Prevalence and Phylogenetic Analysis of Lipoprotein-Gene ragB-1 of Porphyromonas gingivalis—A Pilot Study
Abstract
1. Introduction
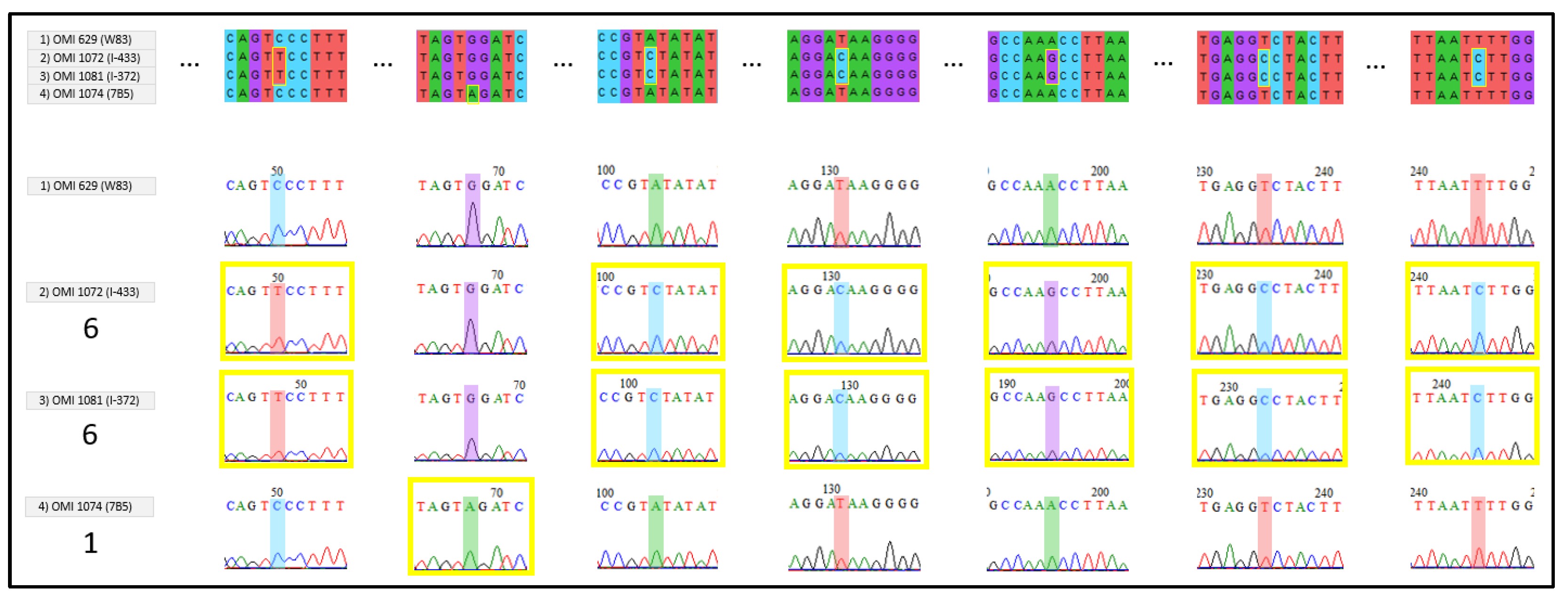
2. Results
3. Materials and Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henry, L.G.; McKenzie, R.M.E.; Robles, A.; Fletcher, H.M. Oxidative stress resistance in Porphyromonas gingivalis. Future Microbiol. 2012, 7, 497–512. [Google Scholar] [CrossRef]
- Dahlén, G.; Gmür, R.; Yoshino, T. Phenotypes, serotypes and antibiotic susceptibility of Swedish Porphyromonas gingivalis isolates from periodontitis and periodontal abscesses. Oral Microbiol. Immunol. 2007, 22, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Amano, A.; Nakagawa, I.; Kataoka, K.; Morisaki, I.; Hamada, S. Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients. J. Clin. Microbiol. 1999, 37, 1426–1430. [Google Scholar] [CrossRef]
- Griffen, A.L.; Lyons, S.R.; Becker, M.R.; Moeschberger, M.L.; Leys, E.J. Porphyromonas gingivalis strain variability and periodontitis. J. Clin. Microbiol. 1999, 37, 4028–4033. [Google Scholar] [CrossRef]
- Missailidis, C.G.; Umeda, J.E.; Ota-Tsuzuki, C.; Anzai, D.; Mayer, M.P.A. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol. Immunol. 2004, 19, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, I.; Amano, A.; Ohara-Nemoto, Y.; Endoh, N.; Morisaki, I.; Kimura, S.; Kawabata, S.; Hamada, S. Identification of a new variant of fimA gene of Porphyromonas gingivalis and its distribution in adults and disabled populations with periodontitis. J. Periodontal Res. 2002, 37, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Hall, L.M.C.; Fawell, S.C.; Shi, X.; Faray-Kele, M.-C.; Aduse-Opoku, J.; Whiley, R.A.; Curtis, M.A. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis. Infect. Immun. 2005, 73, 4253–4262. [Google Scholar] [CrossRef]
- Frandsen, E.V.; Poulsen, K.; Curtis, M.A.; Kilian, M. Evidence of recombination in Porphyromonas gingivalis and random distribution of putative virulence markers. Infect. Immun. 2001, 69, 4479–4485. [Google Scholar] [CrossRef]
- Sawada, K.; Kokeguchi, S.; Hongyo, H.; Sawada, S.; Miyamoto, M.; Maeda, H.; Nishimura, F.; Takashiba, S.; Murayama, Y. Identification by subtractive hybridization of a novel insertion sequence specific for virulent strains of Porphyromonas gingivalis. Infect. Immun. 1999, 67, 5621–5625. [Google Scholar] [CrossRef] [PubMed]
- Potempa, J.; Madej, M.; Scott, D.A. The RagA and RagB proteins of Porphyromonas gingivalis. Mol. Oral Microbiol. 2021, 36, 225–232. [Google Scholar] [CrossRef]
- Kuhn, C. Genetische Polymorphismen in Porphyromonas gingivalis und deren Auswirkung auf dessen Virulenz. Ph.D. Dissertation, Heinrich-Heine-Universität, Düsseldorf, Germany, 2018. [Google Scholar]
- Nagano, K.; Murakami, Y.; Nishikawa, K.; Sakakibara, J.; Shimozato, K.; Yoshimura, F. Characterization of RagA and RagB in Porphyromonas gingivalis: Study using gene-deletion mutants. J. Med. Microbiol. 2007, 56, 1536–1548. [Google Scholar] [CrossRef]
- Bonass, W.A.; Marsh, P.D.; Percival, R.S.; Aduse-Opoku, J.; Hanley, S.A.; Devine, D.A.; Curtis, M.A. Identification of ragAB as a temperature-regulated operon of Porphyromonas gingivalis W50 using differential display of randomly primed RNA. Infect. Immun. 2000, 68, 4012–4017. [Google Scholar] [CrossRef]
- Curtis, M.A.; Hanley, S.A.; Aduse-Opoku, J. The rag locus of Porphyromonas gingivalis: A novel pathogenicity island. J. Periodontal Res. 1999, 34, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Murakami, Y.; Noguchi, T.; Yoshimura, F. Effects of various growth conditions in a chemostat on expression of virulence factors in Porphyromonas gingivalis. Appl. Environ. Microbiol. 2006, 72, 3458–3467. [Google Scholar] [CrossRef] [PubMed]
- Hanley, S.A.; Aduse-Opoku, J.; Curtis, M.A. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect. Immun. 1999, 67, 1157–1171. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Hanley, S.A.; Faray-Kele, M.-C.; Fawell, S.C.; Aduse-Opoku, J.; Whiley, R.A.; Curtis, M.A.; Hall, L.M.C. The rag locus of Porphyromonas gingivalis contributes to virulence in a murine model of soft tissue destruction. Infect. Immun. 2007, 75, 2071–2074. [Google Scholar] [CrossRef]
- Madej, M.; White, J.B.R.; Nowakowska, Z.; Rawson, S.; Scavenius, C.; Enghild, J.J.; Bereta, G.P.; Pothula, K.; Kleinekathoefer, U.; Baslé, A.; et al. Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis. Nat. Microbiol. 2020, 5, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Glenwright, A.J.; Pothula, K.R.; Bhamidimarri, S.P.; Chorev, D.S.; Baslé, A.; Firbank, S.J.; Zheng, H.; Robinson, C.V.; Winterhalter, M.; Kleinekathöfer, U.; et al. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 2017, 541, 407–411. [Google Scholar] [CrossRef]
- Goulas, T.; Garcia-Ferrer, I.; Hutcherson, J.A.; Potempa, B.A.; Potempa, J.; Scott, D.A.; Gomis-Rüth, F.X. Structure of RagB, a major immunodominant outer-membrane surface receptor antigen of Porphyromonas gingivalis. Mol. Oral Microbiol. 2016, 31, 472–485. [Google Scholar] [CrossRef]
- Curtis, M.A.; Slaney, J.M.; Carman, R.J.; Johnson, N.W. Identification of the major surface protein antigens of Porphyromonas gingivalis using IgG antibody reactivity of periodontal case-control serum. Oral Microbiol. Immunol. 1991, 6, 321–326. [Google Scholar] [CrossRef]
- Imai, M.; Murakami, Y.; Nagano, K.; Nakamura, H.; Yoshimura, F. Major outer membrane proteins from Porphyromonas gingivalis: Strain variation, distribution, and clinical significance in periradicular lesions. Eur. J. Oral Sci. 2005, 113, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Zeller, I.; Hutcherson, J.A.; Lamont, R.J.; Demuth, D.R.; Gumus, P.; Nizam, N.; Buduneli, N.; Scott, D.A. Altered antigenic profiling and infectivity of Porphyromonas gingivalis in smokers and non-smokers with periodontitis. J. Periodontol. 2014, 85, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Hutcherson, J.A.; Bagaitkar, J.; Nagano, K.; Yoshimura, F.; Wang, H.; Scott, D.A. Porphyromonas gingivalis RagB is a proinflammatory signal transducer and activator of transcription 4 agonist. Mol. Oral Microbiol. 2015, 30, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Y.; Wang, L.; Guo, Y.; Xiao, S. Prevalence of Porphyromonas gingivalis four rag locus genotypes in patients of orthodontic gingivitis and periodontitis. PLoS ONE 2013, 8, e61028. [Google Scholar] [CrossRef]
- Nelson, K.E.; Fleischmann, R.D.; DeBoy, R.T.; Paulsen, I.T.; Fouts, D.E.; Eisen, J.A.; Daugherty, S.C.; Dodson, R.J.; Durkin, A.S.; Gwinn, M.; et al. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 2003, 185, 5591–5601. [Google Scholar] [CrossRef]
- Xu, J.; Bjursell, M.K.; Himrod, J.; Deng, S.; Carmichael, L.K.; Chiang, H.C.; Hooper, L.V.; Gordon, J.I. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 2003, 299, 2074–2076. [Google Scholar] [CrossRef]
- Chen, T.; Hosogi, Y.; Nishikawa, K.; Abbey, K.; Fleischmann, R.D.; Walling, J.; Duncan, M.J. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 2004, 186, 5473–5479. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Pratten, J.; Wilson, M.; Mullany, P. Transfer of a conjugative transposon, Tn5397 in a model oral biofilm. FEMS Microbiol. Lett. 1999, 177, 63–66. [Google Scholar] [CrossRef]
- Waters, V.L. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 1999, 4, D433–D456. [Google Scholar] [CrossRef]
- Li, Y.H.; Lau, P.C.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 2001, 183, 897–908. [Google Scholar] [CrossRef]
- Li, Y.-H.; Tang, N.; Aspiras, M.B.; Lau, P.C.Y.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 2002, 184, 2699–2708. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Y.; Chi, B.; Kuramitsu, H.K. Genetic exchange between Treponema denticola and Streptococcus gordonii in biofilms. Oral Microbiol. Immunol. 2002, 17, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Stork, M.; Bos, M.P.; Jongerius, I.; de Kok, N.; Schilders, I.; Weynants, V.E.; Poolman, J.T.; Tommassen, J. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 2010, 6, e1000969. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Zheng, D.; She, P.; Ni, P.; Zhu, H.; Xu, H.; Su, Z. Porphyromonas gingivalis B cell Antigen Epitope Vaccine, pIRES-ragB’-mGITRL, Promoted RagB-Specific Antibody Production and Tfh Cells Expansion. Scand. J. Immunol. 2015, 81, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Sun, Q.; Su, Z.; Kong, F.; Shi, X.; Tong, J.; Shen, P.; Peng, T.; Wang, S.; Xu, H. Enhancing specific-antibody production to the ragB vaccine with GITRL that expand Tfh, IFN-γ(+) T cells and attenuates Porphyromonas gingivalis infection in mice. PLoS ONE 2013, 8, e59604. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 1998. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 19 March 2023).
- Fournier, D.; Mouton, C.; Lapierre, P.; Kato, T.; Okuda, K.; Ménard, C. Porphyromonas gulae sp. nov., an anaerobic, gram-negative coccobacillus from the gingival sulcus of various animal hosts. Int. J. Syst. Evol. Microbiol. 2001, 51, 1179–1189. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull Biosci 2011, 2, 60–61. [Google Scholar]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT; Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Dolgilevich, S.; Rafferty, B.; Luchinskaya, D.; Kozarov, E. Genomic comparison of invasive and rare non-invasive strains reveals Porphyromonas gingivalis genetic polymorphisms. J. Oral Microbiol. 2011, 3, 5764. [Google Scholar] [CrossRef]
- Bunte, K.; Kuhn, C.; Walther, C.; Peters, U.; Aarabi, G.; Smeets, R.; Beikler, T. Clinical significance of ragA, ragB, and PG0982 genes in Porphyromonas gingivalis isolates from periodontitis patients. Eur. J. Oral Sci. 2021, 129, e12776. [Google Scholar] [CrossRef]
- Su, Z.; Kong, F.; Wang, S.; Chen, J.; Yin, R.; Zhou, C.; Zhang, Y.; He, Z.; Shi, Y.; Xue, Y.; et al. The rag locus of Porphyromonas gingivalis might arise from Bacteroides via horizontal gene transfer. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 429–437. [Google Scholar] [CrossRef][Green Version]
- Dashper, S.G.; Mitchell, H.L.; Seers, C.A.; Gladman, S.L.; Seemann, T.; Bulach, D.M.; Chandry, P.S.; Cross, K.J.; Cleal, S.M.; Reynolds, E.C. Porphyromonas gingivalis Uses Specific Domain Rearrangements and Allelic Exchange to Generate Diversity in Surface Virulence Factors. Front. Microbiol. 2017, 8, 48. [Google Scholar] [CrossRef]
- Laine, M.L.; van Winkelhoff, A.J. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 1998, 13, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.L.; Salyers, A.A. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 1989, 171, 3199–3204. [Google Scholar] [CrossRef] [PubMed]
- Koropatkin, N.M.; Martens, E.C.; Gordon, J.I.; Smith, T.J. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 2008, 16, 1105–1115. [Google Scholar] [CrossRef] [PubMed]
- Phansopa, C.; Roy, S.; Rafferty, J.B.; Douglas, C.W.I.; Pandhal, J.; Wright, P.C.; Kelly, D.J.; Stafford, G.P. Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem. J. 2014, 458, 499–511. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brunner, J.; Wittink, F.R.A.; Jonker, M.J.; de Jong, M.; Breit, T.M.; Laine, M.L.; Soet, J.J.d.; Crielaard, W. The core genome of the anaerobic oral pathogenic bacterium Porphyromonas gingivalis. BMC Microbiol. 2010, 10, 252. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara-Takahashi, K.; Watanabe, T.; Shimogishi, M.; Shibasaki, M.; Umeda, M.; Izumi, Y.; Nakagawa, I. Phylogenetic diversity in fim and mfa gene clusters between Porphyromonas gingivalis and Porphyromonas gulae, as a potential cause of host specificity. J. Oral Microbiol. 2020, 12, 1775333. [Google Scholar] [CrossRef]





| Strain (OMI) | Species | Original Code | Origin Species | Isolation Year | Country |
|---|---|---|---|---|---|
| 629 | P. gingivalis | W83 | Human | 1991 | Bonn, Germany |
| 1049 | P. gingivalis | AJW5 (VAG 5) | Human | 1991 | Buffalo, NY, USA |
| 1051 | P. gingivalis | 22KN6-12 | Human | Tokushima, Japan | |
| 1060 | P. gingivalis/gulae | OMG 1426 | Monkey | 1989 | Florida, USA |
| 1053 | P. gingivalis | RB22D-1 | Human | Quebec, Canada | |
| 1068 | P. gingivalis | 213Pg2 | Human | 1994 | Indonesia |
| 1071 | P. gingivalis | ATCC49417, RB22D | Human | 1993 | Quebec, Canada |
| 1072 | P. gingivalis/gulae | I-433 | Monkey | 1989 | Florida, USA |
| 1074 | P. gingivalis | 7B5 | Human | Quebec, Canada | |
| 1078 | P. gingivalis | 84Pg1-a | Human | 1994 | Indonesia |
| 1079 | P. gingivalis | 83Pg1-a | Human | 1994 | Indonesia |
| 1081 | P. gingivalis/gulae | I-372 | Monkey | 1989 | Florida, USA |
| 1084 | P. gingivalis | 81Pg1-a | Human | 1994 | Indonesia |
| 1087 | P. gingivalis | 122Pg1-a | Human | 1994 | Indonesia |
| 1101 | P. gingivalis | 83Pg1-b | Human | 2002 | Indonesia |
| 1108 | P. gingivalis | 81Pg1-b | Human | 2002 | Indonesia |
| 1112 | P. gingivalis | A 7436 | Human | Georgia, USA | |
| 1117 | P. gingivalis | W12 | Human | Alabama, USA | |
| 1120 | P. gingivalis | 13JC | Human | Rennes, France | |
| 1122 | P. gingivalis | 17-5 | Human | Minneapolis, MN, USA | |
| 1125 | P. gingivalis | 122Pg1-b | Human | 2002 | Indonesia |
| 1127 | P. gingivalis | 84Pg1-b | Human | 2002 | Indonesia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Böcher, S.; Meyer, H.L.; Dafni, E.; Conrads, G. Prevalence and Phylogenetic Analysis of Lipoprotein-Gene ragB-1 of Porphyromonas gingivalis—A Pilot Study. Antibiotics 2023, 12, 1458. https://doi.org/10.3390/antibiotics12091458
Böcher S, Meyer HL, Dafni E, Conrads G. Prevalence and Phylogenetic Analysis of Lipoprotein-Gene ragB-1 of Porphyromonas gingivalis—A Pilot Study. Antibiotics. 2023; 12(9):1458. https://doi.org/10.3390/antibiotics12091458
Chicago/Turabian StyleBöcher, Sarah, Hendrik L. Meyer, Evdokia Dafni, and Georg Conrads. 2023. "Prevalence and Phylogenetic Analysis of Lipoprotein-Gene ragB-1 of Porphyromonas gingivalis—A Pilot Study" Antibiotics 12, no. 9: 1458. https://doi.org/10.3390/antibiotics12091458
APA StyleBöcher, S., Meyer, H. L., Dafni, E., & Conrads, G. (2023). Prevalence and Phylogenetic Analysis of Lipoprotein-Gene ragB-1 of Porphyromonas gingivalis—A Pilot Study. Antibiotics, 12(9), 1458. https://doi.org/10.3390/antibiotics12091458







