00 *** | 29.81 ± 3.31 *** |
| PvAMP81 | 8.57 ± 4.46 ns | 31.88 ± 2.18 *** | NA | NA |
| PvAMP82 | 64.01 ± 4.28 **** | 74.94 ± 14.27 **** | 43.03 ± 6.37 **** | 43.44 ± 10.24 **** |
| PvAMP84 | 24.29 ± 6.74 **** | 11.29 ± 3.72 ns | NA | NA |
| PvAMP164 | 20.51 ± 4.31 *** | 30.10 ± 3.90 **** | 24.49 ± 10.18 ** | 12.25 ± 1.29 ** |
| PvAMP169 | 13.60 ± 5.92 ns | 55.69 ± 4.65 **** | NA | NA |
| PvAMP172 | 64.78 ± 5.46 **** | 47.43 ± 2.76 **** | NA | NA |
| PvAMP177 | 93.07 ± 7.11 **** | 56.76 ± 3.64 **** | NA | NA |
| PvAMP179 | NA | NA | 50.42 ± 3.56 **** | 10.43 ± 11.21 ns |
| PvAMP183 | 48.26 ± 2.41 **** | 64.66 ± 1.26 **** | 47.36 ± 1.27 **** | 4.71 ± 21.89 ns |
| Peptide | Sequence | Activity Against | Parameter | µM | µg/mL |
|---|---|---|---|---|---|
| PvAMP7 | SLWGMWR-NH2 | E. coli ATCC® 25922 | MIC | 200 | 23.38 |
| PvAMP32 | IIKKIWK-NH2 | E. coli ATCC® 25922 | MIC | 100 | 11.60 |
| MBC | 100 | 11.60 | |||
| IC50 | 94.72 (77.98–118.05) | 10.76 | |||
| S. aureus ATCC® 29213 | MIC | 50 | 5.80 | ||
| IC50 | 107.66 (80.18–142.07) | 12.49 | |||
| P. aeruginosa ATCC® 9027 | MIC | 100 | 11.60 | ||
| MBC | 100 | 11.60 | |||
| IC50 | 123.95 (93.17–160.2) | 14.38 | |||
| PvAMP66 | WKKIKKFF-NH2 | E. coli ATCC® 25922 | MIC | 50 | 7.03 |
| MBC | 50 | 7.03 | |||
| IC50 | 38.32 (32.03–46.14) | 5.39 | |||
| S. aureus ATCC® 29213 | MIC | 25 | 3.51 | ||
| MBC | 50 | 7.03 | |||
| IC50 | 27.55 (21.07–27.55) | 3.87 | |||
| P. aeruginosa ATCC® 9027 | MIC | 12.5 | 177 | ||
| MBC | 12.5 | 1.77 | |||
| IC50 | 12.67 (8.24–16.44) | 1.78 | |||
| K. pneumoniae ATCC® BAA-1705 | MIC | 100 | 14.06 | ||
| IC50 | 45.49 (35.22–60.76) | 6.39 | |||
| PvAMP69 | YRARCVIYC-NH2 | E. coli ATCC® 25922 | MIC | 200 | 28.66 |
| MBC | 200 | 28.66 | |||
| PvAMP82 | GRIFRLLRK-NH2 | E. coli ATCC® 25922 | MIC | 100 | 14.48 |
| MBC | 100 | 14.48 | |||
| P. aeruginosa ATCC® 9027 | MIC | 200 | 28.96 | ||
| MBC | 200 | 28.96 | |||
| PvAMP164 | RSVLKAHCRICRRRG-NH2 | E. coli ATCC® 25922 | MIC | 200 | 45.28 |
| MBC | 200 | 45.28 | |||
| P. aeruginosa ATCC® 9027 | MIC | 100 | 22.64 | ||
| MBC | 100 | 22.64 | |||
| PvAMP172 | CRKLCFRNRCLTYCRGR-NH2 | E. coli ATCC® 25922 | MIC | 50 | 13.43 |
| MBC | 50 | 13.43 | |||
| IC50 | 42.91 (30.34–48.88) | 11.52 | |||
| S. aureus ATCC® 29213 | MIC | 100 | 26.86 | ||
| MBC | 100 | 26.86 | |||
| P. aeruginosa ATCC® 9027 | MIC | 50 | 13.43 | ||
| MBC | 50 | 13.43 | |||
| IC50 | 48.43 (26.93–84.30) | 13.01 | |||
| PvAMP177 | QCRKLCFRNRCLTYCRGR-NH2 | E. coli ATCC® 25922 | MIC | 25 | 7.11 |
| MBC | 50 | 14.23 | |||
| IC50 | 25.32 (12.98–48.25) | 6.64 | |||
| S. aureus ATCC® 29213 | IC50 | 137.91 (116.24–169.69) | 37.82 | ||
| P. aeruginosa ATCC® 9027 | MIC | 50 | 14.23 | ||
| MBC | 50 | 14.23 | |||
| IC50 | 25.93 (14.02–47.01) | 7.38 | |||
| PvAMP183 | QCRKLCFRNRCLTYCRGRG-NH2 | E. coli ATCC® 25922 | MIC | 25 | 6.98 |
| MBC | 50 | 13.96 | |||
| IC50 | 26.89 (14.80–47.84) | 7.51 | |||
| P. aeruginosa ATCC® 9027 | MIC | 50 | 13.96 | ||
| MBC | 100 | 27.92 | |||
| IC50 | 48.28 (26.02–88.63) | 13.48 |
| Stimulus | MIC (µM) | MIC (µg/mL) | IC50 (µM) | Classification at 106 CFU/mL [30] |
|---|---|---|---|---|
| PvAMP66 | 25.00 | 3.51 | 15.80 (11.63–20.47) | NA |
| Gentamicin | 50.00 | 9.31 | 33.38 (30.76–36.27) | Intermediate resistant |
| Chloramphenicol | 100.00 | 4.04 | 56.23 (50.21–63.09) | Susceptible |
| Doxycycline | 50.00 | 3.21 | 38.05 (28.95–51.87) | Susceptible |
| Concentration (µM) | HC50/IC50 | CC50/IC50 | |
|---|---|---|---|
| HC50 | 132.96 (118.82–151.21) | NA | NA |
| CC50 | 172.03 (124.44–ND) | NA | NA |
| IC50 E. coli ATCC® 25922 | 38.32 (32.03–46.14) | 3.47 | 4.49 |
| IC50 S. aureus ATCC® 29213 | 27.55 (21.07–27.55) | 4.83 | 6.24 |
| IC50 P. aeruginosa ATCC® 9027 | 12.67 (8.24–16.44) | 10.49 | 13.58 |
| IC50 K. pneumoniae ATCC® BAA-1705 | 45.49 (35.22–60.76) | 2.92 | 3.78 |
| IC50 MDR K. pneumoniae | 15.80 (11.63–20.47) | 8.42 | 10.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salinas-Restrepo, C.; Naranjo-Duran, A.M.; Quintana, J.; Bueno, J.; Guzman, F.; Hoyos Palacio, L.M.; Segura, C. Short Antimicrobial Peptide Derived from the Venom Gland Transcriptome of Pamphobeteus verdolaga Increases Gentamicin Susceptibility of Multidrug-Resistant Klebsiella pneumoniae. Antibiotics 2024, 13, 6. https://doi.org/10.3390/antibiotics13010006
Salinas-Restrepo C, Naranjo-Duran AM, Quintana J, Bueno J, Guzman F, Hoyos Palacio LM, Segura C. Short Antimicrobial Peptide Derived from the Venom Gland Transcriptome of Pamphobeteus verdolaga Increases Gentamicin Susceptibility of Multidrug-Resistant Klebsiella pneumoniae. Antibiotics. 2024; 13(1):6. https://doi.org/10.3390/antibiotics13010006
Chicago/Turabian StyleSalinas-Restrepo, Cristian, Ana María Naranjo-Duran, Juan Quintana, Julio Bueno, Fanny Guzman, Lina M. Hoyos Palacio, and Cesar Segura. 2024. "Short Antimicrobial Peptide Derived from the Venom Gland Transcriptome of Pamphobeteus verdolaga Increases Gentamicin Susceptibility of Multidrug-Resistant Klebsiella pneumoniae" Antibiotics 13, no. 1: 6. https://doi.org/10.3390/antibiotics13010006
APA StyleSalinas-Restrepo, C., Naranjo-Duran, A. M., Quintana, J., Bueno, J., Guzman, F., Hoyos Palacio, L. M., & Segura, C. (2024). Short Antimicrobial Peptide Derived from the Venom Gland Transcriptome of Pamphobeteus verdolaga Increases Gentamicin Susceptibility of Multidrug-Resistant Klebsiella pneumoniae. Antibiotics, 13(1), 6. https://doi.org/10.3390/antibiotics13010006






