Identification of Acetomycin as an Antifungal Agent Produced by Termite Gut-Associated Streptomycetes against Pyrrhoderma noxium
Abstract
1. Introduction
2. Results and Discussion
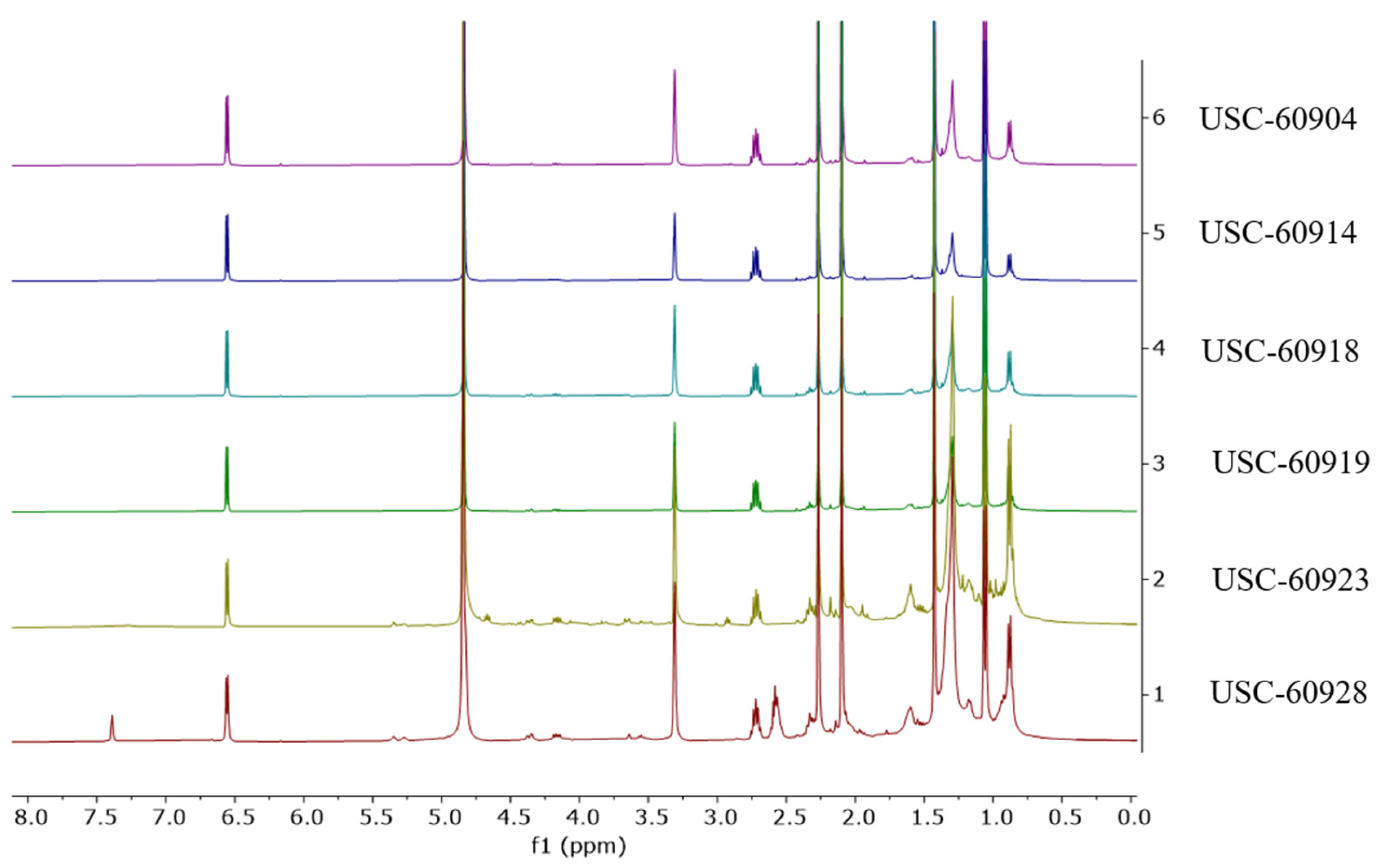
2.1. Data Analysis of NMR Spectra from Fermentation Extracts of Streptomycetes
2.2. Structure Elucidation for Compound 1
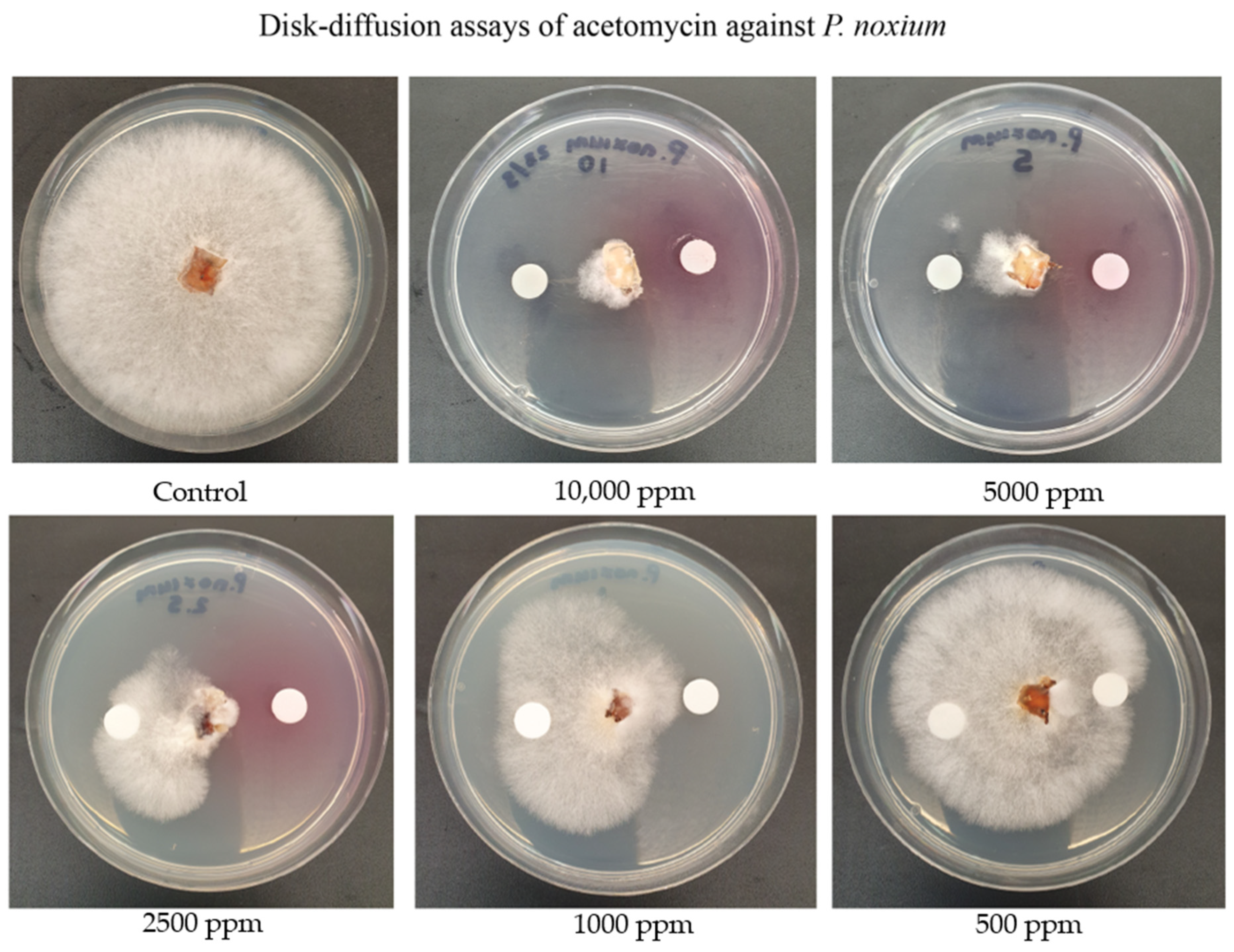
2.3. Assessing the Activity of Acetomycin against P. noxium Using the Disk Diffusion Method
2.4. Assessing Acetomycin for Antifungal Activity against Other Fungal Crop Pathogens
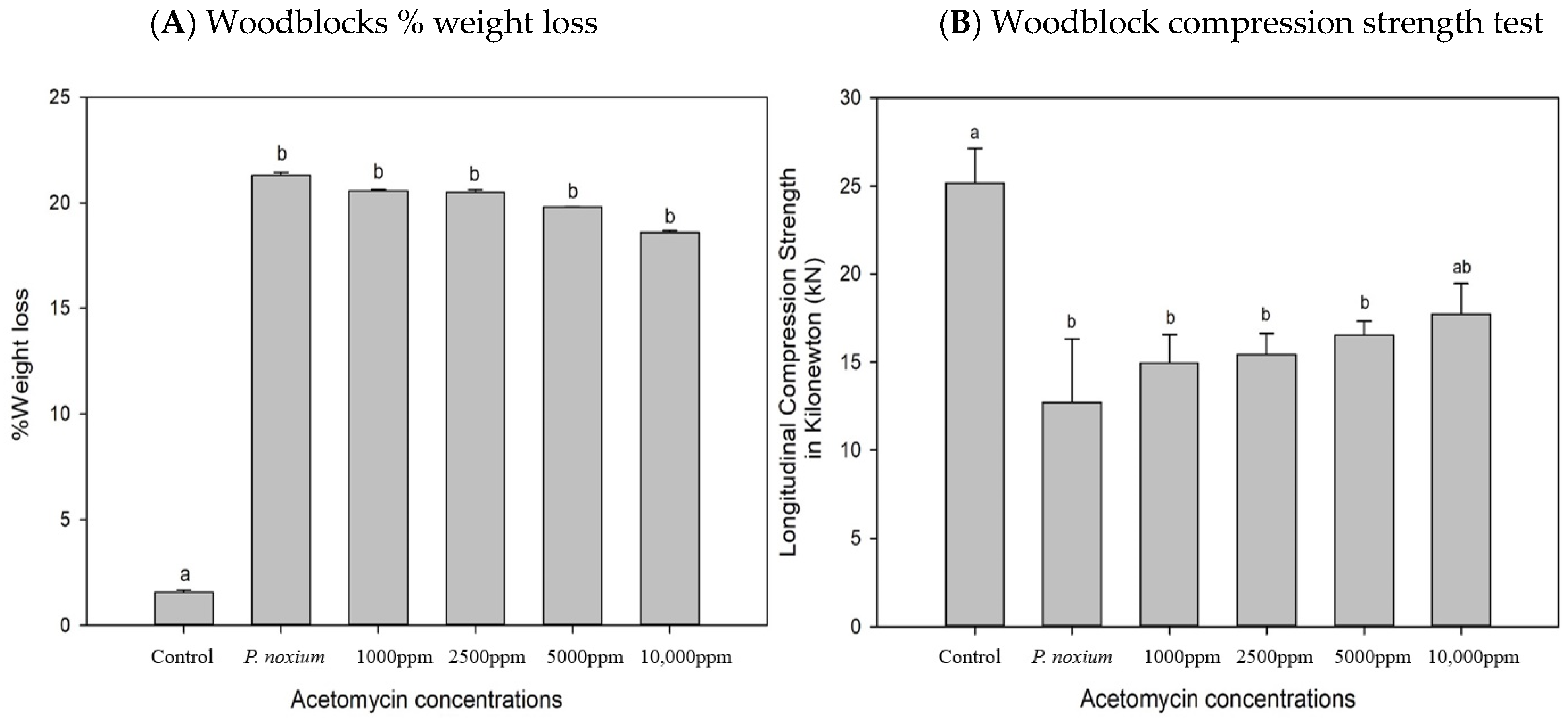
2.5. In Vitro Evaluation of the Inhibitory Potential of Acetomycin on P. noxium Using a Woodblock Test
3. Materials and Methods
3.1. Cultures and Culture Media
3.2. Fermentation and Extraction of Metabolites Produced by Streptomyces Isolates
3.3. Nuclear Magnetic Resonance Analysis
3.4. Crystallographic Data
3.5. Structure Determination
3.6. Assessing Activity from Acetomycin against P. noxium and Other Fungal Crop Pathogens
3.7. Assessing Acetomycin as a Potential Biocontrol against P. noxium Using Woodblocks
3.8. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Savary, S.; Ficke, A.; Aubertot, J.-N.; Hollier, C. Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 2012, 4, 519–537. [Google Scholar] [CrossRef]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The current status of its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int. J. Mol. Sci. 2022, 23, 2329. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.; Kim, M.-S.; Ota, Y.; Sahashi, N.; Hanna, J.; Akiba, M.; Ata, J.; Atibalentja, N.; Brooks, F.; Chung, C.-L. Phylogenetic and population genetic analyses reveal three distinct lineages of the invasive brown root-rot pathogen, Phellinus noxius, and bioclimatic modeling predicts differences in associated climate niches. Eur. J. Plant Pathol. 2020, 156, 751–766. [Google Scholar] [CrossRef]
- Sahashi, N.; Akiba, M.; Ishihara, M.; Ota, Y.; Kanzaki, N. Brown root rot of trees caused by Phellinus noxius in the Ryukyu Islands, subtropical areas of Japan. For. Pathol. 2012, 42, 353–361. [Google Scholar] [CrossRef]
- Ann, P.-J.; Chang, T.-T.; Ko, W.-H. Phellinus noxius brown root rot of fruit and ornamental trees in Taiwan. Plant Dis. 2002, 86, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.-L.; Huang, S.-Y.; Huang, Y.-C.; Tzean, S.-S.; Ann, P.-J.; Tsai, J.-N.; Yang, C.-C.; Lee, H.-H.; Huang, T.-W.; Huang, H.-Y. The genetic structure of Phellinus noxius and dissemination pattern of brown root rot disease in Taiwan. PLoS ONE 2015, 10, e0139445. [Google Scholar] [CrossRef]
- Schwarze, F.W.; Jauss, F.; Spencer, C.; Hallam, C.; Schubert, M. Evaluation of an antagonistic Trichoderma strain for reducing the rate of wood decomposition by the white rot fungus Phellinus noxius. Biol. Control 2012, 61, 160–168. [Google Scholar] [CrossRef]
- Chou, H.; Xiao, Y.-T.; Tsai, J.-N.; Li, T.-T.; Wu, H.-Y.; Liu, L.-y.D.; Tzeng, D.-S.; Chung, C.-L. In vitro and in planta evaluation of Trichoderma asperellum TA as a biocontrol agent against Phellinus noxius, the cause of brown root rot disease of trees. Plant Dis. 2019, 103, 2733–2741. [Google Scholar] [CrossRef]
- Adra, C.; Panchalingam, H.; Kurtböke, D.İ. Termite–gut-associated actinomycetes as biological control agents against phytopathogen Pyrrhoderma noxium. Microbiol. Aust. 2022, 43, 190–193. [Google Scholar] [CrossRef]
- Panchalingam, H.; Powell, D.; Adra, C.; Foster, K.; Tomlin, R.; Quigley, B.L.; Nyari, S.; Hayes, R.A.; Shapcott, A.; Kurtböke, D.İ. Assessing the various antagonistic mechanisms of Trichoderma strains against the brown root rot pathogen Pyrrhoderma noxium infecting heritage fig trees. J. Fungi 2022, 8, 1105. [Google Scholar] [CrossRef]
- Panchalingam, H.; Ashfield-Crook, N.; Naik, V.; Frenken, R.; Foster, K.; Tomlin, R.; Shapcott, A.; Kurtböke, D.İ. Testing the Biocontrol Ability of a Trichoderma-Streptomycetes Consortium against Pyrrhoderma noxium (Corner) LW Zhou and YC Dai in Soil. J. Fungi 2022, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Salman, M.; Jan, S.; Shinwari, Z. Biological control of fungal phytopathogens: A comprehensive review based on Bacillus species. MOJ Biol. Med. 2021, 6, 90–92. [Google Scholar] [CrossRef]
- Carlucci, A.; Sorbo, A.; Colucci, D.; Raimondo, M.L. Biological Control of Severe Fungal Phytopathogens by Streptomyces albidoflavus Strain CARA17 and Its Bioactive Crude Extracts on Lettuce Plants. Plants 2023, 12, 2025. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Li, X.; Li, K.; Zhou, D.; Zhang, M.; Qi, D.; Jing, T.; Zang, X.; Qi, C.; Wang, W.; Xie, J. Biological control of banana wilt disease caused by Fusarium oxyspoum f. sp. Cubense using Streptomyces sp. H4. Biol. Control 2021, 155, 104524. [Google Scholar] [CrossRef]
- LeBlanc, N. Bacteria in the genus Streptomyces are effective biological control agents for management of fungal plant pathogens: A meta-analysis. Biol. Control 2022, 67, 111–121. [Google Scholar] [CrossRef]
- Pacios-Michelena, S.; Aguilar Gonzalez, C.N.; Alvarez-Perez, O.B.; Rodriguez-Herrera, R.; Chávez-González, M.; Arredondo Valdes, R.; Ascacio Valdes, J.A.; Govea Salas, M.; Ilyina, A. Application of Streptomyces antimicrobial compounds for the control of phytopathogens. Front. Sustain. Food Syst. 2021, 5, 696518. [Google Scholar] [CrossRef]
- Sharma, V.; Kaur, R.; Salwan, R. Streptomyces: Host for refactoring of diverse bioactive secondary metabolites. 3 Biotech 2021, 11, 340. [Google Scholar] [CrossRef]
- Donald, L.; Pipite, A.; Subramani, R.; Owen, J.; Keyzers, R.A.; Taufa, T. Streptomyces: Still the biggest producer of new natural secondary metabolites, a current perspective. Microbiol. Res. 2022, 13, 418–465. [Google Scholar] [CrossRef]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Kurtböke, D.İ.; French, J.R.; Hayes, R.A.; Quinn, R.J. Eco-taxonomic insights into actinomycete symbionts of termites for discovery of novel bioactive compounds. In Biotechnological Applications of Biodiversity; Mukherjee, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 147, pp. 111–135. [Google Scholar]
- Zhou, J.; Duan, J.; Gao, M.; Wang, Y.; Wang, X.; Zhao, K. Diversity, roles, and biotechnological applications of symbiotic microorganisms in the gut of termite. Curr. Microbiol. 2019, 76, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, J.; Liu, H.; Liang, S.; Wen, J. Combining metabolomics and network analysis to improve tacrolimus production in Streptomyces tsukubaensis using different exogenous feedings. J. Ind. Microbiol. Biotechnol. 2017, 44, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Amberg, A.; Riefke, B.; Schlotterbeck, G.; Ross, A.; Senn, H.; Dieterle, F.; Keck, M. NMR and MS Methods for Metabolomics. In Drug Safety Evaluation; Gautier, J.C., Ed.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1641, pp. 229–258. [Google Scholar]
- Wishart, D.S.; Cheng, L.L.; Copié, V.; Edison, A.S.; Eghbalnia, H.R.; Hoch, J.C.; Gouveia, G.J.; Pathmasiri, W.; Powers, R.; Schock, T.B. NMR and metabolomics—A roadmap for the future. Metabolites 2022, 12, 678. [Google Scholar] [CrossRef] [PubMed]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Adra, C.; Tran, T.D.; Foster, K.; Tomlin, R.; Kurtböke, D.İ. Untargeted MS-Based Metabolomic Analysis of Termite Gut-Associated Streptomycetes with Antifungal Activity against Pyrrhoderma noxium. Antibiotics 2023, 12, 1373. [Google Scholar] [CrossRef]
- Ettlinger, L.; Gäumann, E.; Hütter, R.; Keller-Schierlein, W.; Kradolfer, F.; Neipp, L.; Prelog, V.; Zähner, H. Stoffwechselprodukte von Actinomyceten. 12. Mitteilung. Über die Isolierung und Charakterisierung von Acetomycin. Helv. Chim. Acta 1958, 41, 216–219. [Google Scholar] [CrossRef]
- Cano, F.; Foces-Foces, C.; Elguero, J. The structure and absolute configuration of acetomycin. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 1988, 44, 919–921. [Google Scholar] [CrossRef]
- Uhr, H.; Zeeck, A.; Clegg, W.; Egert, E.; Fuhrer, H.; Peter, H.H. The Structure of Acetomycin Spectroscopic Characterization and X-Ray Analysis of a Bromo Derivative. J. Antibiot. 1985, 38, 1684–1690. [Google Scholar] [CrossRef]
- Mamber, S.; Mitulski, J.; Borondy, P.; Tunac, J. Biological Effects of Acetomycin II. Inactivation by Esterases in vitro. J. Antibiot. 1987, 40, 77–80. [Google Scholar] [CrossRef]
- Knights, T.; Siddique, K. Manifestation of Botrytis cinerea on chickpeas in Australia. In Update of Research in Progress at the Tamworth Agricultural Institute 2002; NSW Agriculture: Tamworth, Australia, 2003; pp. 12–15. [Google Scholar]
- Su-lin, L.L.; Hocking, A.D.; Scott, E.S. Effect of temperature and water activity on growth and ochratoxin A production by Australian Aspergillus carbonarius and A. niger isolates on a simulated grape juice medium. Int. J. Food Microbiol. 2006, 110, 209–216. [Google Scholar]
- Kumar, V.; Iqbal, N. Post-harvest pathogens and disease management of horticultural crop: A brief review. Plant Arch. 2020, 20, 2054–2058. [Google Scholar]
- Le, D.P.; Gregson, A. Alternaria leaf spot of cotton seedlings grown in New South Wales, Australia is predominantly associated with Alternaria alternata. Australas. Plant Pathol. 2019, 48, 209–216. [Google Scholar] [CrossRef]
- Nallathambi, P.; Umamaheswari, C.; Thakore, B.; More, T. Post-harvest management of ber (Ziziphus mauritiana Lamk) fruit rot (Alternaria alternata Fr. Keissler) using Trichoderma species, fungicides and their combinations. Crop Prot. 2009, 28, 525–532. [Google Scholar] [CrossRef]
- Uenishi, J.i.; Kobayashi, N.; Komine, S.; Okadai, T.; Yonemitsu, O.; Sasaki, T.; Yamada, Y. Total synthesis of (±)-acetomycin and design of esterase-resistant analogs. Chem. Pharm. Bull. 1999, 47, 517–523. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caballero, J.R.I.; Ata, J.P.; Leddy, K.; Glenn, T.C.; Kieran, T.J.; Klopfenstein, N.B.; Kim, M.-S.; Stewart, J.E. Genome comparison and transcriptome analysis of the invasive brown root rot pathogen, Phellinus noxius, from different geographic regions reveals potential enzymes associated with degradation of different wood substrates. Fungal Biol. 2020, 124, 144–154. [Google Scholar] [CrossRef]
- Hattori, T.; Abe, Y.; Usugi, T. Distribution of clones of Phellinus noxius in a windbreak on Ishigaki Island. Eur. J. For. Pathol. 1996, 26, 69–80. [Google Scholar] [CrossRef]
- Joy, J.; Jose, C.; Mathew, P.L.; Thomas, S.; Khalaf, M.N. Biological delignification of biomass. In Green Polymers and Environmental Pollution Control; Apple Academic Press: New York, NY, USA, 2015; p. 271. [Google Scholar]
- Kirk, T.K.; Cullen, D. Enzymology and molecular genetics of wood degradation by white-rot fungi. In Environmentally Friendly Technologies for the Pulp and Paper Industry; Wiley: New York, NY, USA, 1998; pp. 273–307. [Google Scholar]
- Uenishi, J.i.; Muraoka, H.; Nameki, M.; Hata, N. Synthesis of acetomycin bislactone analogues. Synthesis 2004, 2004, 1343–1348. [Google Scholar] [CrossRef]
- Santra, H.K.; Banerjee, D. Natural products as fungicide and their role in crop protection. In Natural Bioactive Products in Sustainable Agriculture; Springer Nature: Singapore, 2020; pp. 131–219. [Google Scholar]
- Thind, T. Changing trends in discovery of new fungicides: A perspective. Indian Phytopathol. 2021, 74, 875–883. [Google Scholar] [CrossRef]
- Liu, W.; Li, Y.; Fan, J.; Tang, L.; Guo, B.; Xue, W.; Fan, L. Natural Products-Based Fungicides: Synthesis and Antifungal Activity against Plant Pathogens of 2H-Chromene Derivatives. Chem. Biodivers. 2022, 19, e202200802. [Google Scholar] [CrossRef]
- Copping, L.G.; Duke, S.O. Natural products that have been used commercially as crop protection agents. Pest Manag. Sci. Former. Pestic. Sci. 2007, 63, 524–554. [Google Scholar] [CrossRef]
- Fukunaga, K.; Misato, T.; Ishii, I.; Asakawa, M. Blasticidin, a new anti-phytopathogenic fungal substance. Part I. J. Agric. Chem. Soc. Jpn. 1955, 19, 181–188. [Google Scholar] [CrossRef]
- Chen, D.; Sprules, T.J.; Lavallée, J.F. Synthesis and in vitro cytotoxicity of (±)-acetomycin and related analogs. Bioorganic Med. Chem. Lett. 1995, 5, 759–762. [Google Scholar] [CrossRef]
- Williams, S.; Wellington, E. Actinomycetes. In Methods of Soil Analysis: Part 2, Chemical and Microbiological Properties, Agronomy Monograph; Page, A.L., Ed.; American Society of Agronomy Inc., Soil Science of America Inc.: Madison, WI, USA, 1983; pp. 969–987. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, A64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, L. WinGX suite for small-molecule single-crystal crystallography. J. Appl. Crystallogr. 1999, 32, 837–838. [Google Scholar] [CrossRef]
- Farrugia, L. ORTEP-3 for Windows—A version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Crystallogr. 1997, 30, 565. [Google Scholar] [CrossRef]
- Hooft, R.W.W.; Straver, L.H.; Spek, A.L. Determination of absolute structure using Bayesian statistics on Bijvoet differences. J. Appl. Crystallogr. 2008, 41, 96–103. [Google Scholar] [CrossRef]
- Spek, A. Single-crystal structure validation with the program PLATON. J. Appl. Crystallogr. 2003, 36, 7–13. [Google Scholar] [CrossRef]
- Bauer, A.W.; Perry, D.M.; Kirby, W.M. Single-disk antibiotic-sensitivity testing of Staphylococci: An analysis of technique and results. AMA Arch. Intern. Med. 1959, 104, 208–216. [Google Scholar] [CrossRef]

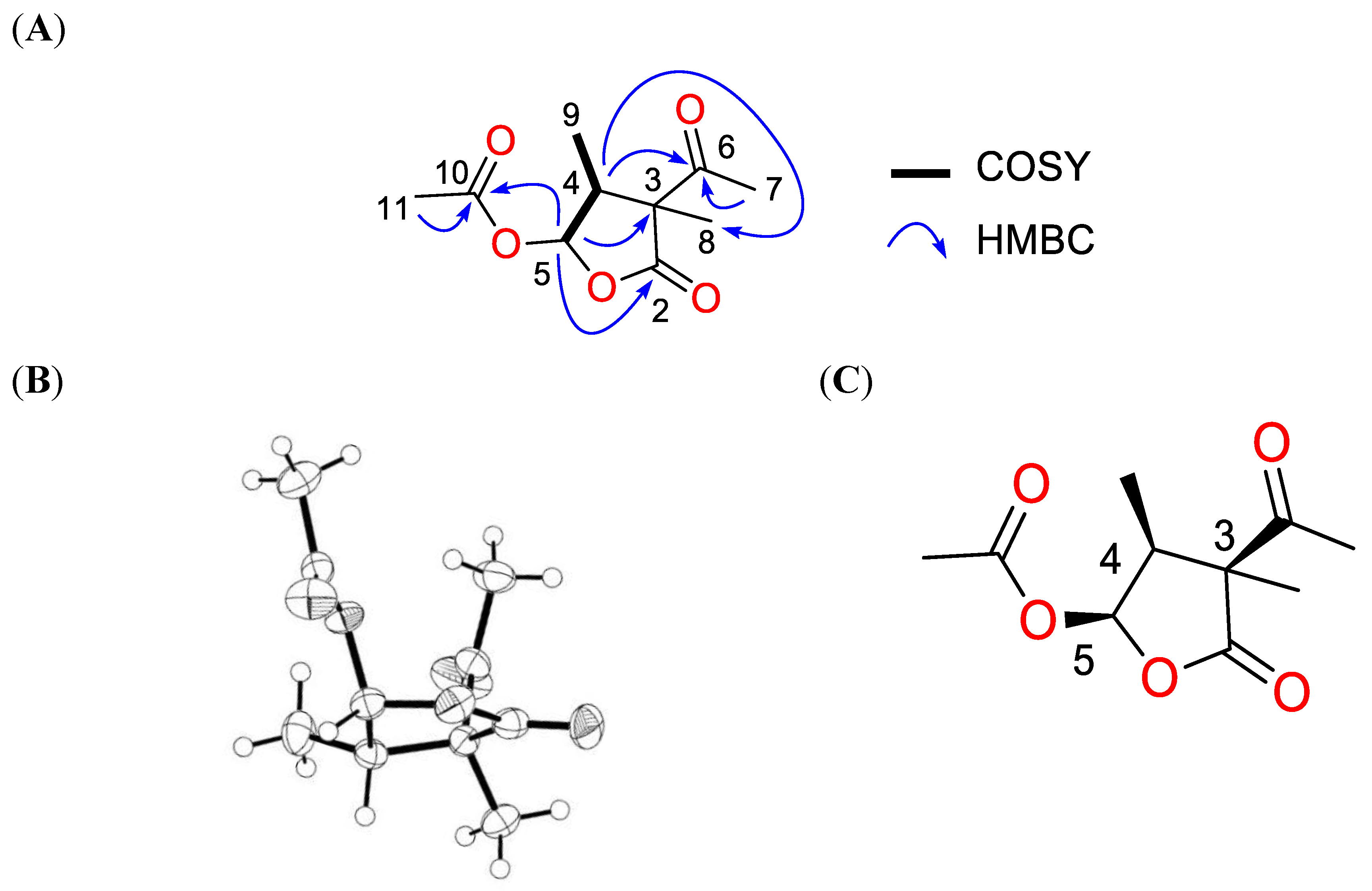

| Position | δC | Multiplet | δH (J in Hz) | HMBC |
|---|---|---|---|---|
| 2 | 178.8 | C | ||
| 3 | 58.7 | C | ||
| 4 | 46.2 | CH | 2.72, m | 3, 5, 6, 8, 9 |
| 5 | 95.7 | CH | 6.56, d (J = 5.2) | 2, 3, 4, 10 |
| 6 | 205.7 | C | ||
| 7 | 28.9 | CH3 | 2.27, s | 3, 6 |
| 8 | 20.8 | CH3 | 1.43, s | 2, 3, 4, 6 |
| 9 | 9.7 | CH3 | 1.06, d (J = 7.6) | 3, 4, 5 |
| 10 | 170.4 | C | ||
| 11 | 20.4 | CH3 | 2.10, s | 10 |
| Fungal Crop Pathogen | 10,000 ppm | 5000 ppm | 2500 ppm | 1000 ppm | 500 ppm |
|---|---|---|---|---|---|
| Botrytis cinerea | +++ | ++ | + | - | - |
| Aspergillus niger | ++ | + | + | + | - |
| Alternaria alternata | +++ | +++ | ++ | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adra, C.; Tran, T.D.; Foster, K.; Tomlin, R.; Kurtböke, D.İ. Identification of Acetomycin as an Antifungal Agent Produced by Termite Gut-Associated Streptomycetes against Pyrrhoderma noxium. Antibiotics 2024, 13, 45. https://doi.org/10.3390/antibiotics13010045
Adra C, Tran TD, Foster K, Tomlin R, Kurtböke Dİ. Identification of Acetomycin as an Antifungal Agent Produced by Termite Gut-Associated Streptomycetes against Pyrrhoderma noxium. Antibiotics. 2024; 13(1):45. https://doi.org/10.3390/antibiotics13010045
Chicago/Turabian StyleAdra, Cherrihan, Trong D. Tran, Keith Foster, Russell Tomlin, and D. İpek Kurtböke. 2024. "Identification of Acetomycin as an Antifungal Agent Produced by Termite Gut-Associated Streptomycetes against Pyrrhoderma noxium" Antibiotics 13, no. 1: 45. https://doi.org/10.3390/antibiotics13010045
APA StyleAdra, C., Tran, T. D., Foster, K., Tomlin, R., & Kurtböke, D. İ. (2024). Identification of Acetomycin as an Antifungal Agent Produced by Termite Gut-Associated Streptomycetes against Pyrrhoderma noxium. Antibiotics, 13(1), 45. https://doi.org/10.3390/antibiotics13010045








