Analyses of Extended-Spectrum-β-Lactamase, Metallo-β-Lactamase, and AmpC-β-Lactamase Producing Enterobacteriaceae from the Dairy Value Chain in India
Abstract
1. Introduction
2. Results
2.1. Isolation of Bacteria
2.2. Antibiotic Susceptibility Testing (AST)
2.3. Polymerase Chain Reaction (PCR) Detection of Resistance Genes
2.4. Comparison between Results
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Sample Collection
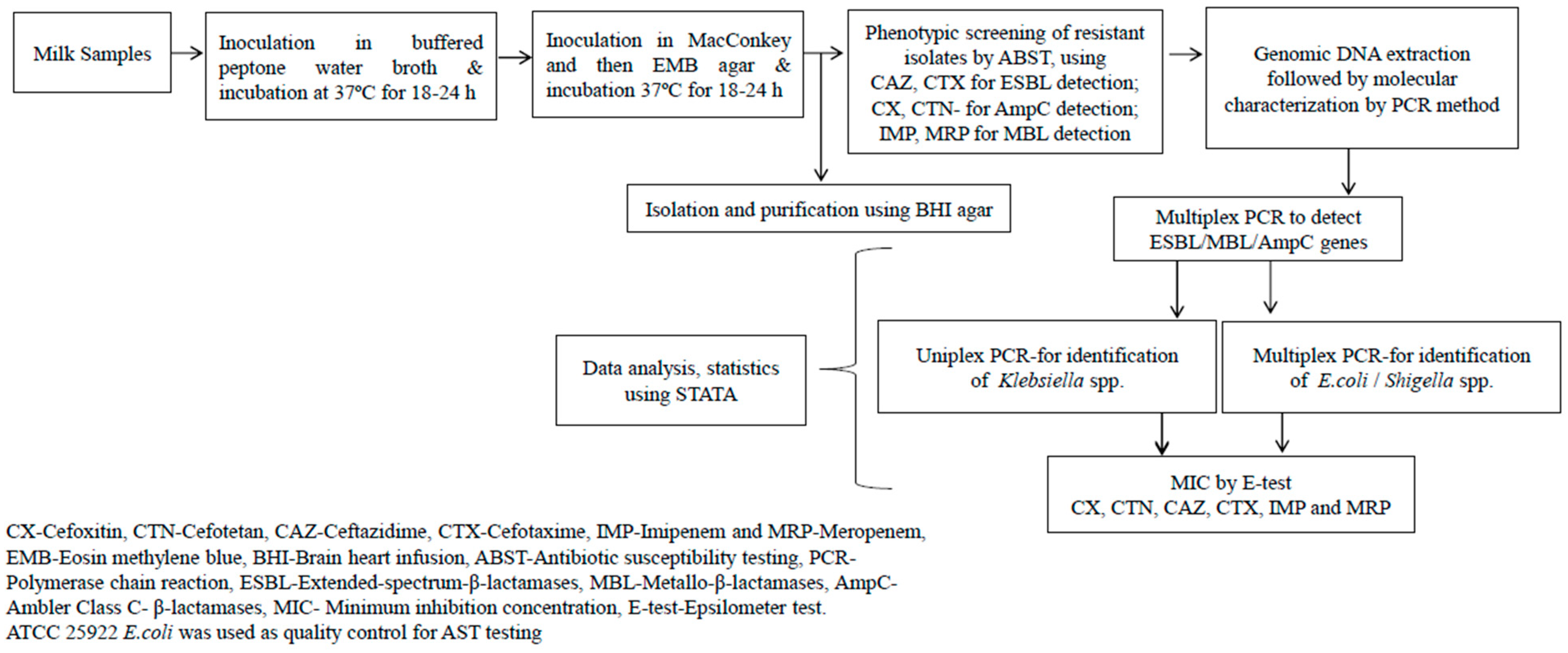
4.3. Isolation of Bacteria
4.4. Antibiotic Susceptibility Testing (AST)
4.5. Molecular Detection of Resistance Genes by PCR
4.6. Epsilometer Test
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Ministry of Fisheries, Animal Husbandry & Dairying. 2023. Available online: https://pib.gov.in/PressReleaseIframePage.aspx?PRID=1897084 (accessed on 27 April 2023).
- FAO. Dairy Production and Products. Food and Agriculture Organization of the United Nations. 2022. Available online: https://www.fao.org/dairy-production-products/products/en/ (accessed on 30 September 2022).
- Patil, B.R. Dynamics of Livestock Development in Gujarat, India: Experiences of an Indian NGO; Wageningen University and Research: Wageningen, The Netherlands, 2006. [Google Scholar]
- Staal, S.J.; Pratt, A.N.; Jabbar, M.A. A Comparison of Dairy Policies and Development in South Asia and East Africa; ILRI: Nairobi, Kenya, 2010. [Google Scholar]
- Rao, C.K.; Bachhman, F.; Sharma, V.; Venkataramaiah, P.; Panda, J.; Rathinam, R. Smallholder Dairy Value Chain Development in India and Select States (Assam and Bihar): Situation Analysis and Trends. ILRI Proj. Rep. 2014, 10–32. [Google Scholar]
- Deka, R.P.; Bayan, B.; Baltenweck, I.; Grace, D. Mapping of Informal Dairy Value Chain Actors in Selected Districts of Assam; The International Livestock Research Institute: Nairobi, Kenya, 2019; pp. 1–2. [Google Scholar] [CrossRef]
- Sharma, S.; Sharma, D.K. Mapping of Milk Processing Units in Organized Sector: A Case Study for Haryana. Int. J. Environ. Agric. Res. 2020, 6, 1–6. [Google Scholar]
- Karthikeyan, R.; Gunasekaran, P.; Rajendhran, J. Molecular Serotyping and Pathogenic Potential of Listeria Monocytogenes Isolated from Milk and Milk Products in Tamil Nadu, India. Foodborne Pathog. Dis. 2015, 12, 522–528. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ); Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernández Escámez, P.S.; Girones, R.; Koutsoumanis, K.; Lindqvist, R.; et al. Risk for the Development of Antimicrobial Resistance (AMR) Due to Feeding of Calves with Milk Containing Residues of Antibiotics. EFSA J. 2017, 15, 4665. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Kirchhelle, C. Pharming Animals: A Global History of Antibiotics in Food Production (1935–2017). Palgrave Commun. 2018, 4, 1–13. [Google Scholar] [CrossRef]
- Paramasivam, R.; Gopal, D.R.; Dhandapani, R.; Subbarayalu, R.; Elangovan, M.P.; Prabhu, B.; Veerappan, V.; Nandheeswaran, A.; Paramasivam, S.; Muthupandian, S. Is AMR in Dairy Products a Threat to Human Health? An Updated Review on the Origin, Prevention, Treatment, and Economic Impacts of Subclinical Mastitis. Infect. Drug Resist. 2023, 16, 155–178. [Google Scholar] [CrossRef]
- Sharma, C.; Rokana, N.; Chandra, M.; Singh, B.P.; Gulhane, R.D.; Gill, J.P.S.; Ray, P.; Puniya, A.K.; Panwar, H. Antimicrobial Resistance: Its Surveillance, Impact, and Alternative Management Strategies in Dairy Animals. Front. Vet. Sci. 2018, 4, 1–27. [Google Scholar] [CrossRef]
- Wolfenson, K.D.M. Coping with the Food and Agriculture Challenge: Smallholders’ Agenda; Food and Agriculture Organisation of the United Nations: Rome, Italy, 2013. [Google Scholar]
- WHO. Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014; Available online: https://apps.who.int/iris/handle/10665/112642 (accessed on 10 December 2022).
- Ruegg, P.L. Making Antibiotic Treatment Decisions for Clinical Mastitis. Vet. Clin. Food Anim. Pract. 2018, 34, 413–425. [Google Scholar] [CrossRef]
- Bergšpica, I.; Kaprou, G.; Alexa, E.A.; Prieto, M.; Alvarez-Ordóñez, A. Extended Spectrum β-Lactamase (ESBL) Producing Escherichia coli in Pigs and Pork Meat in the European Union. Antibiotics 2020, 9, 678. [Google Scholar] [CrossRef] [PubMed]
- Rawat, D.; Nair, D. Extended-Spectrum β-Lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263. [Google Scholar] [CrossRef]
- Tamang, M.D.; Nam, H.-M.; Jang, G.-C.; Kim, S.-R.; Chae, M.H.; Jung, S.-C.; Byun, J.-W.; Park, Y.H.; Lim, S.-K. Molecular Characterization of Extended-Spectrum-β-Lactamase- Producing and Plasmid-Mediated AmpC β-Lactamase-Producing Escherichia coli Isolated from Stray Dogs in South Korea. Antimicrob. Agents Chemother. 2012, 56, 2705–2712. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G. Carbapenem Resistance: Overview of the Problem and Future Perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kaye, K.S.; Jason, M.P. Infections Caused by Resistant Gram-negative Bacteria: Epidemiology and Management. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2015, 35, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Chiotos, K.; Han, J.H.; Tamma, P.D. Carbapenem-Resistant Enterobacteriaceae Infections in Children. Curr. Infect. Dis. Rep. 2016, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liebana, E.; Carattoli, A.; Coque, T.M.; Hasman, H.; Magiorakos, A.-P.; Mevius, D.; Peixe, L.; Poirel, L.; Schuepbach-Regula, G.; Torneke, K.; et al. Public Health Risks of Enterobacterial Isolates Producing Extended-Spectrum β-Lactamases or AmpC β-Lactamases in Food and Food-Producing Animals: An EU Perspective of Epidemiology, Analytical Methods, Risk Factors, and Control Options. Clin. Infect. Dis. 2013, 56, 1030–1037. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Govinden, U.; Bester, L.A.; Essack, S.Y. Colistin and Tigecycline Resistance in Carbapenemase-producing Gram-negative Bacteria: Emerging Resistance Mechanisms and Detection Methods. J. Appl. Microbiol. 2016, 121, 601–617. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Saleh, S.E.; Alshahrani, M.Y.; Aboshanab, K.M.; Aboulwafa, M.M.; Hassouna, N.A. OXA-48 Carbapenemase-Encoding Transferable Plasmids of Klebsiella pneumoniae Recovered from Egyptian Patients Suffering from Complicated Urinary Tract Infections. Biology 2021, 10, 889. [Google Scholar] [CrossRef]
- Lucey, J.A. Raw Milk Consumption: Risks and Benefits. Nutr. Today 2015, 50, 189–193. Available online: https://journals.lww.com/nutritiontodayonline/Fulltext/2015/07000/Raw_Milk_Consumption__Risks_and_Benefits.10.aspx (accessed on 11 September 2023). [CrossRef]
- Oliver, S.P.; Murinda, S.E.; Jayarao, B.M. Impact of Antibiotic Use in Adult Dairy Cows on Antimicrobial Resistance of Veterinary and Human Pathogens: A Comprehensive Review. Foodborne Pathog. Dis. 2011, 8, 337–355. [Google Scholar] [CrossRef] [PubMed]
- Sobur, M.A.; Sabuj, A.A.M.; Sarker, R.; Rahman, A.M.M.T.; Kabir, S.M.L.; Rahman, M.T. Antibiotic-resistant Escherichia coli and Salmonella spp. associated with dairy cattle and farm environment having public health significance. Vet. World 2019, 12, 984. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, M.M.; Elkenany, R.M.; Zakaria, A.I.; Badawy, B.M. Epidemiological study on Listeria monocytogenes in Egyptian dairy cattle farms’ insights into genetic diversity of multi-antibiotic-resistant strains by ERIC-PCR. Environ. Sci. Pollut. Res. 2022, 29, 54359–54377. [Google Scholar] [CrossRef]
- Serwecińska, L. Antimicrobials and antibiotic-resistant bacteria: A risk to the environment and to public health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Hassani, S.; Moosavy, M.-H.; Gharajalar, S.N.; Khatibi, S.A.; Hajibemani, A.; Barabadi, Z. High prevalence of antibiotic resistance in pathogenic foodborne bacteria isolated from bovine milk. Sci. Rep. 2022, 12, 3878. [Google Scholar] [CrossRef] [PubMed]
- Montso, K.P.; Dlamini, S.B.; Kumar, A.; Ateba, C.N. Antimicrobial resistance factors of extended-spectrum beta-lactamases producing Escherichia coli and Klebsiella pneumoniae isolated from Cattle Farms and Raw Beef in North-West Province, South Africa. Biomed Res. Int. 2019. [Google Scholar] [CrossRef] [PubMed]
- Resende, J.A.; Silva, V.L.; de Oliveira, T.L.R.; de Oliveira Fortunato, S.; da Costa Carneiro, J.; Otenio, M.H.; Diniz, C.G. Prevalence and persistence of potentially pathogenic and antibiotic resistant bacteria during anaerobic digestion treatment of cattle manure. Bioresour. Technol. 2014, 153, 284–291. [Google Scholar] [CrossRef]
- Bhatt, V.; Ahir, V.; Koringa, P.; Jakhesara, S.; Rank, D.; Nauriyal, D.; Kunjadia, A.; Joshi, C. Milk Microbiome Signatures of Subclinical Mastitis-affected Cattle Analysed by Shotgun Sequencing. J. Appl. Microbiol. 2012, 112, 639–650. [Google Scholar] [CrossRef]
- Ewers, C.; Klotz, P.; Scheufen, S.; Leidner, U.; Göttig, S.; Semmler, T. Genome Sequence of OXA-23 Producing Acinetobacter baumannii IHIT7853, a Carbapenem-Resistant Strain from a Cat Belonging to International Clone IC1. Gut Pathog. 2016, 8, 1–7. [Google Scholar] [CrossRef][Green Version]
- de Souza, Z.N.; de Moura, D.F.; Campos, L.A.d.A.; Córdula, C.R.; Cavalcanti, I.M.F. Antibiotic Resistance Profiles on Pathogenic Bacteria in the Brazilian Environments. Arch. Microbiol. 2023, 205, 185. [Google Scholar] [CrossRef]
- Niewiadomska, A.M.; Jayabalasingham, B.; Seidman, J.C.; Willem, L.; Grenfell, B.; Spiro, D.; Viboud, C. Population-level mathematical modeling of antimicrobial resistance: A systematic review. BMC Med. 2019, 17, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Sinha, B.; Kumari, R.; Vineeth, M.R.; Shrivastava, K.; Verma, A.; Gupta, I.D. Udder and teat morphometry in relation to clinical mastitis in dairy cows. Trop. Anim. Health Prod. 2022, 54, 99. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Manimaran, A.; Kumaresan, A.; Jeyakumar, S.; Sreela, L.; Mooventhan, P.; Sivaram, M. Mastitis effects on reproductive performance in dairy cattle: A review. Trop. Anim. Health Prod. 2017, 49, 663–673. [Google Scholar] [CrossRef]
- Bangar, Y.C.; Singh, B.; Dohare, A.K.; Verma, M.R. A systematic review and meta-analysis of prevalence of subclinical mastitis in dairy cows in India. Trop. Anim. Health Prod. 2015, 47, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Kunjadia, P.; Koringa, P.; Joshi, C.; Kunjadiya, A. Microbiological profiles in clinical and subclinical cases of mastitis in milking Jafarabadi buffalo. Res. Vet. Sci. 2019, 125, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Awandkar, S.P.; Kulkarni, M.B.; Khode, N.V. Bacteria from bovine clinical mastitis showed multiple drug resistance. Vet. Res. Commun. 2022, 46, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Deb, R.; Kumar, A.; Chakraborty, S.; Verma, A.K.; Tiwari, R.; Dhama, K.; Singh, U.; Kumar, S. Trends in diagnosis and control of bovine mastitis: A review. Pak. J. Biol. Sci. 2013, 16, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, P.; Goudar, A.L.; Suresh, K.P.; Roy, P. Global and countrywide prevalence of subclinical and clinical mastitis in dairy cattle and buffaloes by systematic review and meta-analysis. Res. Vet. Sci. 2021, 136, 561–586. [Google Scholar] [CrossRef]
- CLSI M100-S25; Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015; Volume 32.
- Tekiner, H.; Özpınar, H. Occurrence and Characteristics of Extended Spectrum Beta-Lactamases-Producing Enterobacteriaceae from Foods of Animal Origin. Braz. J. Microbiol. 2016, 47, 444–451. [Google Scholar] [CrossRef]
- Diab, M.; Hamze, M.; Bonnet, R.; Saras, E.; Madec, J.Y.; Haenni, M. OXA-48 and CTX-M-15 Extended-Spectrum Beta-Lactamases in Raw Milk in Lebanon: Epidemic Spread of Dominant Klebsiella pneumoniae Clones. J. Med. Microbiol. 2017, 66, 1688–1691. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and Abundant Antibiotic Resistance Genes in Chinese Swine Farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Kamaruzzaman, E.A.; Aziz, S.A.; Bitrus, A.A.; Zakaria, Z.; Hassan, L. Occurrence and Characteristics of Extended-Spectrum β-Lactamase-Producing Escherichia Coli from Dairy Cattle, Milk, and Farm Environments in Peninsular Malaysia. Pathogens 2020, 9, 1007. [Google Scholar] [CrossRef] [PubMed]
- Dey, T.K.; Shome, B.R.; Bandyopadhyay, S.; Goyal, N.K.; Lundkvist, Å.; Deka, R.P.; Shome, R.; Venugopal, N.; Grace, D.; Sharma, G.; et al. Molecular Characterization of Methicillin-Resistant Staphylococci from the Dairy Value Chain in Two Indian States. Pathogens 2023, 12, 344. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N.; Abboud, M.; Durand, J.P.; Brouqui, P.; De Lamballerie, X. Dual Infection by Dengue Virus and Shigellas onnei in Patient Returning from India. Emerg. Infect. Dis. 2003, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Shimamoto, T. Molecular Characterization of Multidrug-Resistant Shigella spp. of Food Origin. Int. J. Food Microbiol. 2015, 194, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Pakbin, B.; Amani, Z.; Allahyari, S.; Mousavi, S.; Mahmoudi, R.; Brück, W.M.; Peymani, A. Genetic Diversity and Antibiotic Resistance of Shigella spp. Isolates from Food Products. Food Sci. Nutr. 2021, 9, 6362–6371. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Pandey, R. Viral Infectious Diseases Severity: Co-Presence of Transcriptionally Active Microbes (TAMs) Can Play an Integral Role for Disease Severity. Front. Immunol. 2022, 13, 1056036. [Google Scholar] [CrossRef] [PubMed]
- Baylis, C.; Uyttendaele, M.; Joosten, H.; Davies, A. The Enterobacteriaceae and Their Significance to the Food Industry. In The Enterobacteriaceae and Their Significance to the Food Industry; ILSI Europe: Brussels, Belgium, 2011. [Google Scholar]
- Coutinho, R.L.; Visconde, M.F.; Descio, F.J.; Nicoletti, A.G.; Pinto, F.C.; da Silva, A.C.R.; Rodrigues-Costa, F.; Gales, A.C.; Furtado, G.H. Community-Acquired Invasive Liver Abscess Syndrome Caused by a K1 Serotype Klebsiella pneumoniae Isolate in Brazil: A Case Report of Hypervirulent ST23. Memórias Inst. Oswaldo Cruz 2014, 109, 970–971. [Google Scholar] [CrossRef]
- Koovapra, S.; Bandyopadhyay, S.; Das, G.; Bhattacharyya, D.; Banerjee, J.; Mahanti, A.; Samanta, I.; Nanda, P.; Kumar, A.; Mukherjee, R.; et al. Molecular Signature of Extended Spectrum β-Lactamase Producing Klebsiella pneumoniae Isolated from Bovine Milk in Eastern and North-Eastern India. Infect. Genet. Evol. 2016, 44, 395–402. [Google Scholar] [CrossRef]
- Odenthal, S.; Akineden, Ö.; Usleber, E. Extended-Spectrum β-Lactamase Producing Enterobacteriaceae in Bulk Tank Milk from German Dairy Farms. Int. J. Food Microbiol. 2016, 238, 72–78. [Google Scholar] [CrossRef]
- Madec, J.-Y.; Marissa, H.; Patrice, N.; Laurent, P. Extended-Spectrum β-Lactamase/AmpC-and Carbapenemase-Producing Enterobacteriaceae in Animals: A Threat for Humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Desai, S.; Bagchi, A.; Singh, P.; Joshi, M.; Joshi, C.; Patankar, J.; Maheshwari, G.; Rajni, E.; Shah, M.; et al. Diversity and Distribution of β-Lactamase Genes Circulating in Indian Isolates of Multidrug-Resistant Klebsiella pneumoniae. Antibiotics 2023, 12, 449. [Google Scholar] [CrossRef] [PubMed]
- Thomson, K.S. Extended-Spectrum-β-Lactamase, AmpC, and Carbapenemase Issues. J. Clin. Microbiol. 2010, 48, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Ghatak, S.; Singha, A.; Sen, A.; Guha, C.; Ahuja, A.; Bhattacharjee, U.; Das, S.; Pradhan, N.R.; Puro, K.; Jana, C.; et al. Detection of New Delhi Metallo-Beta-Lactamase and Extended-Spectrum Beta-Lactamase Genes in Escherichia coli Isolated from Mastitic Milk Samples. Transbound. Emerg. Dis. 2013, 60, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Paghdar, D.; Nayak, J.; Mathakiya, R.A.; Parmar, B.C.; Gida, H.K.; Bhavsar, P.P. Isolation and Molecular Characterization of Extended Spectrum Beta Lactamase Producing Escherichia coli from Milk. J. Anim. Res. 2020, 10, 143–148. [Google Scholar] [CrossRef]
- Agrawal, S.; Singh, A.P.; Singh, R.; Saikia, R.; Choudhury, S.; Shukla, A.; Prabhu, S.N.; Agrawal, J. Molecular Characterization of Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolated from Postpartum Uterine Infection in Dairy Cattle in India. Vet. World 2021, 14, 200–209. [Google Scholar] [CrossRef]
- Deb, R.; Chaudhary, P.; De, S. CRISPR/Cas9 Cassette Targeting Escherichia coli BlaCTX-M Specific Gene of Mastitis Cow Milk Origin Can Alter the Antibiotic Resistant Phenotype for Cefotaxime. Anim. Biotechnol. 2022, 1–6. [Google Scholar] [CrossRef]
- Joseph, J.; Sudha, K. Occurrence of Multiple Drug-Resistant Shiga Toxigenic Escherichia coli in Raw Milk Samples Collected from Retail Outlets in South India. J. Food Sci. Technol. 2022, 59, 2150–2159. [Google Scholar] [CrossRef]
- Maurya, N.; Jangra, M.; Tambat, R.; Nandanwar, H. Alliance of efflux pumps with β-lactamases in multidrug-resistant Klebsiella pneumoniae isolates. Microb. Drug Resist. 2019, 25, 1155–1163. [Google Scholar] [CrossRef]
- Diagbouga, S.; Salah, F.D.; Sadji, A.Y.; Dabire, A.M.; Nadembega, C.; Kere, A.B.; Soubeiga, S.T.; Ouattar, A.K.; Zohoncon, T.; Belemgnegre, M.; et al. Detection of high prevalence of TEM. SHV/CTX-M Genes in ESBL Producing and Multidrug Resistant Klebsiella pneumoniae and Klebsiella oxytoca. J. Clin. Diagn. Res. 2016, 4, 1–7. [Google Scholar]
- Hassoubah, S.A. Genetic analysis of multidrug-resistant and AmpC-producing Citrobacter freundii. Eur. Rev. Med. Pharmacol. Sci. 2023, 27. [Google Scholar]
- Dandachi, I.; Chabou, S.; Daoud, Z.; Rolain, J.M. Prevalence and emergence of extended-spectrum cephalosporin-, carbapenem- and colistin-resistant gram negative bacteria of animal origin in the Mediterranean basin. Front. Microbiol. 2018, 9, 1–26. [Google Scholar] [CrossRef]
- Karkaba, A. Occurrence and distribution of extended spectrum β-lactamase and AmpC-β-lactamase-producing Enterobacteriaceae and methicillin resistant Staphylococcus aureus in companion animals in New Zealand: A thesis presented in partial fulfilment of the requirement. Ph.D. Thesis, Massey University, North Island, New Zealand, 2017. [Google Scholar]
- Wawira, C.M. Phenotypic and Genotypic Characterization of Uropathogenic Escherichia Coli Isolates from Patients in Selected Hospitals in Kenya. Ph.D. Thesis, Maseno University, Maseno, Kenya, 2020. [Google Scholar]
- Taneja, N.; Singh, G.; Singh, M.; Madhup, S.; Pahil, S.; Sharma, M. High occurrence of blaCMY-1 AmpC lactamase producing Escherichia coli in cases of complicated urinary tract infection (UTI) from a tertiary health care centre in north India. Indian J. Med. Res. 2012, 136, 289. [Google Scholar] [PubMed]
- Matloko, K.; Fri, J.; Ateba, T.P.; Molale-Tom, L.G.; Ateba, C.N. Evidence of potentially unrelated AmpC beta-lactamase producing Enterobacteriaceae from cattle, cattle products and hospital environments commonly harboring the bla ACC resistance determinant. PLoS ONE 2021, 16, 253647. [Google Scholar] [CrossRef] [PubMed]
- Winokur, P.L.; Vonstein, D.L.; Hoffman, L.J.; Uhlenhopp, E.K.; Doern, G.V. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 2001, 45, 2716–2722. [Google Scholar] [CrossRef] [PubMed]
- Dolejska, M.; Papagiannitsis, C.C. Plasmid-mediated resistance is going wild. Plasmid 2018, 99, 99–111. [Google Scholar] [CrossRef]
- Sudarwanto, M.; Akineden, Ö.; Odenthal, S.; Gross, M.; Usleber, E. Extended-spectrum β-lactamase (ESBL)–producing Klebsiella pneumoniae in bulk tank milk from dairy farms in Indonesia. Foodborne Pathog. Dis. 2015, 12, 585–590. [Google Scholar] [CrossRef]
- Parajuli, N.P.; Acharya, S.P.; Mishra, S.K.; Parajuli, K.; Rijal, B.P.; Pokhrel, B.M. High burden of antimicrobial resistance among gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob. Resist. Infect. Control. 2017, 6, 1–9. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Sepulcri, C.; Giacobbe, D.R.; Peghin, M. Treatment of bloodstream infections due to Gram-negative bacteria with difficult-to-treat resistance. Antibiotics. 2020, 9, 632. [Google Scholar] [CrossRef]
- Wang, T.Z.; Kodiyanplakkal, R.P.L.; Calfee, D.P. Antimicrobial resistance in nephrology. Nat. Rev. Nephrol. 2019, 15, 463–481. [Google Scholar] [CrossRef]
- Giriyapur, R.S.; Namratha, W.N.; Krishna, B.V.S.; Asha, B.P.; Chandrasekhar, M.R. Comparison of Disc Diffusion Methods for the Detection of Extended-Spectrum Beta Lactamase-Producing Enterobacteriaceae. J. Lab. Physicians 2011, 3, 33–36. [Google Scholar] [CrossRef]
- Reuland, E.A.; Hays, J.P.; de Jongh, D.M.C.; Abdelrehim, E.; Willemsen, I.; Kluytmans, J.A.J.W.; Savelkoul, P.H.M.; Vandenbroucke-Grauls, C.M.J.E.; al Naiemi, N. Detection and occurrence of plasmid-mediated AmpC in highly resistant gram-negative rods. PLoS ONE 2014, 9, 91396. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Ranjit, R.C. Antibiotic Resistance in India: Drivers and Opportunities for Action. PLoS Med. 2016, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.; Carvalho, I.; Currie, C.; Sousa, M.; Igrejas, G.; Poeta, P. Extended-Spectrum-β-Lactamase and Carbapenemase-Producing Enterobacteriaceae in Food-Producing Animals in Europe: An Impact on Public Health? Antibiot. Drug Resist. 2019, 261–273. [Google Scholar]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Pokharel, S.; Raut, S.; Adhikari, B. Tackling Antimicrobial Resistance in Low-Income and Middle-Income Countries. BMJ Glob. Health 2019, 4, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Willis, L.D.; Chandler, C. Quick Fix for Care, Productivity, Hygiene and Inequality: Reframing the Entrenched Problem of Antibiotic Overuse. BMJ Glob. Health 2019, 4, 1590. [Google Scholar]
- EFSA. Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in Salmonella, Campylobacter and indicator Escherichia coli and Enterococcus spp. bacteria transmitted through food. Eur. Food Saf. Auth. J. 2012, 10, 2742. [Google Scholar] [CrossRef]
- de Alcântara Rodrigues, I.; Ferrari, R.G.; Panzenhagen, P.H.N.; Mano, S.B.; Conte-Junior, C.A. Antimicrobial Resistance Genes in Bacteria from Animal-Based Foods. Adv. Appl. Microbiol. 2020, 112, 143–183. [Google Scholar] [CrossRef]
- Harakeh, S.; Saleh, I.; Zouhairi, O.; Baydoun, E.; Barbour, E.; Alwan, N. Antimicrobial Resistance of Listeria Monocytogenes Isolated from Dairy-Based Food Products. Sci. Total Environ. 2009, 407, 4022–4027. [Google Scholar] [CrossRef] [PubMed]
- Alegbeleye, O.O.; Guimarães, J.T.; Cruz, A.G.; Sant’Ana, A.S. Hazards of a ‘Healthy’ Trend? An Appraisal of the Risks of Raw Milk Consumption and the Potential of Novel Treatment Technologies to Serve as Alternatives to Pasteurization. Trends Food Sci. Technol. 2018, 82, 148–166. [Google Scholar] [CrossRef]
- FDA. Antimicrobials Sold or Distributed for Use in Food-Producing Animals; US Food and Drug Administration: Silver Spring, MD, USA, 2015. Available online: https://www.fda.gov/files/aboutfda/published/2015-Summary-Report-on-Antimicrobials-Sold-or-Distributed-for-Use-in-Food-Producing-Animals.pdf (accessed on 22 December 2022).
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet Health 2017, 1, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.M.; Griffiths, M.W.; McEwen, S.A.; McNab, W.B.; Yee, A.J. Antimicrobial drug residues in milk and meat: Causes, concerns, prevalence, regulations, tests, and test performance. J. Food Prot. 1998, 61, 742–756. [Google Scholar] [CrossRef] [PubMed]
- Nyokabi, S.N. Biosecurity measures in meat and milk value chains: A study in Bura Sub-county, Kenya. Ph.D. Thesis, University of Hohenheim, Stuttgart, Germany, 2015. [Google Scholar]
- DAHD. Department of Animal Husbandry, Dairying and Fisheries. Ministry of Agriculture. 20th Livestock Census, All India Report 2019. Available online: http://dahd.nic.in/dahd/writeData/livestock (accessed on 5 November 2022).
- Naing, L.; Winn, T.; Rusli, B.N. Practical Issues in Calculating the Sample Size for Prevalence Studies. Arch. Orofac. Sci. 2006, 1, 9–14. [Google Scholar]
- Hudzicki, J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol Author Information. Am. Soc. Microbiol. 2009, 1–13. Available online: https://www.asm.org/Protocols/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Pro (accessed on 11 September 2023).
- Kagira, J.M.; Ngotho, M.; Mugo, E.; Kiplimo, M.; Maina, N. Occurrence of Antibiotic Resistance in Bacteria Isolated from Milk of Dairy Cows in Small-Holder Farms in Juja Sub-County, Kenya. Asian J. Res. Anim. Vet. Sci. 2022, 9, 37–46. [Google Scholar]
- Kojima, A.; Ishii, Y.; Ishihara, K.; Esaki, H.; Asai, T.; Oda, C.; Tamura, Y.; Takahashi, T.; Yamaguchi, K. Extended-Spectrum-β-Lactamase-Producing Escherichia coli Strains Isolated from Farm Animals from 1999 to 2002: Report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 2005, 49, 3533–3537. [Google Scholar] [CrossRef]
- Semedo, T.; Santos, M.A.; Martins, P.; Lopes, M.F.S.; Marques, J.J.F.; Tenreiro, R.; Crespo, M.T.B. Comparative Study Using Type Strains and Clinical and Food Isolates to Examine Hemolytic Activity and Occurrence of the Cyl Operon in Enterococci. J. Clin. Microbiol. 2003, 41, 2569–2576. [Google Scholar] [CrossRef]
- Johann, D.D.P.; Ashfaque, H.; Nancy, D.H. Phenotypic and Molecular Detection of CTX-M-Beta-Lactamases Produced by E. coli and Klebsiella spp. J. Clin. Microbiol. 2004, 42, 5715–5721. [Google Scholar]
- Mendes, R.E.; Kiyota, K.A.; Monteiro, J.; Castanheira, M.; Andrade, S.S.; Gales, A.C.; Pignatari, A.C.C.; Tufik, S. Rapid Detection and Identification of Metallo-β-Lactamase-Encoding Genes by Multiplex Real-Time PCR Assay and Melt Curve Analysis. J. Clin. Microbiol. 2007, 45, 544–547. [Google Scholar] [CrossRef]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef]
- Modak, R.; Das Mitra, S.; Krishnamoorthy, P.; Bhat, A.; Banerjee, A.; Gowsica, B.; Bhuvana, M.; Dhanikachalam, V.; Natesan, K.; Shome, R.; et al. Histone H3K14 and H4K8 Hyperacetylation Is Associated with Escherichia coli Induced Mastitis in Mice. Epigenetics 2012, 7, 492–501. [Google Scholar] [CrossRef]
- Mitra, S.D.; Wilfred, A.D.; Tewari, R.I.; Nimita, V.C.; Bhuvana, M.; Krithiga, N.; Shome, B.R.; Rahman, H.A. Duplex PCR for Specific Detection of Escherichia coli and Its Differentiation from Other Enterobacteriaceae. Indian J. Anim. Sci. 2015, 85, 832–835. [Google Scholar]
- Chander, Y.; Ramakrishnan, M.A.; Jindal, N.; Hanson, K.; Goyal, M. Differentiation of Klebsiella pneumoniae and K. oxytoca by Multiplex Polymerase Chain Reaction. Int. J. Appl. Res. Vet. Med. 2011, 9, 138. [Google Scholar]
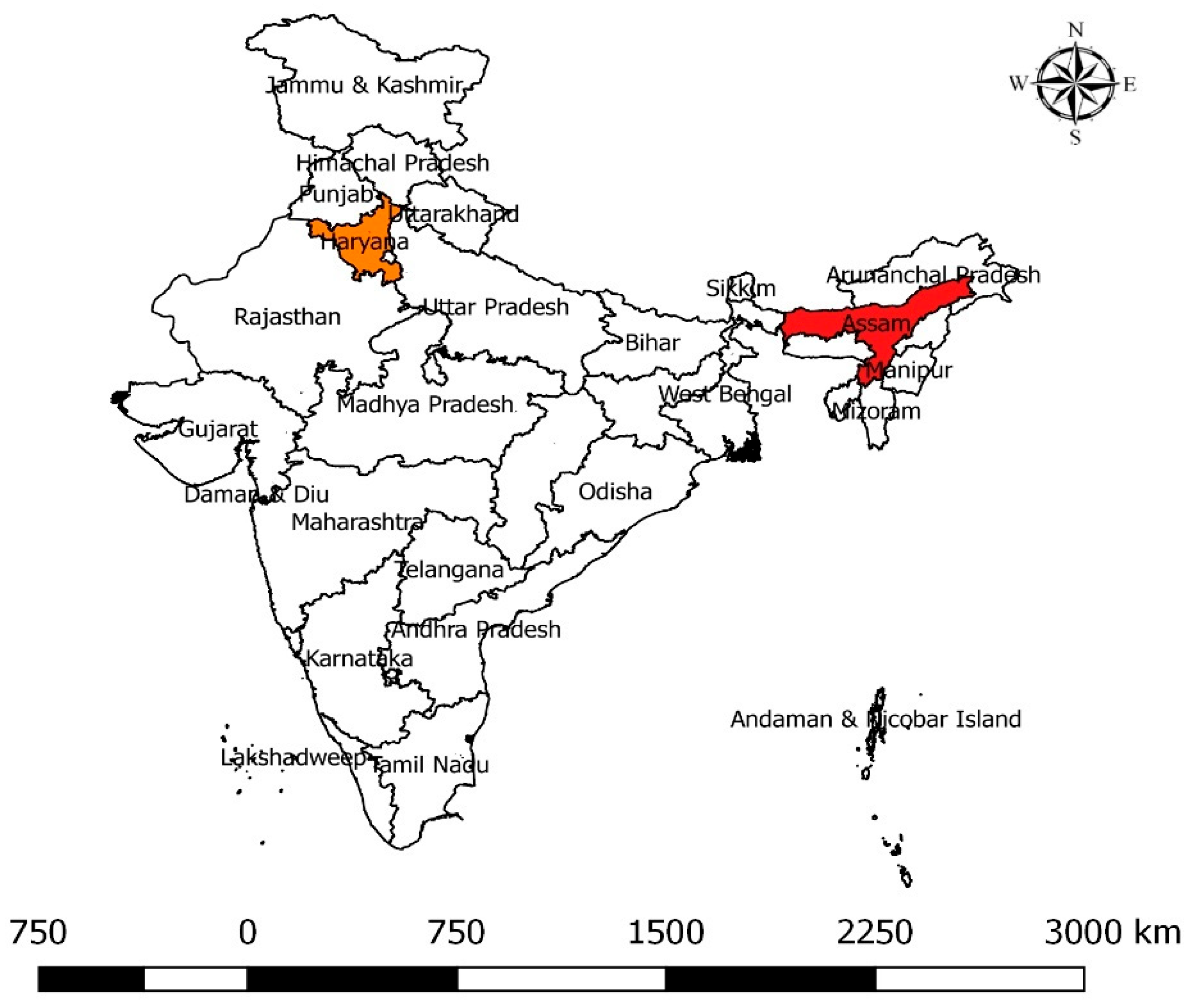
| Milk Source | Sample Type | Assam | Haryana | Total |
|---|---|---|---|---|
| Milk from dairy farmer | Raw milk | 116 | 126 | 242 |
| Sample positive * | 113 | 112 | 225 | |
| Isolate | 148 | 115 | 263 | |
| Milk from dairy vendor | Raw milk | 63 | 74 | 137 |
| Sample positive * | 63 | 73 | 136 | |
| Isolate | 63 | 73 | 136 | |
| Pasteurized milk | 0 | 22 | 22 | |
| Sample positive * | 0 | 22 | 22 | |
| Isolates | 0 | 22 | 22 | |
| Total sample | 179 | 222 | 401 | |
| Total positive * | 176 | 207 | 383 | |
| Total isolate | 211 | 210 | 421 |
| Total Isolates (n = 421) | Isolates from Haryana (n = 210) | Isolates from Assam (n = 211) | p-Value | Isolates from Farmers (n = 263) | Isolates from Vendors (n = 158) | p-Value | |
|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | |||
| Resistant isolates | 295 (70.07) | 133 (63.33) | 162 (76.78) | 0.003 | 183 (69.58) | 112 (70.89) | 0.826 |
| Non-resistant isolates | 126 (29.93) | 77 (36.67) | 49 (23.22) | 80 (30.42) | 46 (29.11) |
| Milk Sources | Isolates | ||||||
|---|---|---|---|---|---|---|---|
| Resistant to 6 Antibiotics | Resistant to 5 Antibiotics | Resistant to 4 Antibiotics | Resistant to 3 Antibiotics | Resistant to 2 Antibiotics | Resistant to 1 Antibiotics | Sensitive Isolates | |
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Assam (n = 211) | 0 | 4 (1.90) | 20 (9.48) | 31 (14.69) | 49 (23.22) | 58 (27.49) | 49 (23.22) |
| Haryana (n = 210) | 3 (1.43) | 2 (0.95) | 15 (7.14) | 29 (13.81) | 38 (18.10) | 46 (21.90) | 77 (36.67) |
| Farmer (n = 263) | 2 (0.76) | 3 (1.14) | 18 (6.84) | 33 (12.55) | 57 (21.67) | 70 (26.62) | 80 (30.42) |
| Vendor (n = 158) | 1 (0.63) | 3 (1.90) | 17 (10.76) | 27 (17.09) | 30 (18.99) | 34 (21.52) | 46 (29.11) |
| Total (n = 421) | 3 (0.71) | 6 (1.43) | 35 (8.31) | 60 (14.25) | 87 (20.67) | 104 (24.70) | 126 (29.93) |
| Milk Source | β-Lactamases Positive | * ESBL Genes Positive | * MBL Genes Positive | * AmpC Genes Positive | AMR Genes in Combination (n = 421) | ||
|---|---|---|---|---|---|---|---|
| AmpC + MBL | ESBL + AmpC | ESBL + MBL | |||||
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| Assam (n = 211) | 15 (7.11) | 3 (1.42) | 6 (2.84) | 7 (3.31) | 1 (0.23) | 0 | 0 |
| Haryana (n = 210) | 28 (13.33) | 6 (2.86) | 5 (2.38) | 21 (10.00) | 2 (0.47) | 1 (0.23) | 1 (0.23) |
| Total (n = 421) | 43 (10.21) | 9 (2.13) | 11 (2.61) | 28 (6.65) | 3 (0.71) | 1 (0.23) | 1 (0.23) |
| Farmer (n = 263) | 28 (10.65) | 6 (2.28) | 10 (3.80) | 16 (6.08) | 3 (0.71) | 1 (0.23) | 1 (0.23) |
| Vendor (n = 158) | 15 (10.13) | 3 (1.89) | 1 (0.63) | 12 (7.59) | 0 | 0 | 0 |
| Total (n = 421) | 43 (10.21) | 9 (2.13) | 11 (2.61) | 28 (6.65) | 3 (0.71) | 1 (0.23) | 1 (0.23) |
| Milk Source | E. coli | Shigella spp. | Klebsiella spp. | Other Enterobacteriaceae | Total Isolates |
|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n | |
| Assam (n = 15) | 0 | 10 (66.67) | 0 | 5 (33.33) | 15 |
| Haryana (n = 28) | 2 (7.14) | 2 (7.14) | 6 (21.43) | ** 18 (64.29) | 28 |
| Total (n = 43) | 2 (4.65) | 12 (27.90) | 6 (13.95) | 23 (53.48) | 43 |
| Milk source | E. coli | Shigella spp. | Klebsiella spp. | Other Enterobacteriaceae | Total isolates |
| n (%) | n (%) | n (%) | n (%) | n | |
| Farmer’s (n = 28) | 2 (7.14) | 12 (42.86) | 4 (14.29) | 10 (35.71) | 28 |
| Vendor’s (n = 15) | 0 | 0 | 2 (13.33) | ** 13 (86.67) | 15 |
| Total (n = 43) | 2 (4.65) | 12 (27.90) | 6 (13.95) | 23 (53.48) | 43 |
| β-Lactamases | E. coli | Shigella spp. | Klebsiella spp. | Other Enterobacteriaceae | * Total Genes Identified |
| n (%) | n (%) | n (%) | n (%) | n | |
| AmpC genes * (n = 28) | 0 | 7 (25.00) | 5 (17.85) | 17 ** (60.71) | 28 |
| MBL genes * (n = 11) | 1 (9.09) | 6 (54.54) | 0 | 4 (36.36) | 11 |
| ESBL genes * (n = 9) | 1 (11.11) | 1 (11.11) | 2 (18.18) | 5 (55.55) | 9 |
| Antibiotic Classes | Phenotypic Resistance by DDT | ESBL Positive Genes # (n = 9) | MBL Positive Genes * (n = 11) | AmpC Positive Genes $ (n = 28) | Phenotypic Resistance by E-Test | ESBL Positive Genes # (n = 9) | MBL Positive Genes * (n = 11) | AmpC Positive Genes $ (n = 28) |
|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |||
| 2nd generation cephalosporin (AmpC Resistance $) | Resistant to cefoxitin | 5 (55.56) | 6 (54.54) | 19 (67.86) | Resistant to cefoxitin | 3 (33.33) | 3 (27.27) | 20 (71.43) |
| Resistant to cefotetan | 4 (44.44) | 4 (36.36) | 16 (57.14) | Resistant to cefotetan | 3 (33.33) | 3 (27.27) | 13 (46.43) | |
| 3rd generation cephalosporin (ESBL Resistance #) | Resistant to cefotaxime | 6 (66.67) | 6 (54.55) | 6 (21.43) | Resistant to cefotaxime | 4 (44.44) | 3 (27.27) | 12 (42.86) |
| Resistant to ceftazidime | 4 (44.44) | 4 (36.36) | 9 (32.14) | Resistant to ceftazidime | 3 (33.33) | 0 | 12 (42.86) | |
| Carbapenem-β-lactam (MBL Resistance *) | Resistant to imipenem | 1 (11.11) | 1 (9.09) | 1 (3.57) | Resistant to imipenem | 1 (11.11) | 1 (9.09) | 1 (3.57) |
| Resistant to meropenem | 1 (11.11) | 0 | 0 | Resistant to meropenem | 1 (11.11) | 1 (9.09) | 7 (25) |
| Milk Source | Isolates | ||
|---|---|---|---|
| PCR Positive # | Resistant by DDT | Resistant by E-Test | |
| n (%) | n (%) | n (%) | |
| Assam | 15/43 (34.88) | 13/15 (86.67) | 8/15 (53.33) |
| Haryana | 28/43 (65.11) | 20/28 (71.43) | 21/28 (75.00) |
| Total | 43/43 (100.00) | 33/43 (76.74) | 29/43 (67.44) |
| β-lactamases | Isolates | ||
| AMR genes * | Resistant by DDT | Resistant by E-test | |
| n | n (%) | n (%) | |
| ESBL genes (n = 9) | 9 | 6 (66.67) | 7 (77.78) |
| MBL genes (n = 11) | 11 | 8 (72.73) | 4 (36.36) |
| AmpC genes (n = 28) | 28 | 22 (78.57) | 21 (75.00) |
| Total | 43 | 36 (83.72) | 32 (74.41) |
| Classes | β-Lactam Antibiotics | Concentration of Antibiotic Disc (in µg) | Breakpoints for Resistance by DDT (mm) | Concentration of Antibiotic Strips (µg/mL) | Breakpoints for Resistance by E-Test (µg/mL) | |
|---|---|---|---|---|---|---|
| 3rd generation cephalosporins | For ESBL detection | Cefotaxime Ceftazidime | 30 30 | ≤22 ≤17 | 0.016–256 0.016–256 | >4 >16 |
| 2nd generation cephalosporins | For AmpC detection | Cefoxitin Cefotetan | 30 30 | ≤14 ≤12 | 0.016–256 0.016–256 | >32 >64 |
| Carbapenem-β-lactams | For MBL detection | Imipenem Meropenem | 30 30 | ≤19 ≤19 | 0.002–32 0.002–32 | ≥4 >4 |
| Gene | Forward (5′-3′) | Reverse (5′-3′) | Length (* bp) | * AT | * Ref. |
|---|---|---|---|---|---|
| ESBL genes | 60 °C | [102,103,104] | |||
| blaTEM | ATGAGTATTCAACATTTTCG | TTACCAATGCTTAATCAGTG | 861 | ||
| blaSHV | ATGCGTTATATTCGCCTGTG | TTAGCGTTGCCAGTGCTCGA | 860 | ||
| blaCTXM1 | GACGATGTCACTGGCTGAGC | AGCCGCCGACGCTAATACA | 499 | ||
| blaCTXM2 | GCGACCTGGTTAACTACAATCC | CGGTAGTATTGCCCTTAAGCC | 351 | ||
| blaCTXM3 | CGCTTTGCCATGTGCAGCACC | GCTCAGTACGATCGAGCC | 307 | ||
| blaCTXM4 | GCTGGAGAAAAGCAGCGGAG | GTAAGCTGACGCAACGTCTG | 474 | ||
| MBL genes | 53 °C | [105] | |||
| blaIMP | GAATAG(A/G)(A/G)TGGCTTAA(C/T)TCTC | CCAAAC(C/T ACTA(G/C)GTTATC | 188 | ||
| blaVIM | GTTTGGTCGCATATCGCAAC | AATGCGCAGCACCAGGATAG | 382 | ||
| blaGIM | TCAATTAGCTCTTGGGCTGAC | CGGAACGACCATTTGAATGG | 72 | ||
| blaSIM | GTACAAGGGATTCGGCATCG | TGGCCTGTTCCCATGTGAG | 569 | ||
| blaSPM | CTAAATCGAGAGCCCTGCTTG | CCTTTTCCGCGACCTTGATC | 798 | ||
| AmpC genes | 64 °C | [106] | |||
| blaFOX | AACATGGGGTATCAGGGAGATG | CAAAGCGCGTAACCGGATTGG | 190 | ||
| blaMOX | GCTGCTCAAGGAGCACAGGAT | CACATTGACATAGGTGTGGTGC | 520 | ||
| blaEBC | TCGGTAAAGCCGATGTTGCGG | CTTCCACTGCGGCTGCCAGTT | 302 | ||
| blaACC | AACAGCCTCAGCAGCCGGTTA | TTCGCCGCAATCATCCCTAGC | 346 | ||
| blaDHA | AACTTTCACAGGTGTGCTGGGT | CCGTACGCATACTGGCTTTGC | 405 | ||
| blaCMY | TGGCCAGAACTGACAGGCAAA | R TTTCTCCTGAACGTGGCTGGC | 462 | ||
| E. coli | 60 °C | [107,108] | |||
| lacY | CTACCGGTGAACAGGGTAGC | GTCGCTGAAAAACGCACTTC | 289 | ||
| lacZ | ATGAAAGCTGGCTACAGGAAGG | CTCCACACAGTTTCGGGTTTTC | 517 | ||
| cyd | CCGTATCATGGTGGCGTGTGG | GCCGGCTGAGTAGTCGTGGAAG | 398 | ||
| uidA | CGCCGATGCAGATATTCG | GCTGTGACGCACAGTTCATAG | 603 | ||
| phoA | GGTAACGTTTCACCGCAGAGTTG | CAGGGTTGGTACACTGTCATTACG | 468 | ||
| Shigella spp. | |||||
| phoA | GGTAACGTTTCACCGCAGAGTTG | CAGGGTTGGTACACTGTCATTACG | 468 | ||
| Klebsiella spp. | 62 °C | [109] | |||
| gyrA | CGCGTACTATACGCCATGAACGTA | ACCGTTGATCACTTCGGTCAGG | 441 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, T.K.; Lindahl, J.F.; Lundkvist, Å.; Grace, D.; Deka, R.P.; Shome, R.; Bandyopadhyay, S.; Goyal, N.K.; Sharma, G.; Shome, B.R. Analyses of Extended-Spectrum-β-Lactamase, Metallo-β-Lactamase, and AmpC-β-Lactamase Producing Enterobacteriaceae from the Dairy Value Chain in India. Antibiotics 2023, 12, 1449. https://doi.org/10.3390/antibiotics12091449
Dey TK, Lindahl JF, Lundkvist Å, Grace D, Deka RP, Shome R, Bandyopadhyay S, Goyal NK, Sharma G, Shome BR. Analyses of Extended-Spectrum-β-Lactamase, Metallo-β-Lactamase, and AmpC-β-Lactamase Producing Enterobacteriaceae from the Dairy Value Chain in India. Antibiotics. 2023; 12(9):1449. https://doi.org/10.3390/antibiotics12091449
Chicago/Turabian StyleDey, Tushar Kumar, Johanna Frida Lindahl, Åke Lundkvist, Delia Grace, Ram Pratim Deka, Rajeswari Shome, Samiran Bandyopadhyay, Naresh Kumar Goyal, Garima Sharma, and Bibek Ranjan Shome. 2023. "Analyses of Extended-Spectrum-β-Lactamase, Metallo-β-Lactamase, and AmpC-β-Lactamase Producing Enterobacteriaceae from the Dairy Value Chain in India" Antibiotics 12, no. 9: 1449. https://doi.org/10.3390/antibiotics12091449
APA StyleDey, T. K., Lindahl, J. F., Lundkvist, Å., Grace, D., Deka, R. P., Shome, R., Bandyopadhyay, S., Goyal, N. K., Sharma, G., & Shome, B. R. (2023). Analyses of Extended-Spectrum-β-Lactamase, Metallo-β-Lactamase, and AmpC-β-Lactamase Producing Enterobacteriaceae from the Dairy Value Chain in India. Antibiotics, 12(9), 1449. https://doi.org/10.3390/antibiotics12091449







