Anti-Salmonella Defence and Intestinal Homeostatic Maintenance In Vitro of a Consortium Containing Limosilactobacillus fermentum 3872 and Ligilactobacillus salivarius 7247 Strains in Human, Porcine, and Chicken Enterocytes
Abstract
1. Introduction
2. Results
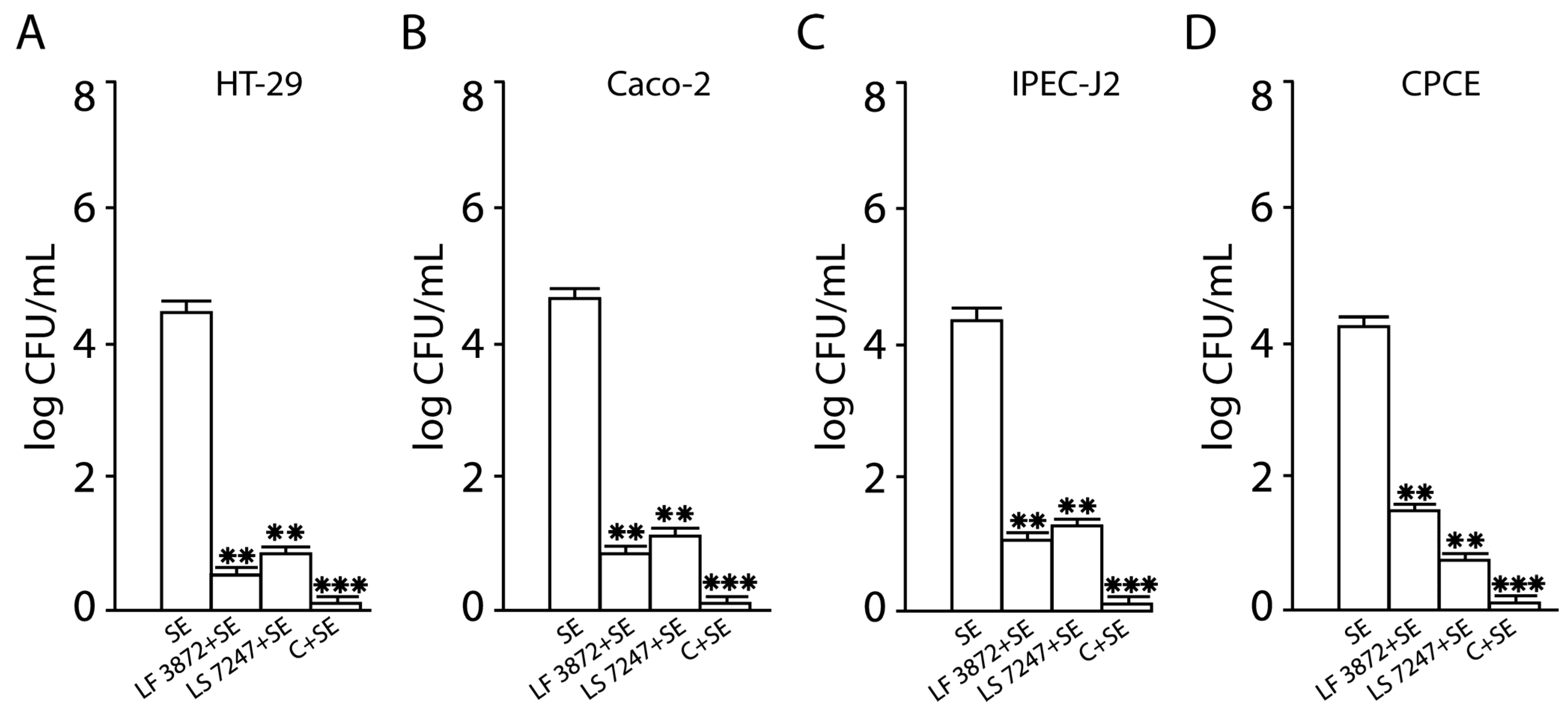
2.1. Inhibition of SE Adhesion to Intact Human and Animal Enterocytes by LF3872 and LS7247 Strains and Their Consortium
2.2. Inhibition of SE Invasion into Intact Human and Animal Enterocytes by LF3872 and LS7247 Strains and Their Consortium
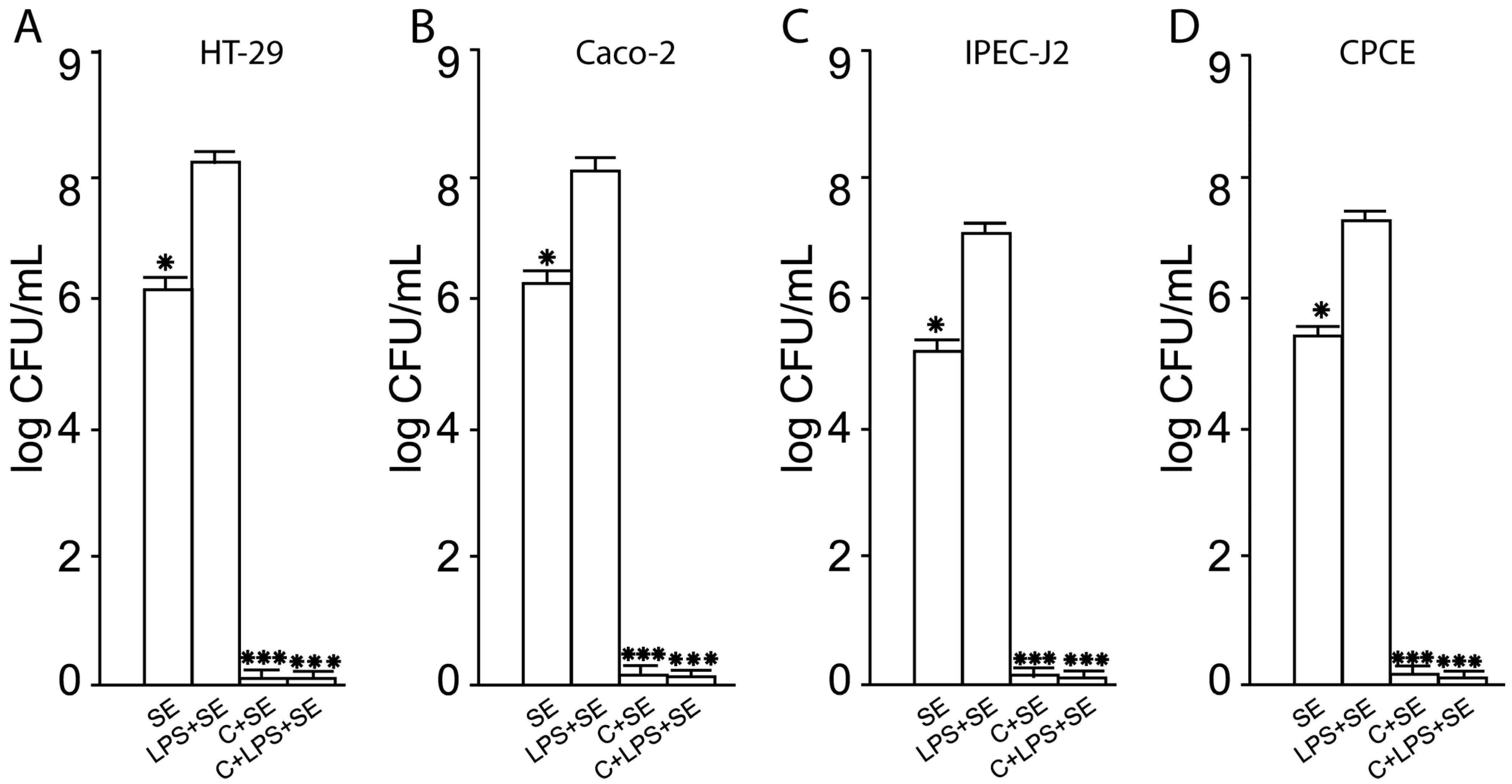
2.3. Inhibition of SE Adhesion to Activated Human and Animal Enterocytes by Consortium of LF3872 and LS7247 Strain
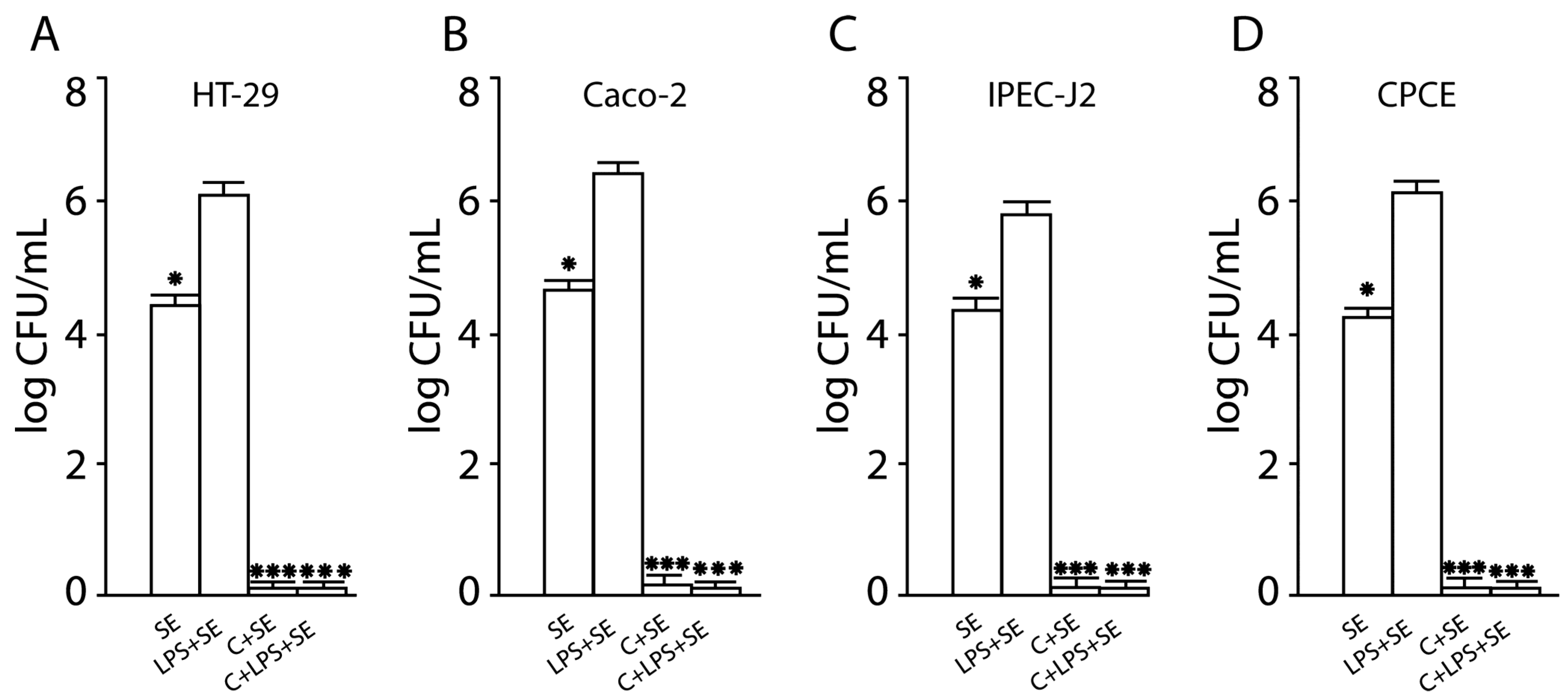
2.4. Inhibition of SE Invasion into Activated Human and Animal Enterocytes by Consortium of LF3872 and LS7247 Strains
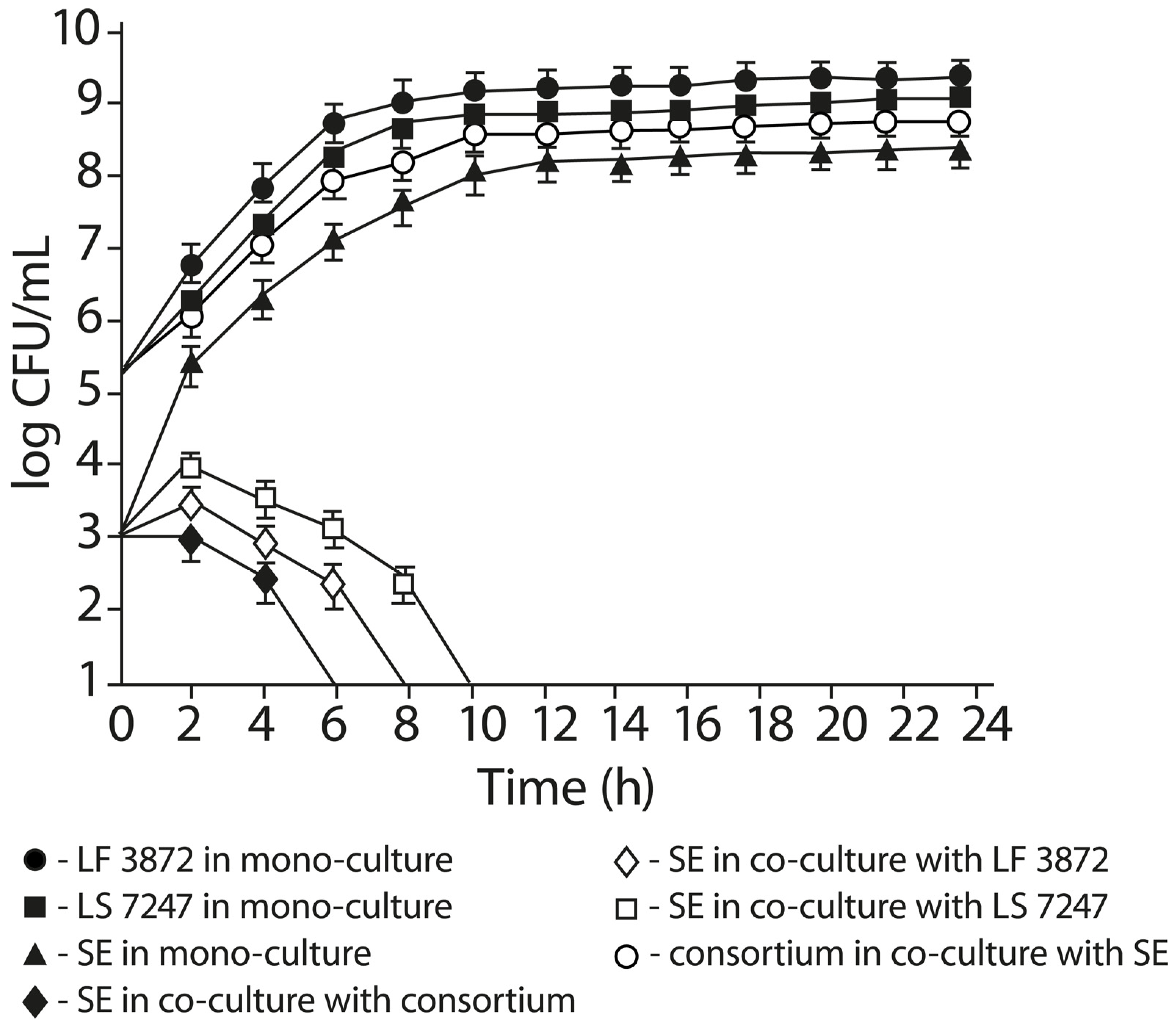
2.5. Anti-Salmonella Activity of LF3872 and LS7247 Consortium in Co-Culture Test
2.6. The Effect of Cell-Free Supernatant from LF3872 and LS7247 Consortium on the Viability of Human HT-29 and Caco-2, Porcine IPEC-J2, and Chicken CPCE Cells
2.7. The Effect of CFS from LF3872 and LS7247 Consortium on the Expression of SE Genes Responsible for Intestinal Colonization and Virulence
2.8. Consortium of LF3872 and LS7247 Strains Suppressed SE-Induced Production of IL-8, TNF-α, and Il-1β in Human HT-29 and Caco-2, Porcine IPEC-J2, and Chicken CPCE Cells
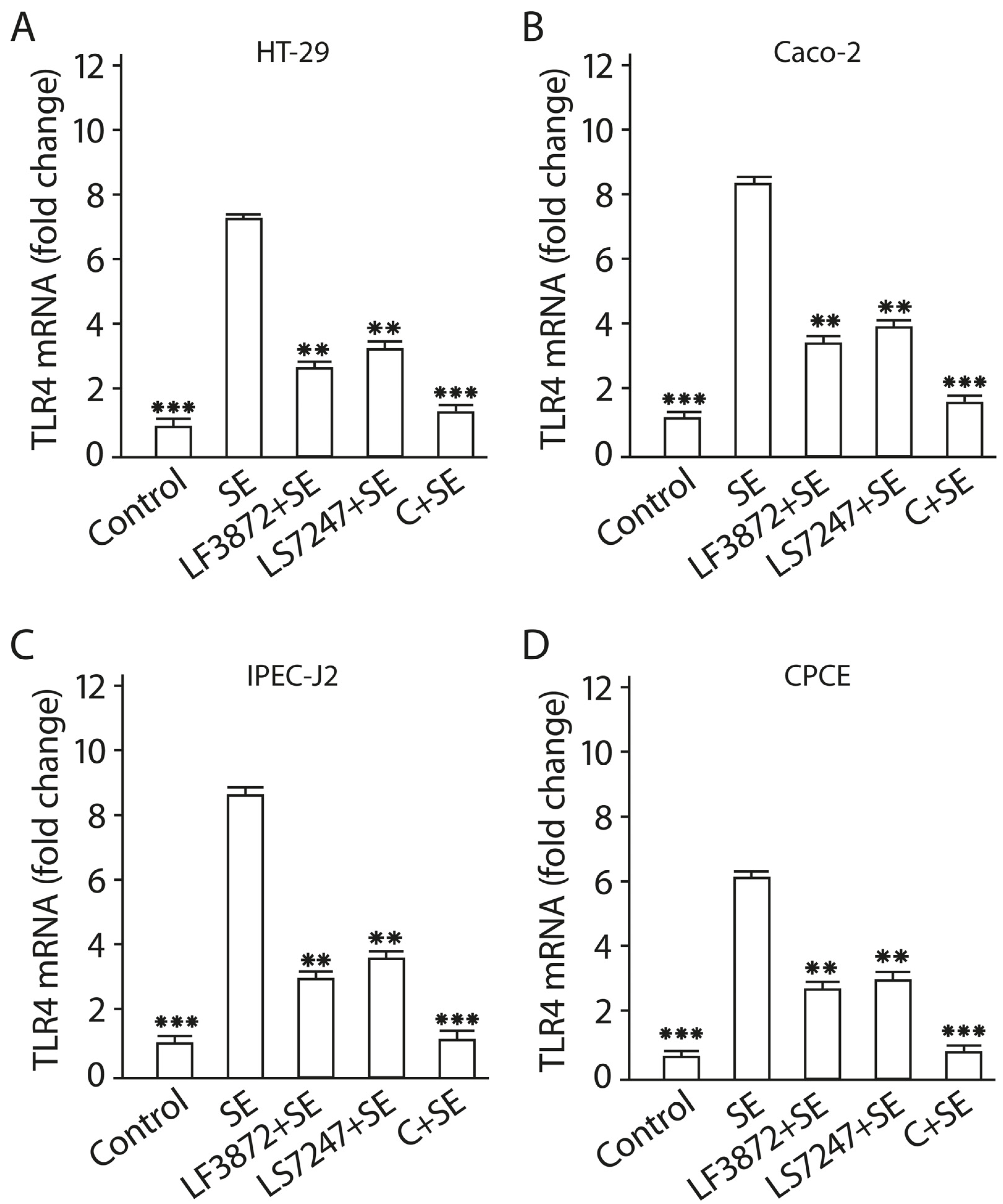
2.9. Consortium of LF3872 and LS7247 Strains Suppressed SE-Induced Expression of TLR4 and Stimulated TLR9 Expression in Human HT-29 and Caco-2 and Porcine IPEC-J2 Cells, and TLR21 in Chicken CPCE Cells
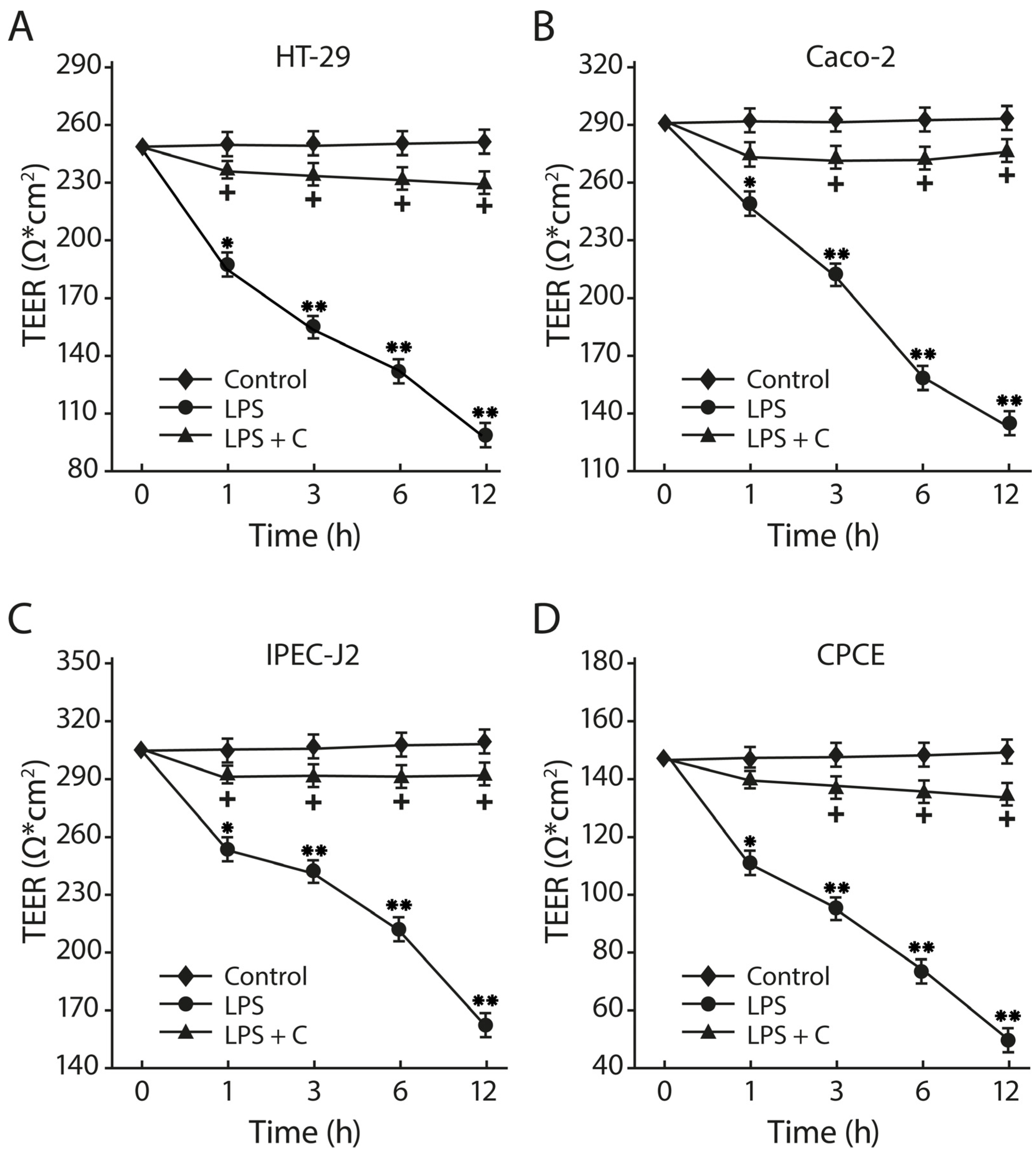
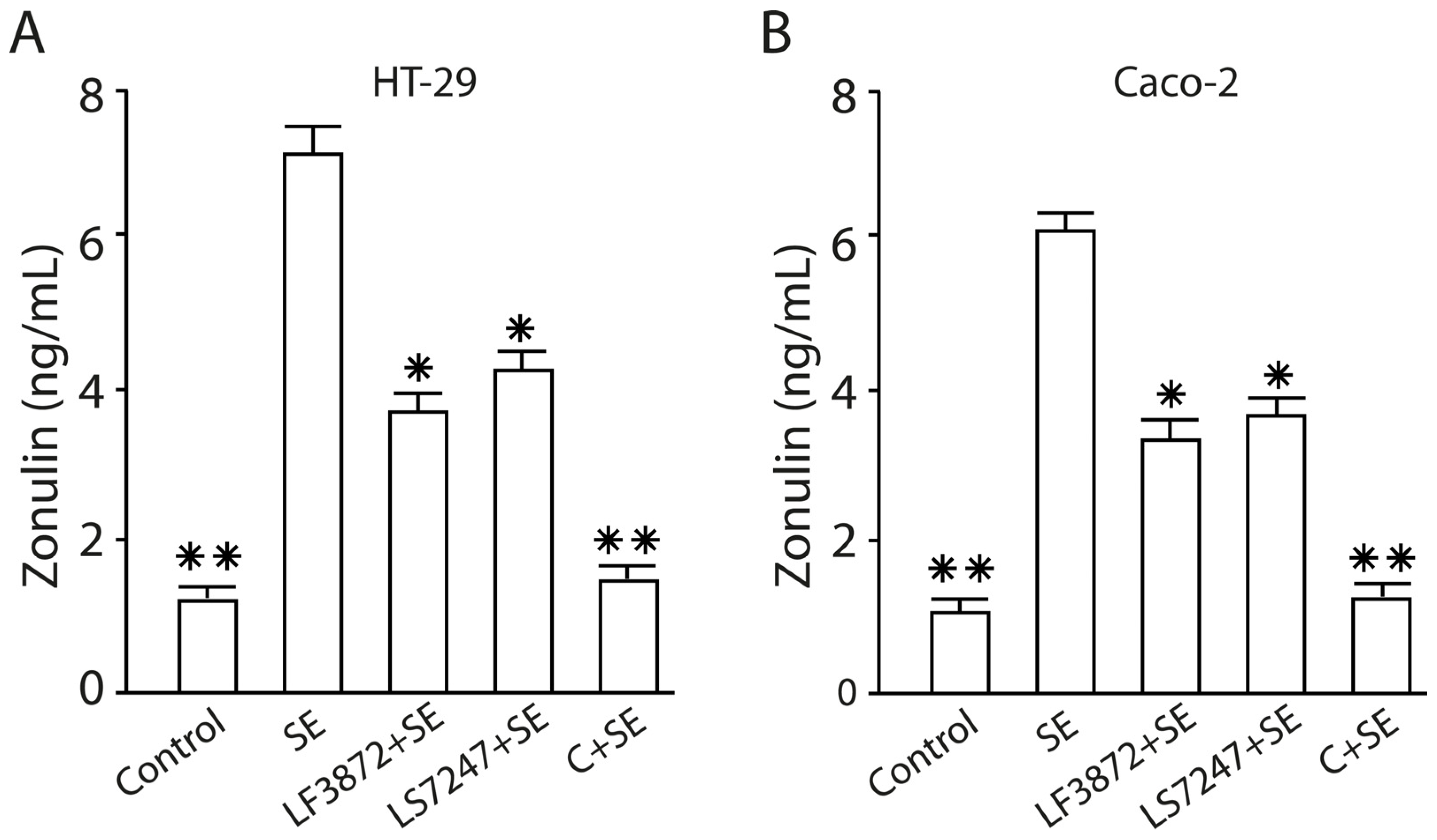
2.10. The LF3872 and LS7247 Consortium Protected the Intestinal Barrier and Reduced LPS-Induced Paracellular Permeability in Human HT-29 and Caco-2, Porcine IPEC-J2, and Chicken CPCE Cells
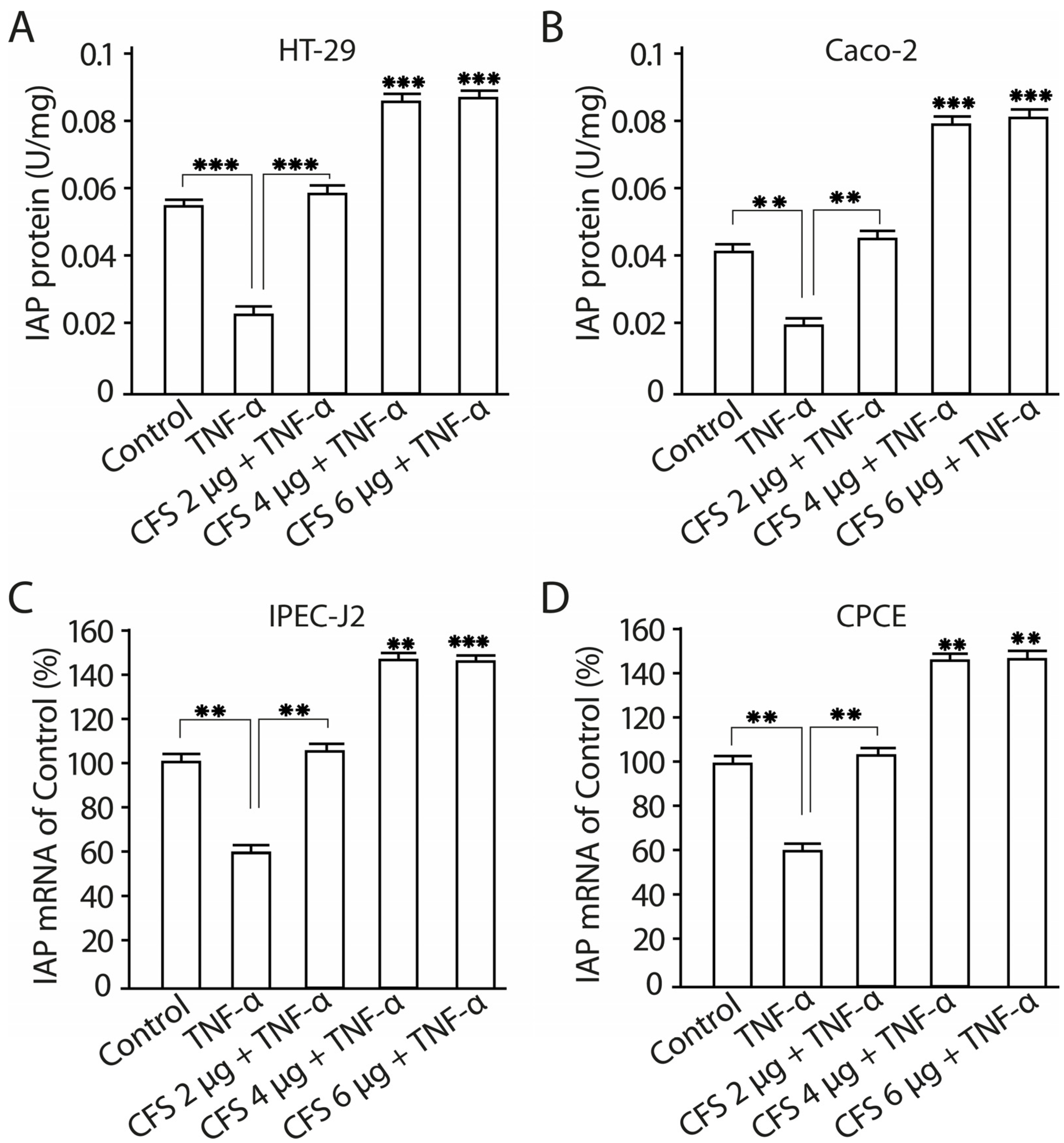
2.11. The Effect of CFS from LF3872 and LS7247 Consortium on the Intestinal Alkaline Phosphatase Production and mRNA Expression in Human HT-29 and Caco-2 Enterocytes
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Intestinal Epithelial Cells and Growth Conditions
4.2.1. Human Intestinal Epithelial Cells
4.2.2. Porcine Intestinal Epithelial Cells
4.2.3. Chicken Primary Cecal Enterocytes
4.3. Adhesion Assay
4.4. Invasion Assay
4.5. Determination of Anti-Salmonella Activity of LF3872 and LS7247 Strains and Their Consortium by Co-Cultivation Method in a Liquid Medium
4.6. Preparation of CFS from Consortium of LF3872 and LS7247 Strains
4.7. Determination of SIC of CFS of LF3872 and LS7247 Consortium
4.8. Effect of CFS on Viability of Human HT29 and Caco-2, Porcine IPEC-J2, and Chicken CPCE Cells
4.9. RNA Extraction and cDNA Synthesis
4.10. Quantitative Real-Time Polymerase Chain Reaction
4.11. Analysis of Cytokines in Culture Supernatants of Human, Porcine, and Chicken Enterocytes
4.12. Co-Culture of LF3872, LS7247, Their Consortium, and SE with Human HT29 and Caco-2, Porcine IPEC-J2, and Chicken CPCE Cells
4.13. TEER Measurements
4.14. Permeability Studies
- Papp = apparent permeability coefficient [cm/s];
- dQ/dt = rate of appearance of FD4 on the basolateral side [µg/s];
- A = surface area of the monolayer [cm2];
- C0 = initial FD4 concentration in the apical side [µg/mL].
4.15. Quantification of Tight Junction Regulator Zonulin
4.16. Activity of IAP Measurement
4.17. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Steve Yan, S.; Pendrak, M.L.; Abela-Ridder, B.; Punderson, J.W.; Fedorko, D.P.; Foley, S.L. An overview of Salmonella typing. Clin. Appl. Immunol. Rev. 2004, 4, 189–204. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Mattheus, W.; Vanhoof, R.; Bertrand, S. Trends in serotype distribution and antimicrobial susceptibility in Salmonella enterica isolates from humans in Belgium, 2009 to 2013. Antimicrob. Agents Chemother. 2015, 59, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Afshari, A.; Baratpour, A.; Khanzade, S.; Jamshidi, A. Salmonella Enteritidis and Salmonella Typhimorium identification in poultry carcasses. Iran. J. Microbiol. 2018, 10, 45–50. Available online: http://www.ncbi.nlm.nih.gov/pubmed/29922418 (accessed on 20 October 2023). [PubMed]
- Verbrugghe, E.; Dhaenens, M.; Leyman, B.; Boyen, F.; Shearer, N.; Van Parys, A.; Haesendonck, R.; Bert, W.; Favoreel, H.; Deforce, D.; et al. Host Stress Drives Salmonella Recrudescence. Sci. Rep. 2016, 6, 20849. [Google Scholar] [CrossRef] [PubMed]
- Torgerson, P.R.; Devleesschauwer, B.; Praet, N.; Speybroeck, N.; Willingham, A.L.; Kasuga, F.; Rokni, M.B.; Zhou, X.-N.; Fèvre, E.M.; Sripa, B.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 11 Foodborne Parasitic Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001920. [Google Scholar] [CrossRef]
- Rabsch, W.; Hargis, B.M.; Tsolis, R.M.; Kingsley, R.A.; Hinz, K.H.; Tschäpe, H.; Bäumler, A.J. Competitive exclusion of Salmonella Enteritidis by Salmonella Gallinarum in poultry. Emerg. Infect. Dis. 2000, 6, 443–448. [Google Scholar] [CrossRef]
- Hogue, A.; White, P.; Guard-Petter, J.; Schlosser, W.; Gast, R.; Ebel, E.; Farrar, J.; Gomez, T.; Madden, J.; Madison, M.; et al. Epidemiology and control of egg-associated Salmonella Enteritidis in the United States of America. Rev. Sci. Tech. 1997, 16, 542–553. [Google Scholar] [CrossRef]
- Guard-Petter, J. The chicken, the egg and Salmonella Enteritidis. Environ. Microbiol. 2001, 3, 421–430. [Google Scholar] [CrossRef]
- Poppe, C. Salmonella enteritidis in Canada. Int. J. Food Microbiol. 1994, 21, 1–5. [Google Scholar] [CrossRef]
- Braden, C.R. Salmonella enterica serotype Enteritidis and eggs: A national epidemic in the United States. Clin. Infect. Dis. 2006, 43, 512–517. [Google Scholar] [CrossRef]
- Betancor, L.; Pereira, M.; Martinez, A.; Giossa, G.; Fookes, M.; Flores, K.; Barrios, P.; Repiso, V.; Vignoli, R.; Cordeiro, N.; et al. Prevalence of Salmonella enterica in poultry and eggs in Uruguay during an epidemic due to Salmonella enterica serovar Enteritidis. J. Clin. Microbiol. 2010, 48, 2413–2423. [Google Scholar] [CrossRef] [PubMed]
- Galarce, N.E.; Bravo, J.L.; Robeson, J.P.; Borie, C.F. Bacteriophage cocktail reduces Salmonella enterica serovar Enteritidis counts in raw and smoked salmon tissues. Rev. Argent. Microbiol. 2014, 46, 333–337. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. Eur. Food Saf. Auth. 2019, 17, e05598. [Google Scholar] [CrossRef]
- Feasey, N.A.; Dougan, G.; Kingsley, R.A.; Heyderman, R.S.; Gordon, M.A. Invasive non-typhoidal salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 2012, 379, 2489–2499. [Google Scholar] [CrossRef] [PubMed]
- Oneko, M.; Kariuki, S.; Muturi-Kioi, V.; Otieno, K.; Otieno, V.O.; Williamson, J.M.; Folster, J.; Parsons, M.B.; Slutsker, L.; Mahon, B.E.; et al. Emergence of Community-Acquired, Multidrug-Resistant Invasive Nontyphoidal Salmonella Disease in Rural Western Kenya, 2009–2013. Clin. Infect. Dis. 2015, 61 (Suppl. S4), S310–S316. [Google Scholar] [CrossRef] [PubMed]
- GBD 2017 Non-Typhoidal Salmonella Invasive Disease Collaborators. The global burden of non-typhoidal salmonella invasive disease: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. Infect. Dis. 2019, 19, 1312–1324. [Google Scholar] [CrossRef]
- Aung, K.T.; Khor, W.C.; Ong, K.H.; Tan, W.L.; Wong, Z.N.; Oh, J.Q.; Wong, W.K.; Tan, B.Z.Y.; Maiwald, M.; Tee, N.W.S.; et al. Characterisation of Salmonella Enteritidis ST11 and ST1925 Associated with Human Intestinal and Extra-Intestinal Infections in Singapore. Int. J. Environ. Res. Public Health 2022, 19, 5671. [Google Scholar] [CrossRef]
- De Reu, K.; Grijspeerdt, K.; Messens, W.; Heyndrickx, M.; Uyttendaele, M.; Debevere, J.; Herman, L. Eggshell factors influencing eggshell penetration and whole egg contamination by different bacteria, including Salmonella enteritidis. Int. J. Food Microbiol. 2006, 112, 253–260. [Google Scholar] [CrossRef]
- Arnold, M.E.; Martelli, F.; McLaren, I.; Davies, R.H. Estimation of the rate of egg contamination from Salmonella-infected chickens. Zoonoses Public Health 2014, 61, 18–27. [Google Scholar] [CrossRef]
- Aung, K.T.; ManLing, C.; KengWai, M.; Lim, N.; JiaQuan, O.; SuLin, K.; YiHua, L.; Goh, T.L.V.; Yap, H.M.; Gutiérrez, R.A.; et al. Microbiological assessment of chicken meat sold at chicken rice stalls in Singapore. Southeast Asian J. Trop. Med. Public Health 2018, 49, 1043–1052. [Google Scholar]
- Aung, K.T.; Chen, H.J.; Chau, M.L.; Yap, G.; Lim, X.F.; Humaidi, M.; Chua, C.; Yeo, G.; Yap, H.M.; Oh, J.Q.; et al. Salmonella in Retail Food and Wild Birds in Singapore-Prevalence, Antimicrobial Resistance, and Sequence Types. Int. J. Environ. Res. Public Health 2019, 16, 4235. [Google Scholar] [CrossRef]
- Huston, C.L.; Wittum, T.E.; Love, B.C.; Keen, J.E. Prevalence of fecal shedding of Salmonella spp. in dairy herds. J. Am. Vet. Med. Assoc. 2002, 220, 645–649. [Google Scholar] [CrossRef]
- Adetoye, A.; Pinloche, E.; Adeniyi, B.A.; Ayeni, F.A. Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 2018, 18, 96. [Google Scholar] [CrossRef]
- Casaux, M.L.; Neto, W.S.; Schild, C.O.; Costa, R.A.; Macías-Rioseco, M.; Caffarena, R.D.; Silveira, C.S.; Aráoz, V.; Díaz, B.D.; Giannitti, F.; et al. Epidemiological and clinicopathological findings in 15 fatal outbreaks of salmonellosis in dairy calves and virulence genes in the causative Salmonella enterica Typhimurium and Dublin strains. Braz. J. Microbiol. 2023, 54, 475–490. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. Eur. Food Saf. Auth. 2023, 21, e07867. [Google Scholar] [CrossRef]
- Card, R.M.; Chisnall, T.; Begum, R.; Sarker, M.S.; Hossain, M.S.; Sagor, M.S.; Mahmud, M.A.; Uddin, A.S.M.A.; Karim, M.R.; Lindahl, J.F.; et al. Multidrug-resistant non-typhoidal Salmonella of public health significance recovered from migratory birds in Bangladesh. Front. Microbiol. 2023, 14, 1162657. [Google Scholar] [CrossRef]
- Dias de Oliveira, S.; Siqueira Flores, F.; dos Santos, L.R.; Brandelli, A. Antimicrobial resistance in Salmonella enteritidis strains isolated from broiler carcasses, food, human and poultry-related samples. Int. J. Food Microbiol. 2005, 97, 297–305. [Google Scholar] [CrossRef]
- Rodríguez, I.; Rodicio, M.R.; Guerra, B.; Hopkins, K.L. Potential international spread of multidrug-resistant invasive Salmonella enterica serovar Enteritidis. Emerg. Infect. Dis. 2012, 18, 1173–1176. [Google Scholar] [CrossRef]
- Wang, X.; Biswas, S.; Paudyal, N.; Pan, H.; Li, X.; Fang, W.; Yue, M. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered from the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016. Front. Microbiol. 2019, 10, 985. [Google Scholar] [CrossRef]
- Qamar, A.; Ismail, T.; Akhtar, S. Prevalence and antibiotic resistance of Salmonella spp. in South Punjab-Pakistan. PLoS ONE 2020, 15, e0232382. [Google Scholar] [CrossRef]
- Saleh, S.; Van Puyvelde, S.; Staes, A.; Timmerman, E.; Barbé, B.; Jacobs, J.; Gevaert, K.; Deborggraeve, S. Salmonella Typhi, Paratyphi A, Enteritidis and Typhimurium core proteomes reveal differentially expressed proteins linked to the cell surface and pathogenicity. PLoS Negl. Trop. Dis. 2019, 13, e0007416. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Hammad, A.M.; Shimamoto, T. Towards a compatible probiotic-antibiotic combination therapy: Assessment of antimicrobial resistance in the Japanese probiotics. J. Appl. Microbiol. 2010, 109, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- Heider, L.C.; Funk, J.A.; Hoet, A.E.; Meiring, R.W.; Gebreyes, W.A.; Wittum, T.E. Identification of Escherichia coli and Salmonella enterica organisms with reduced susceptibility to ceftriaxone from fecal samples of cows in dairy herds. Am. J. Vet. Res. 2009, 70, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Alcaine, S.D.; Warnick, L.D.; Wiedmann, M. Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 2007, 70, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Isolauri, E.; Salminen, S.; Sanz, Y. The impact of probiotic on gut health. Curr. Drug Metab. 2009, 10, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Rishi, P.; Mavi, S.K.; Bharrhan, S.; Shukla, G.; Tewari, R. Protective efficacy of probiotic alone or in conjunction with a prebiotic in Salmonella-induced liver damage. FEMS Microbiol. Ecol. 2009, 69, 222–230. [Google Scholar] [CrossRef]
- Das, J.K.; Mishra, D.; Ray, P.; Tripathy, P.; Beuria, T.K.; Singh, N.; Suar, M. In vitro evaluation of anti-infective activity of a Lactobacillus plantarum strain against Salmonella enterica serovar Enteritidis. Gut Pathog. 2013, 5, 11. [Google Scholar] [CrossRef]
- Nami, Y.; Haghshenas, B.; Abdullah, N.; Barzegari, A.; Radiah, D.; Rosli, R.; Yari Khosroushahi, A. Probiotics or antibiotics: Future challenges in medicine. J. Med. Microbiol. 2015, 64, 137–146. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; de Oliveira Coelho, B.; Magalhães Júnior, A.I.; Thomaz-Soccol, V.; Soccol, C.R. How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv. 2018, 36, 2060–2076. [Google Scholar] [CrossRef]
- Rocha, T.S.; Baptista, A.A.S.; Donato, T.C.; Milbradt, E.L.; Okamoto, A.S.; Rodrigues, J.C.Z.; Coppola, M.P.; Andreatti Filho, R.L. Evaluation of in vitro and in vivo adhesion and immunomodulatory effect of Lactobacillus species strains isolated from chickens. Poult. Sci. 2012, 91, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, L.; Zhou, L.; Yang, X.; Zhao, X. Using In Vitro Immunomodulatory Properties of Lactic Acid Bacteria for Selection of Probiotics against Salmonella Infection in Broiler Chicks. PLoS ONE 2016, 11, e0147630. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-M.; Jang, H.-M.; Jeong, J.-J.; Han, M.J.; Kim, D.-H. Lactobacillus johnsonii CJLJ103 attenuates colitis and memory impairment in mice by inhibiting gut microbiota lipopolysaccharide production and NF-κB activation. J. Funct. Foods 2017, 34, 359–368. [Google Scholar] [CrossRef]
- Plaza-Diaz, J.; Gomez-Llorente, C.; Fontana, L.; Gil, A. Modulation of immunity and inflammatory gene expression in the gut, in inflammatory diseases of the gut and in the liver by probiotics. World J. Gastroenterol. 2014, 20, 15632–15649. [Google Scholar] [CrossRef] [PubMed]
- Delneste, Y.; Beauvillain, C.; Jeannin, P. Innate immunity: Structure and function of TLRs. Med. Sci. 2007, 23, 67–73. [Google Scholar] [CrossRef]
- Kumar, V. Toll-Like Receptors in Adaptive Immunity. Handb. Exp. Pharmacol. 2022, 276, 95–131. [Google Scholar] [CrossRef]
- Villena, J.; Kitazawa, H. Modulation of Intestinal TLR4-Inflammatory Signaling Pathways by Probiotic Microorganisms: Lessons Learned from Lactobacillus jensenii TL2937. Front. Immunol. 2014, 4, 512. [Google Scholar] [CrossRef]
- Bermudez-Brito, M.; Muñoz-Quezada, S.; Gomez-Llorente, C.; Matencio, E.; Bernal, M.J.; Romero, F.; Gil, A. Human intestinal dendritic cells decrease cytokine release against Salmonella infection in the presence of Lactobacillus paracasei upon TLR activation. PLoS ONE 2012, 7, e43197. [Google Scholar] [CrossRef]
- Villena, J.; Suzuki, R.; Fujie, H.; Chiba, E.; Takahashi, T.; Tomosada, Y.; Shimazu, T.; Aso, H.; Ohwada, S.; Suda, Y.; et al. Immunobiotic Lactobacillus jensenii modulates the Toll-like receptor 4-induced inflammatory response via negative regulation in porcine antigen-presenting cells. Clin. Vaccine Immunol. 2012, 19, 1038–1053. [Google Scholar] [CrossRef]
- Soltan Dallal, M.M.; Mojarrad, M.; Baghbani, F.; Raoofian, R.; Mardaneh, J.; Salehipour, Z. Effects of probiotic Lactobacillus acidophilus and Lactobacillus casei on colorectal tumor cells activity (CaCo-2). Arch. Iran. Med. 2015, 18, 167–172. Available online: http://www.ncbi.nlm.nih.gov/pubmed/25773690 (accessed on 20 October 2023).
- Chen, C.-Y.; Tsen, H.-Y.; Lin, C.-L.; Yu, B.; Chen, C.-S. Oral administration of a combination of select lactic acid bacteria strains to reduce the Salmonella invasion and inflammation of broiler chicks. Poult. Sci. 2012, 91, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.; Adams, M.; La Ragione, R.M.; Woodward, M.J. Colonisation of poultry by Salmonella Enteritidis S1400 is reduced by combined administration of Lactobacillus salivarius 59 and Enterococcus faecium PXN-33. Vet. Microbiol. 2017, 199, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Youk, S.; Song, C.-S. Effectiveness of Administering a Mixture of Lactic Acid Bacteria to Control Salmonella ser. Enteritidis Infections in Broilers. Animals 2022, 12, 374. [Google Scholar] [CrossRef] [PubMed]
- Abramov, V.M.; Kosarev, I.V.; Machulin, A.V.; Priputnevich, T.V.; Chikileva, I.O.; Deryusheva, E.I.; Abashina, T.N.; Donetskova, A.D.; Panin, A.N.; Melnikov, V.G.; et al. Limosilactobacillus fermentum Strain 3872: Antibacterial and Immunoregulatory Properties and Synergy with Prebiotics against Socially Significant Antibiotic-Resistant Infections of Animals and Humans. Antibiotics 2022, 11, 1437. [Google Scholar] [CrossRef]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Lactobacillus fermentum 3872 genome sequencing reveals plasmid and chromosomal genes potentially involved in a probiotic activity. FEMS Microbiol. Lett. 2015, 362, fnv068. [Google Scholar] [CrossRef] [PubMed]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Potential probiotic-associated traits revealed from completed high quality genome sequence of Lactobacillus fermentum 3872. Stand. Genom. Sci. 2017, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Lactobacillus fermentum 3872 as a potential tool for combatting Campylobacter jejuni infections. Virulence 2017, 8, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Karlyshev, A.V.; Gould, S. Ligilactobacillus salivarius 2102-15 complete genome sequence data. Data Br. 2023, 50, 109564. [Google Scholar] [CrossRef]
- Abramov, V.M.; Kosarev, I.V.; Machulin, A.V.; Priputnevich, T.V.; Deryusheva, E.I.; Nemashkalova, E.L.; Chikileva, I.O.; Abashina, T.N.; Panin, A.N.; Melnikov, V.G.; et al. Limosilactobacillus fermentum 3872 That Produces Class III Bacteriocin Forms Co-Aggregates with the Antibiotic-Resistant Staphylococcus aureus Strains and Induces Their Lethal Damage. Antibiotics 2023, 12, 471. [Google Scholar] [CrossRef]
- Kogut, M.H.; Chiang, H.-I.; Swaggerty, C.L.; Pevzner, I.Y.; Zhou, H. Gene Expression Analysis of Toll-Like Receptor Pathways in Heterophils from Genetic Chicken Lines that Differ in Their Susceptibility to Salmonella enteritidis. Front. Genet. 2012, 3, 121. [Google Scholar] [CrossRef]
- Lee, M.B.; Greig, J.D. A review of nosocomial Salmonella outbreaks: Infection control interventions found effective. Public Health 2013, 127, 199–206. [Google Scholar] [CrossRef]
- Rodrigues, D.; Cerca, N.; Teixeira, P.; Oliveira, R.; Ceri, H.; Azeredo, J. Listeria monocytogenes and Salmonella enterica Enteritidis biofilms susceptibility to different disinfectants and stress-response and virulence gene expression of surviving cells. Microb. Drug Resist. 2011, 17, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Arguello, H.; Carvajal, A.; Collazos, J.A.; García-Feliz, C.; Rubio, P. Prevalence and serovars of Salmonella enterica on pig carcasses, slaughtered pigs and the environment of four Spanish slaughterhouses. Food Res. Int. 2012, 45, 905–912. [Google Scholar] [CrossRef]
- Wang, H.; Wu, N.; Jiang, Y.; Ye, K.; Xu, X.; Zhou, G. Response of long-term acid stress to biofilm formation of meat-related Salmonella Enteritidis. Food Control 2016, 69, 214–220. [Google Scholar] [CrossRef]
- Guard-Petter, J. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl. Environ. Microbiol. 1998, 64, 2166–2172. [Google Scholar] [CrossRef]
- Guard-Petter, J.; Lakshmi, B.; Carlson, R.; Ingram, K. Characterization of lipopolysaccharide heterogeneity in Salmonella enteritidis by an improved gel electrophoresis method. Appl. Environ. Microbiol. 1995, 61, 2845–2851. [Google Scholar] [CrossRef]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide Epidemiology of Salmonella Serovars in Animal-Based Foods: A Meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef]
- Fàbrega, A.; Vila, J. Salmonella enterica serovar Typhimurium skills to succeed in the host: Virulence and regulation. Clin. Microbiol. Rev. 2013, 26, 308–341. [Google Scholar] [CrossRef]
- Stoll, B.J.; Hansen, N.I.; Bell, E.F.; Walsh, M.C.; Carlo, W.A.; Shankaran, S.; Laptook, A.R.; Sánchez, P.J.; Van Meurs, K.P.; Wyckoff, M.; et al. Trends in Care Practices, Morbidity, and Mortality of Extremely Preterm Neonates, 1993–2012. JAMA 2015, 314, 1039–1051. [Google Scholar] [CrossRef]
- Crump, J.A.; Sjölund-Karlsson, M.; Gordon, M.A.; Parry, C.M. Epidemiology, Clinical Presentation, Laboratory Diagnosis, Antimicrobial Resistance, and Antimicrobial Management of Invasive Salmonella Infections. Clin. Microbiol. Rev. 2015, 28, 901–937. [Google Scholar] [CrossRef]
- Hoshino, K.; Takeuchi, O.; Kawai, T.; Sanjo, H.; Ogawa, T.; Takeda, Y.; Takeda, K.; Akira, S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol. 1999, 162, 3749–3752. Available online: http://www.ncbi.nlm.nih.gov/pubmed/10201887 (accessed on 20 October 2023). [CrossRef]
- Nakaira-Takahagi, E.; Golim, M.A.; Bannwart, C.F.; Puccia, R.; Peraçoli, M.T.S. Interactions between TLR2, TLR4, and mannose receptors with gp43 from Paracoccidioides brasiliensis induce cytokine production by human monocytes. Med. Mycol. 2011, 49, 694–703. [Google Scholar] [CrossRef] [PubMed]
- Lehto, E.M.; Salminen, S.J. Inhibition of Salmonella typhimurium adhesion to Caco-2 cell cultures by Lactobacillus strain GG spent culture supernate: Only a pH effect? FEMS Immunol. Med. Microbiol. 1997, 18, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.H.; Pinkner, J.S.; Roth, R.; Heuser, J.; Nicholes, A.V.; Abraham, S.N.; Hultgren, S.J. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 1995, 92, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Ofek, I.; Mirelman, D.; Sharon, N. Adherence of Escherichia coli to human mucosal cells mediated by mannose receptors. Nature 1977, 265, 623–625. [Google Scholar] [CrossRef] [PubMed]
- Firon, N.; Ofek, I.; Sharon, N. Carbohydrate-binding sites of the mannose-specific fimbrial lectins of enterobacteria. Infect. Immun. 1984, 43, 1088–1090. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, J.; Mackenzie, J.; de Paz, J.L.; Chipwaza, B.; Choudhury, D.; Zavialov, A.; Mannerstedt, K.; Anderson, J.; Piérard, D.; Wyns, L.; et al. The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of Escherichia coli pathotypes. Mol. Microbiol. 2006, 61, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Rosen, D.A.; Pinkner, J.S.; Walker, J.N.; Elam, J.S.; Jones, J.M.; Hultgren, S.J. Molecular variations in Klebsiella pneumoniae and Escherichia coli FimH affect function and pathogenesis in the urinary tract. Infect. Immun. 2008, 76, 3346–3356. [Google Scholar] [CrossRef]
- Coconnier, M.H.; Bernet, M.F.; Kernéis, S.; Chauvière, G.; Fourniat, J.; Servin, A.L. Inhibition of adhesion of enteroinvasive pathogens to human intestinal Caco-2 cells by Lactobacillus acidophilus strain LB decreases bacterial invasion. FEMS Microbiol. Lett. 1993, 110, 299–305. [Google Scholar] [CrossRef][Green Version]
- Coconnier, M.H.; Liévin, V.; Lorrot, M.; Servin, A.L. Antagonistic activity of Lactobacillus acidophilus LB against intracellular Salmonella enterica serovar Typhimurium infecting human enterocyte-like Caco-2/TC-7 cells. Appl. Environ. Microbiol. 2000, 66, 1152–1157. [Google Scholar] [CrossRef]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [PubMed]
- Notti, R.Q.; Stebbins, C.E. The Structure and Function of Type III Secretion Systems. Microbiol. Spectr. 2016, 4, 4 . [Google Scholar] [CrossRef]
- Raffatellu, M.; Wilson, R.P.; Chessa, D.; Andrews-Polymenis, H.; Tran, Q.T.; Lawhon, S.; Khare, S.; Adams, L.G.; Bäumler, A.J. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect. Immun. 2005, 73, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Thiennimitr, P.; Winter, S.E.; Winter, M.G.; Xavier, M.N.; Tolstikov, V.; Huseby, D.L.; Sterzenbach, T.; Tsolis, R.M.; Roth, J.R.; Bäumler, A.J. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. USA 2011, 108, 17480–17485. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Song, L.; Chen, Y.; Jiao, X.; Pan, Z. Salmonella Enteritidis activates inflammatory storm via SPI-1 and SPI-2 to promote intracellular proliferation and bacterial virulence. Front. Cell. Infect. Microbiol. 2023, 13, 1158888. [Google Scholar] [CrossRef]
- Bang, I.-S.; Frye, J.G.; McClelland, M.; Velayudhan, J.; Fang, F.C. Alternative sigma factor interactions in Salmonella: Sigma and sigma promote antioxidant defences by enhancing sigma levels. Mol. Microbiol. 2005, 56, 811–823. [Google Scholar] [CrossRef]
- Crouch, M.-L.; Becker, L.A.; Bang, I.-S.; Tanabe, H.; Ouellette, A.J.; Fang, F.C. The alternative sigma factor sigma is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol. Microbiol. 2005, 56, 789–799. [Google Scholar] [CrossRef]
- Richardson, A.R.; Soliven, K.C.; Castor, M.E.; Barnes, P.D.; Libby, S.J.; Fang, F.C. The Base Excision Repair system of Salmonella enterica serovar typhimurium counteracts DNA damage by host nitric oxide. PLoS Pathog. 2009, 5, e1000451. [Google Scholar] [CrossRef]
- Winter, S.E.; Thiennimitr, P.; Winter, M.G.; Butler, B.P.; Huseby, D.L.; Crawford, R.W.; Russell, J.M.; Bevins, C.L.; Adams, L.G.; Tsolis, R.M.; et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 2010, 467, 426–429. [Google Scholar] [CrossRef]
- Abhisingha, M.; Dumnil, J.; Pitaksutheepong, C. Selection of Potential Probiotic Lactobacillus with Inhibitory Activity Against Salmonella and Fecal Coliform Bacteria. Probiotics Antimicrob. Proteins 2018, 10, 218–227. [Google Scholar] [CrossRef]
- Hallstrom, K.; McCormick, B.A. Salmonella Interaction with and Passage through the Intestinal Mucosa: Through the Lens of the Organism. Front. Microbiol. 2011, 2, 88. [Google Scholar] [CrossRef]
- Pico-Rodríguez, J.T.; Martínez-Jarquín, H.; Gómez-Chávez, J.d.J.; Juárez-Ramírez, M.; Martínez-Chavarría, L.C. Effect of Salmonella pathogenicity island 1 and 2 (SPI-1 and SPI-2) deletion on intestinal colonization and systemic dissemination in chickens. Vet. Res. Commun. 2023. [Google Scholar] [CrossRef]
- Marongiu, L.; Gornati, L.; Artuso, I.; Zanoni, I.; Granucci, F. Below the surface: The inner lives of TLR4 and TLR9. J. Leukoc. Biol. 2019, 106, 147–160. [Google Scholar] [CrossRef]
- Moshiri, M.; Dallal, M.M.S.; Rezaei, F.; Douraghi, M.; Sharifi, L.; Noroozbabaei, Z.; Gholami, M.; Mirshafiey, A. The Effect of Lactobacillus acidophilus PTCC 1643 on Cultured Intestinal Epithelial Cells Infected with Salmonella enterica serovar Enteritidis. Osong Public Health Res. Perspect. 2017, 8, 54–60. [Google Scholar] [CrossRef]
- Thomalla, M.; Schmid, A.; Neumann, E.; Pfefferle, P.I.; Müller-Ladner, U.; Schäffler, A.; Karrasch, T. Evidence of an anti-inflammatory toll-like receptor 9 (TLR 9) pathway in adipocytes. J. Endocrinol. 2019, 240, 325–343. [Google Scholar] [CrossRef]
- Lee, J.; Mo, J.-H.; Katakura, K.; Alkalay, I.; Rucker, A.N.; Liu, Y.-T.; Lee, H.-K.; Shen, C.; Cojocaru, G.; Shenouda, S.; et al. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat. Cell Biol. 2006, 8, 1327–1336. [Google Scholar] [CrossRef]
- Fasano, A. Zonulin and its regulation of intestinal barrier function: The biological door to inflammation, autoimmunity, and cancer. Physiol. Rev. 2011, 91, 151–175. [Google Scholar] [CrossRef]
- Vancamelbeke, M.; Vermeire, S. The intestinal barrier: A fundamental role in health and disease. Expert Rev. Gastroenterol. Hepatol. 2017, 11, 821–834. [Google Scholar] [CrossRef]
- Han, H.; You, Y.; Cha, S.; Kim, T.-R.; Sohn, M.; Park, J. Multi-Species Probiotic Strain Mixture Enhances Intestinal Barrier Function by Regulating Inflammation and Tight Junctions in Lipopolysaccharides Stimulated Caco-2 Cells. Microorganisms 2023, 11, 656. [Google Scholar] [CrossRef]
- Malo, M.S.; Biswas, S.; Abedrapo, M.A.; Yeh, L.; Chen, A.; Hodin, R.A. The pro-inflammatory cytokines, IL-1beta and TNF-alpha, inhibit intestinal alkaline phosphatase gene expression. DNA Cell Biol. 2006, 25, 684–695. [Google Scholar] [CrossRef]
- Malo, M.S.; Alam, S.N.; Mostafa, G.; Zeller, S.J.; Johnson, P.V.; Mohammad, N.; Chen, K.T.; Moss, A.K.; Ramasamy, S.; Faruqui, A.; et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut 2010, 59, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wandler, A.M.; Postlethwait, J.H.; Guillemin, K. Dynamic Evolution of the LPS-Detoxifying Enzyme Intestinal Alkaline Phosphatase in Zebrafish and Other Vertebrates. Front. Immunol. 2012, 3, 314. [Google Scholar] [CrossRef]
- Malo, M.S.; Moaven, O.; Muhammad, N.; Biswas, B.; Alam, S.N.; Economopoulos, K.P.; Gul, S.S.; Hamarneh, S.R.; Malo, N.S.; Teshager, A.; et al. Intestinal alkaline phosphatase promotes gut bacterial growth by reducing the concentration of luminal nucleotide triphosphates. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G826–G838. [Google Scholar] [CrossRef] [PubMed]
- Lallès, J.-P. Recent advances in intestinal alkaline phosphatase, inflammation, and nutrition. Nutr. Rev. 2019, 77, 710–724. [Google Scholar] [CrossRef] [PubMed]
- Santos, G.M.; Ismael, S.; Morais, J.; Araújo, J.R.; Faria, A.; Calhau, C.; Marques, C. Intestinal Alkaline Phosphatase: A Review of This Enzyme Role in the Intestinal Barrier Function. Microorganisms 2022, 10, 746. [Google Scholar] [CrossRef] [PubMed]
- Dullens, S.P.J.; Mensink, R.P.; Mariman, E.C.M.; Plat, J. Differentiated CaCo-2 cells as an in-vitro model to evaluate de-novo apolipoprotein A-I production in the small intestine. Eur. J. Gastroenterol. Hepatol. 2009, 21, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Le Bivic, A.; Hirn, M.; Reggio, H. HT-29 cells are an in vitro model for the generation of cell polarity in epithelia during embryonic differentiation. Proc. Natl. Acad. Sci. USA 1988, 85, 136–140. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, M.; Zihler Berner, A.; Chervet, N.; Chassard, C.; Lacroix, C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J. Microbiol. Methods 2013, 94, 274–279. [Google Scholar] [CrossRef]
- Dudík, B.; Kiňová Sepová, H.; Bilka, F.; Pašková, Ľ.; Bilková, A. Mucin pre-cultivated Lactobacillus reuteri E shows enhanced adhesion and increases mucin expression in HT-29 cells. Antonie Van Leeuwenhoek 2020, 113, 1191–1200. [Google Scholar] [CrossRef]
- Schierack, P.; Nordhoff, M.; Pollmann, M.; Weyrauch, K.D.; Amasheh, S.; Lodemann, U.; Jores, J.; Tachu, B.; Kleta, S.; Blikslager, A.; et al. Characterization of a porcine intestinal epithelial cell line for in vitro studies of microbial pathogenesis in swine. Histochem. Cell Biol. 2006, 125, 293–305. [Google Scholar] [CrossRef]
- Abramov, V.M.; Kosarev, I.V.; Machulin, A.V.; Deryusheva, E.I.; Priputnevich, T.V.; Panin, A.N.; Chikileva, I.O.; Abashina, T.N.; Manoyan, A.M.; Ahmetzyanova, A.A.; et al. Ligilactobacillus salivarius 7247 Strain: Probiotic Properties and Anti-Salmonella Effect with Prebiotics. Antibiotics 2023, 12, 1535. [Google Scholar] [CrossRef] [PubMed]
- Rath, N.C.; Liyanage, R.; Gupta, A.; Packialakshmi, B.; Lay, J.O. A method to culture chicken enterocytes and their characterization. Poult. Sci. 2018, 97, 4040–4047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Dupont, A.; Torow, N.; Gohde, F.; Leschner, S.; Lienenklaus, S.; Weiss, S.; Brinkmann, M.M.; Kühnel, M.; Hensel, M.; et al. Age-dependent enterocyte invasion and microcolony formation by Salmonella. PLoS Pathog. 2014, 10, e1004385. [Google Scholar] [CrossRef] [PubMed]
- Coman, M.M.; Verdenelli, M.C.; Cecchini, C.; Silvi, S.; Orpianesi, C.; Boyko, N.; Cresci, A. In vitro evaluation of antimicrobial activity of Lactobacillus rhamnosus IMC 501(®), Lactobacillus paracasei IMC 502(®) and SYNBIO(®) against pathogens. J. Appl. Microbiol. 2014, 117, 518–527. [Google Scholar] [CrossRef]
- Taweechotipatr, M.; Iyer, C.; Spinler, J.K.; Versalovic, J.; Tumwasorn, S. Lactobacillus saerimneri and Lactobacillus ruminis: Novel human-derived probiotic strains with immunomodulatory activities. FEMS Microbiol. Lett. 2009, 293, 65–72. [Google Scholar] [CrossRef]
- Jung, J.-W.; Cho, S.-D.; Ahn, N.-S.; Yang, S.-R.; Park, J.-S.; Jo, E.-H.; Hwang, J.-W.; Jung, J.-Y.; Kim, S.-H.; Kang, K.-S.; et al. Ras/MAP kinase pathways are involved in Ras specific apoptosis induced by sodium butyrate. Cancer Lett. 2005, 225, 199–206. [Google Scholar] [CrossRef]
- Sakurazawa, T.; Ohkusa, T. Cytotoxicity of organic acids produced by anaerobic intestinal bacteria on cultured epithelial cells. J. Gastroenterol. 2005, 40, 600–609. [Google Scholar] [CrossRef]
- Noda, S.; Yamada, A.; Nakaoka, K.; Goseki-Sone, M. 1-alpha,25-Dihydroxyvitamin D3 up-regulates the expression of 2 types of human intestinal alkaline phosphatase alternative splicing variants in Caco-2 cells and may be an important regulator of their expression in gut homeostasis. Nutr. Res. 2017, 46, 59–67. [Google Scholar] [CrossRef]





| Genes | Fold Change * | |
|---|---|---|
| 2.0 μg/mL CFS | 4.0 μg/mL CFS | |
| sopB | −5.7 ± 0.5 *** | −7.2 ± 0.4 *** |
| invH | −3.9 ± 0.2 ** | −5.6 ± 0.5 *** |
| sipB | −5.4 ± 0.3 *** | −6.9 ± 0.4 *** |
| pipB | −3.6 ± 0.2 ** | −5.2 ± 0.5 *** |
| orf245 | −5.6 ± 0.5 *** | −7.8 ± 0.3 *** |
| sipA | −7.9 ± 0.6 *** | −9.5 ± 0.4 *** |
| ssaV | −5.7 ± 0.3 *** | −6.4 ± 0.5 *** |
| spvB | −1.4 ± 0.2 * | −1.8 ± 0.3 * |
| mgtC | −4.2 ± 0.5 ** | −7.5 ± 0.4 *** |
| sodC | −3.7 ± 0.4 ** | −5.3 ± 0.2 *** |
| tatA | −1.8 ± 0.2 * | −2.6 ± 0.2 * |
| hflK | −1.3 ± 0.2 * | −1.5 ± 0.2 * |
| ompR | −8.4 ± 0.5 *** | −13.7 ± 0.6 *** |
| mrr1 | −2.6 ± 0.4 * | −4.8 ± 0.3 ** |
| lrp | −1.3 ± 0.2 * | −1.7 ± 0.2 * |
| xthA | −8.4 ± 0.5 *** | −15.6 ± 0.4 *** |
| rpoS | −2.7 ± 0.4 * | −4.2 ± 0.3 ** |
| ssrA | −1.9 ± 0.2 * | −2.6 ± 0.2 * |
| rfbH | −9.4 ± 0.6 *** | −11.8 ± 0.5 *** |
| Micro-organism | Strain | Collection | Antibiotic Resistance | Growth Conditions |
|---|---|---|---|---|
| L. fermentum | LF 3872 1 | IIE a | MRS b 37 °C in CO2 incubator at anaerobic atmosphere (5% hydrogen, 10% carbon dioxide, 85% nitrogen) 24–48 h | |
| L. salivarius | LS 7247 2 | IIE | The same | |
| S. Enteritidis | Egg 6235 3 | IIE | TET, CHL, LVF, CIP, NAL, SMZ, TMP, AMP | BHI c 37 °C aerobically 18 h |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abramov, V.M.; Kosarev, I.V.; Machulin, A.V.; Deryusheva, E.I.; Priputnevich, T.V.; Panin, A.N.; Chikileva, I.O.; Abashina, T.N.; Manoyan, A.M.; Akhmetzyanova, A.A.; et al. Anti-Salmonella Defence and Intestinal Homeostatic Maintenance In Vitro of a Consortium Containing Limosilactobacillus fermentum 3872 and Ligilactobacillus salivarius 7247 Strains in Human, Porcine, and Chicken Enterocytes. Antibiotics 2024, 13, 30. https://doi.org/10.3390/antibiotics13010030
Abramov VM, Kosarev IV, Machulin AV, Deryusheva EI, Priputnevich TV, Panin AN, Chikileva IO, Abashina TN, Manoyan AM, Akhmetzyanova AA, et al. Anti-Salmonella Defence and Intestinal Homeostatic Maintenance In Vitro of a Consortium Containing Limosilactobacillus fermentum 3872 and Ligilactobacillus salivarius 7247 Strains in Human, Porcine, and Chicken Enterocytes. Antibiotics. 2024; 13(1):30. https://doi.org/10.3390/antibiotics13010030
Chicago/Turabian StyleAbramov, Vyacheslav M., Igor V. Kosarev, Andrey V. Machulin, Evgenia I. Deryusheva, Tatiana V. Priputnevich, Alexander N. Panin, Irina O. Chikileva, Tatiana N. Abashina, Ashot M. Manoyan, Anna A. Akhmetzyanova, and et al. 2024. "Anti-Salmonella Defence and Intestinal Homeostatic Maintenance In Vitro of a Consortium Containing Limosilactobacillus fermentum 3872 and Ligilactobacillus salivarius 7247 Strains in Human, Porcine, and Chicken Enterocytes" Antibiotics 13, no. 1: 30. https://doi.org/10.3390/antibiotics13010030
APA StyleAbramov, V. M., Kosarev, I. V., Machulin, A. V., Deryusheva, E. I., Priputnevich, T. V., Panin, A. N., Chikileva, I. O., Abashina, T. N., Manoyan, A. M., Akhmetzyanova, A. A., Blumenkrants, D. A., Ivanova, O. E., Papazyan, T. T., Nikonov, I. N., Suzina, N. E., Melnikov, V. G., Khlebnikov, V. S., Sakulin, V. K., Samoilenko, V. A., ... Karlyshev, A. V. (2024). Anti-Salmonella Defence and Intestinal Homeostatic Maintenance In Vitro of a Consortium Containing Limosilactobacillus fermentum 3872 and Ligilactobacillus salivarius 7247 Strains in Human, Porcine, and Chicken Enterocytes. Antibiotics, 13(1), 30. https://doi.org/10.3390/antibiotics13010030








