Abstract
In this study, we sought to profile the abundances and drivers of antibiotic resistance genes in an urban river impacted by combined sewage overflow (CSO) events. Water samples were collected weekly during the summer for two years; then, quantitative PCR was applied to determine the abundance of resistance genes associated with tetracycline, quinolones, and β-lactam antibiotics. In addition to sampling a CSO-impacted site near the city center, we also sampled a less urban site ~12 km upstream with no proximal sewage inputs. The tetracycline genes tetO and tetW were rarely found upstream, but were common at the CSO-impacted site, suggesting that the primary source was untreated sewage. In contrast, ampC was detected in all samples indicating a more consistent and diffuse source. The two other genes, qnrA and blaTEM, were present in only 40–50% of samples and showed more nuanced spatiotemporal patterns consistent with upstream agricultural inputs. The results of this study highlight the complex sources of ARGs in urban riverine ecosystems, and that interdisciplinary collaborations across diverse groups of stakeholders are necessary to combat the emerging threat of antibiotic resistance through anthropogenic pollution.
Keywords:
antibiotics; ARGs; combined sewage overflow; CSO; wastewater; sewage; tetracycline; ß-lactams; quinolones; tetO; tetW; blaTEM; qnrA; ampC 1. Introduction
Antibiotic resistance is an emerging global public health concern [1,2]. In 2019, 1.3 million deaths were directly attributable to antibiotic resistance, and an estimated total of 5 million deaths were associated with resistant bacteria [3]. Antibiotic resistance genes (ARGs), which confer resistance to specific antibiotics through a variety of mechanisms, occur naturally to mediate resource competition among microorganisms. However, the increased use of antibiotics as clinical agents has radically increased the evolution and spread of ARGs. Although most efforts to contain the antibiotic resistance problem are focused on biomedical settings, ARGs are rapidly emerging as environmental contaminants. There has been a particular focus on urban rivers as environmental reservoirs because they are a vital source of fresh drinking water and widely used for recreation. Sources of ARGs in these waterways include municipal wastewater systems [4], effluent from hospitals [5], pharmaceutical manufacturing [6,7], and upstream agricultural runoff [8].
Urban waterways and rivers are well-known hotspots for ARGs [9,10]. This is often attributed to fecal contamination, the introduction of antibiotics or metals that apply selective pressure to microbial communities, and horizontal gene transfer of the mobile genetic elements conferring antibiotic resistance [11]. Even in urban environments with extensive sanitation systems, the fact that wastewater treatment plants (WWTP) lack antibiotic-targeted treatment processes means that receiving waterbodies can be reservoirs of residual antibiotics and ARGs. In many older cities, the problem is compounded by the discharge of untreated wastewater from combined sewage–stormwater systems that overflow during heavy rain events. These combined sewage overflow (CSO) events appear to be a major source of antibiotics, resistance genes, and resistant organisms to urban rivers [12,13,14]. Since these rivers can act as conduits of antibiotics and ARGs to downstream aquatic and coastal systems, these wastewater inputs can have lasting repercussions across broad spatial scales.
A recent global meta-analysis found sanitation infrastructure to be one of the strongest drivers of elevated antimicrobial resistance, having a much greater effect than antibiotic consumption rates [15]. Although numerous studies have considered the impact of wastewater on the distribution and diversity of ARGs in urban rivers [4,16,17,18,19], the specific impact of combined sewage systems has received comparatively less attention [20,21]. Combined sewage systems are common in older cities around the world, especially in Europe and the United States. For example, the most recent estimates from the U.S. Environmental Protection Agency are that 40 million Americans live in municipalities with combined sewage–stormwater systems, which collectively discharge 850 billion gallons of contaminated wastewater each year [22]. Understanding the role of CSOs in the environmental dissemination of ARGs is becoming increasingly important as urbanization and climate change can lead to even more frequent and intense overflow events [23,24].
The purpose of this study was to document the prevalence of ARGs and their relationship with water quality parameters in an urbanized segment of the James River (Virginia, USA) that is frequently impacted by CSO events. We compared a site upstream of major urbanization (“HUG”) to one located near a large CSO outfall at the city center (“CSO”). The abundances of five ARGs were determined using quantitative polymerase chain reaction (qPCR) and compared with changes in water quality and hydrological conditions to identify potential sources of ARG within the watershed. We focused on summer when recreational contact, and thus the potential for human health impacts, is highest.
2. Results and Discussion
Fecal contamination is one of the main sources of ARGs in urban aquatic environments [25], in part due to increased population density and high levels of impervious surfaces [26,27]. Combined sewage overflow systems in urban settings are known to be a major source of ARGs and resistant bacteria to receiving waterbodies [12,14,28]. Our results suggest that while CSO events are likely a large source of ARGs in the river, non-point sources are also important sources of ARGs in the river. Overall, ARGs were more abundant at the downstream CSO site than the upstream site (HUG) (Figure 1).
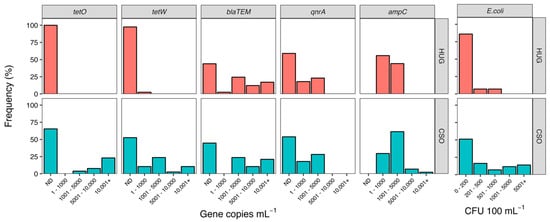
Figure 1.
Abundance frequency histograms for ARG (left) and E. coli (right). Data are presented as the relative frequency (% of samples) in each abundance interval during the two-year study period. The top row (pink bars) shows data from the upstream site (HUG) and the bottom row (blue bars) shows the downstream site (CSO) near the city center. The not detected (ND) category represents samples that did not produce amplicons.
Tetracycline-resistance genes showed the most striking differences across sites. The tetO gene was never detected at the upstream site and the tetW gene was only detected in ~2% of samples, whereas these genes were detected in 35% (tetO) to 48% (tetW) of samples from the CSO site. Mann–Whitney tests confirmed elevated abundances of both genes at the CSO site (tetO: U = 243, p = 0.001; tetW: U = 424, p < 0.0001). Correlation analysis showed abundances of both tetracycline genes were most strongly related to E. coli abundance (Table 1), suggesting fecal contamination is a major source of these ARGs in the river. The increase in abundance of tetO at the CSO site also correlated with an increase in TN (ρ = 0.60) and TP (ρ = 0.67). Since increased concentrations of N and P have been well documented in association with CSO events [29,30], this correlation provides further support that tetO derived from fecal contamination due to sewage overflow. In contrast, the increase in abundance of tetW at the CSO site was correlated with precipitation, suggesting non-point sources of fecal contamination via runoff or leaky sewerage [31].

Table 1.
Spearman’s correlation coefficients (ρ) for ARG abundances and environmental parameters at the CSO site. Bold values represent statistically significant coefficients (α = 0.01), and asterisks reflect degree of significance (* 0.001 < p ≤ 0.01, ** 0.001 < p).
Tetracycline resistance genes (tet) are some of the most commonly detected and diverse ARGs in aquatic habitats, with over 20 types identified thus far [32]. These genes are frequently found in animal and human feces [33], are highly abundant in sewage [34,35,36], and their presence in surface waters has been linked to fecal contamination at the national level in the United States [37]. The abundance of tet genes has also been found to be elevated downstream of WWTPs [38]. Our findings are consistent with those prior studies: both of the tet genes we considered were found in high abundance at the CSO site when the river was contaminated with fecal material due to either sewage overflow events (tetO) or runoff (tetW)). Matsui and Miki [13] also studied multiple tet genes in an urban river and, like us, found some to be linked with rainfall and non-point discharge (tetM), whereas others were not (tetA and tetB). Their study also demonstrated that increasing the storage of CSO systems can decrease the spread of tet genes. Our data suggest that similar actions would decrease the spread of tetO but may not impact tetW. This highlights the need for studies such as ours that seek to differentiate point and non-point sources of ARG pollution within the urban landscape, as each may ultimately require different management approaches.
In addition to being common in sewage, tet genes are a major component of soil resistomes [39,40]. Given this, it was somewhat surprising to find both tetracycline genes to be essentially absent at the upstream site. The widespread distribution of tet genes in soil is likely due, at least in part, to the fact that many code for efflux pumps that can serve diverse functions in addition to conferring antibiotic resistance [32]. However, tetO and tetW are unusual in that they code for ribosomal protection proteins; thus, their presence/absence is likely more reflective of tetracycline exposure compared to other tet genes. Our failure to find tetW and tetO at the upstream site suggests there is not a significant proximal upstream source of human waste or any nearby agricultural operations that utilize tetracycline. Although tetracyclines are one of the primarily antibiotics groups used for agricultural purposes [41], most of the nearby farms are equine, and the use of tetracycline antibiotics to treat horses is discouraged because doing so can cause severe and even fatal diarrhea [42,43].
The blaTEM gene, which encodes for the TEM-1 β-lactamase, confers resistance to β-lactam antibiotics such as the penicillins and early cephalosporins. This gene was found in ~55% of all samples and showed similar abundances across the two sites (U = 766, p = 0.90). Of the five ARGs we surveyed, only blaTEM was abundant at the upstream site (Figure 1). Approximately ~30% of HUG samples had > 6000 copies mL−1 of blaTEM (15% had > 10,000 copies mL−1), whereas the abundance of the four other ARGs never exceeded 4500 copies mL−1 at the upstream site. The high overall abundance and broad distribution of the blaTEM gene were not surprising, as this gene appears to be one of the most widespread ARGs across multiple environmental compartments [44,45]. This is further supported by a recent survey of 2000 streams and rivers across the USA, which found blaTEM to be ubiquitous across all ecoregions of the country, despite different land-use regimes and environmental conditions [37]. This widespread distribution of blaTEM is likely due to the fact ß-lactams were the first major class of antibiotics to be discovered and account for approximately two-thirds, by weight, of all antibiotics prescribed to humans [46]. Like tetW, we found blaTEM abundances to be correlated with E. coli (ρ = 0.48) and prior-day precipitation (ρ = 0.46). This suggests upstream or catchment sources of blaTEM in the river, likely linked to fecal contamination being introduced into the river during rain events via runoff. Similar research examining the drivers of ARGs in a riverine system found a strong link between ARG abundances and precipitation, suggesting the catchment area is a source of ARGs in the river [16]. The remaining two genes, ampC and qnrA, did not show a direct link with fecal contamination or sewage overflow indicators, and were similarly distributed across the two sites (Figure 1). The ampC gene, which also confers resistance to multiple members of the ß-lactams class of antibiotics, was the only ARG that we detected in every sample. Although the Mann–Whitney test indicated elevated abundance of ampC at the CSO site (U = 656, p = 0.01), the difference was relatively small (median and SE-median for HUG: 919 ± 211; for CSO: 1412 ± 421), especially compared to the abundance of other ARGs. The abundance of ampC was positively correlated with overall bacterial abundance, but did not appear to be related to any of the water quality or hydrologic parameters we measured. Coertze and Bezuidenhout [47,48] also found it challenging to correlate environmental factors or land use as a predictor of ampC abundance, and speculated that there must be a pervasive source or reservoir of this ARG in the rivers they studied in South Africa. Something similar could be occurring in our system, though the positive correlation of ampC and bacterial abundance suggests that ampC is simply widespread in the James River bacterial community. Our results indicate that ampC is a highly dispersed gene, consistent with reports of broad global distribution of the gene across terrestrial [39,49] and aquatic [50] ecosystems, including relatively pristine or unimpacted sites [51,52].
Of the five ARGs we considered, qnrA, which confers resistance to fluoroquinolones, was the least common overall. The qnrA gene was only detected in ~40% of samples, and abundance never exceeded 3000 copies mL−1 (Figure 1). No differences were detected across sites (U = 697, p = 0.48), and qnrA abundances were not correlated with either E. coli abundance or TN and TP concentrations (Table 1). Together, these results suggest that sewage was not a significant reservoir of qnrA, despite the fact that fluoroquinolones are frequently prescribed to humans. This finding may be due to the fact that we sampled exclusively during summer. Seasonal variation in fluoroquinolone concentration has repeatedly been found in freshwater rivers [53], WWTP influent [54], and sludge from WWTPs [55], with levels being consistently higher in winter. This indicates changing prescription and usage levels of fluoroquinolones in human medicine [56], and suggests that our qnrA results could be very different if we sampled during winter months.
Correlation analysis did reveal a positive relationship between qnrA abundance and rainfall the day prior to sampling (ρ = 0.43), which suggests that precipitation mobilizes an upstream or catchment-level source of this resistance gene to the James River. The watershed upstream of the HUG site is approximately 10% pasture land [57], and there are multiple equestrian centers, with the closest being ~10 km upstream, making this a plausible source of qnrA genes or fluoroquinolones for the river. Antimicrobial resistance is prevalent in bacteria from horses [58], and qnr genes have previously been detected in the feces of horses and in environmental samples from equine clinics and horseback riding centers [59,60,61]. Our hypothesis that equestrian facilities are the most likely agricultural source of qnrA is also supported by the fact that fluoroquinolones are widely utilized in animal husbandry and in equestrian activities/rearing [59,62], but rarely used in food-producing animals (~0.1% of U.S. domestic antibiotic sales during the study period [63]).
Interestingly, of the ARGs that correlated with precipitation, the qnrA gene was the only one that was not also linked with E. coli abundance. This could suggest that the fluoroquinolone-resistant bacteria survive longer in the riverine system than the fecal indicator bacteria, or that the plasmid containing this ARG is especially persistent. This persistence could be due to the selective pressure of fluoroquinolones in the river water. For example, Stanton et al. [64] documented that ciprofloxacin can select for resistance persistence even at the relatively low concentrations reported in aquatic environments. This effect may be compounded by the fact that fluoroquinolones can remain stable in river water for at least two weeks [65]. Future work is needed to determine fluoroquinolone concentrations in the James River, but these prior studies suggest that low antibiotic concentrations may be decreasing the rate at which resistance genes disappear from the environment. These findings are particularly concerning, because the increased persistence of resistant bacteria or resistance genes is likely to associated with higher human exposure risk and greater impact on environmental microbiomes.
3. Conclusions
Taken together, our efforts show that over a relatively small spatial scale (~12 km), abundances of ARGs can differ by 3–5 orders of magnitude. Our multi-year dataset encompassing a variety of weather conditions shows diverse sources, including urban runoff, CSO events, and upstream land use, contribute to ARG abundances in urban rivers. Furthermore, our results show spatiotemporal considerations should be incorporated into investigations aiming to elucidate sources of ARGs to urban rivers or waterways. Future work that considers additional genes such as intI1, which has been used as a proxy indicator of anthropogenic pollution [37], could help further distinguish among the potential ARG sources and would provide valuable information regarding the potential for ARG spread by horizontal gene transfer.
While efforts to improve wastewater treatment technology and infrastructure will certainly reduce ARG abundance and antibiotic pollution in urban aquatic ecosystems, our results suggest that levels of select ARGs could remain elevated due to non-point sources within the urban area and from the surrounding landscape. Such a complex and multifaceted public health concern requires collaboration between environmental scientists, public health officials, wastewater management authorities, agricultural stakeholders, and policy makers in order to address the emerging threat of antibiotic resistance in aquatic environments.
4. Materials and Methods
4.1. Site Description
Richmond, Virginia (USA) is a moderately sized city with approximately 226,000 residents and a population density of ~10,000 people per km2 (United States Census Bureau, 2016). It relies on a combined sewage–stormwater system, which overflows and discharges untreated wastewater into the James River during heavy rainfall or snow melt. Richmond’s combined sewer system is the largest in the state of Virginia, and services approximately one-third of the city (~50 km2).
This study compared two sites along the James River as it flows through Richmond (Figure 2). The first was located within the Huguenot Flatwater area of James River Park (HUG; 37.560471, −77.545801), approximately 12 km upstream of the city center. This site was selected to assess water quality before significant urbanization and prior to any known CSO outfalls. The watershed upstream of HUG is forested and agricultural land, but also includes the cities of Lynchburg (~225 km upstream with 80,000 residents in 2016) and Charlottesville (~150 km upstream with 47,000 residents) and several smaller municipalities.

Figure 2.
Map of the James River along Richmond, Virginia (USA). Red circles mark the upstream (HUG, left panel) and downstream (CSO, right panel) sampling sites, which are separated by ~12 km. The black insert box, expanded on the right, shows the extensive urbanization surrounding the downstream CSO site.
The second site was located near the city center, adjacent to the city’s largest CSO outflow (37.529486, −77.429382), which is referred to as CSO-006 by the Richmond Department of Public Utilities. Long-term monitoring of Escherichia coli (E. coli) abundance indicates consistently higher levels of fecal contamination at this site compared to HUG. During the two-year sampling period of this study, E. coli concentrations exceeded 200 CFU 100 mL−1 in 50% of CSO samples versus <25% of HUG samples (Figure 1). The maximum E. coli abundance observed at the CSO site was approximately four-times greater than at HUG (32,200 versus 800 CFU 100 mL−1).
4.2. Water Sampling
Approximately every week during the summers (May 1st through October 15th) of 2015 and 2016, surface water samples (n = 44) were collected from each site using a bucket thrown from shore. Water was transferred into sterile plastic 1 L bottles and transported back to lab on ice within two hours. Upon return to the lab, samples were immediately processed to determine E. coli abundance following the EPA Method #1603 [66]. Plate counts were performed using modified mTEC agar (BD Difco, Sparks, MD, USA), and the results are reported as colony-forming units (CFU) 100 mL−1 of river water. In addition, aliquots (300 mL) of water were filtered using 0.2-µm pore-size polycarbonate membranes (Millipore, Molsheim, France) to isolate the microbial community for genetic analysis. Filters were stored at −20 °C until DNA extraction could be performed.
4.3. Environmental Data
Total nitrogen (TN) and total phosphorus (TP) concentrations (Figure 3) were determined as part of a long-term monitoring program using previously published analytical methods [67]. Discharge data (Figure 4) were obtained from the United States Geological Survey (USGS) gaging station at site 02,037,500 (37.563055, −77.547222), which is adjacent the HUG sampling site. Precipitation data (Figure 5) were obtained from the National Climatic Data Center using the Richmond International Airport site, “KRIC”.
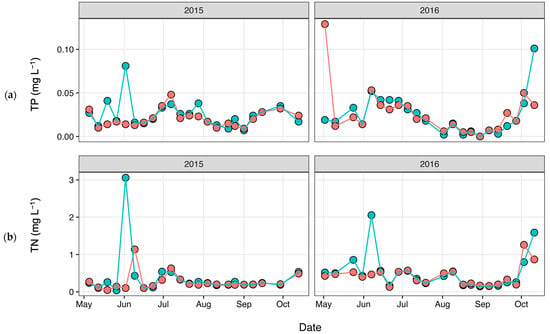
Figure 3.
(a) Total phosphorus (TP) and (b) total nitrogen (TN) concentrations measured during each sampling event, with pink corresponding to HUG and blue corresponding to CSO.
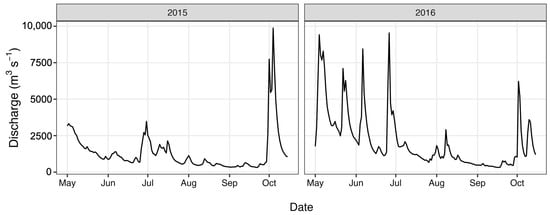
Figure 4.
River discharge values during the study period.
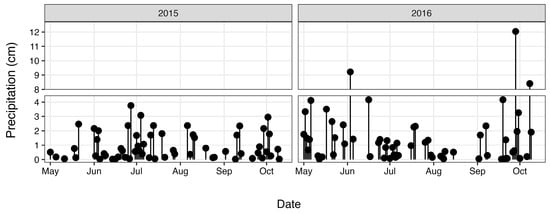
Figure 5.
Precipitation during the study period.
4.4. DNA Extractions
Extractions were performed using the PowerWater DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA, USA). The following modifications were made to the manufacturer’s protocol to increase extraction efficiency. First, each filter was torn into small pieces using sterile forceps prior to being inserted into the PowerWater Bead tube. Next, to minimize DNA shearing, all vortex speeds were reduced to the lowest possible speed that still allowed for mixing. The incubation step for the removal of non-DNA organic and inorganic matter was extended to 10 min. Lastly, the elution step, normally one 100 µL elution with no incubation, was divided into two 50 µL elutions with an additional 5-min incubation at room temperature before each centrifugation. Successful extraction was determined using agarose gel electrophoresis (1.5%) and ethidium bromide staining, and DNA concentration was measured using Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA).
4.5. Quantitative Polymerase Chain Reaction (qPCR)
Total bacterial abundance was determined by quantifying the 16S rRNA gene with the primers Eub338/Eub518 following previously reported reaction conditions [68]. We quantified the abundance of five ARGs: tetO and tetW for resistance to tetracyclines, blaTEM and ampC for β-lactam antibiotics, and qnrA for quinolones. These ARGs confer resistance to some the top prescribed antibiotics in the United States [69], and bacteria with resistance to the respective antibiotics have previously been detected at the two sites [12]. Each qPCR run included an appropriate standard curve that covered at least eight orders of magnitude, with the lowest starting point being 88 (tetO), 32 (tetW), 44 (blaTEM), 51 (ampC), and 156 (qnrA) gene copies per reaction. For the 16S rRNA gene assay, the curve was constructed using genomic DNA extracted from E. coli Strain NCTC 9001 (ATCC, Manassas, Virginia, USA). For the ARGs, standard curves were constructed using plasmid DNA extracted using the Zyppy Plasmid Miniprep Kit (Zymo Research Corp, Irvine, California, USA). Sources were: SpyTag-β-Lactamase-Spycatcher (pET28a) (Addgene, Cambridge, Massachusetts, USA) for blaTEM; pTrcHis + qnrA in E. coli J53 (Thermo Fisher Scientific, Waltham, Massachusetts, USA), and pCR®2.1-TOPO + either tetW, tetO, or ampC in DH5α E. coli (Invitrogen, Carlsbad, CA, USA). Each qPCR run also included two types of negative control: (i) a “negative template control”, which replaced sample DNA with nuclease-free water; and (ii) a “gene-free control”, which replaced sample DNA with genomic DNA from Methanococcus voltae (DSM #1537, DSMZ, Braunschweig, Germany).
Each qPCR assay was performed in triplicate (qnrA performed in quadruplicate) using a CFX384 Real-Time System (Bio-Rad, Hercules, CA, USA) and Bio-Rad SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). Reaction mixtures (15 µL) also contained 5 ng of template DNA and the appropriate concentration of forward and reverse primer (Integrated DNA Technologies, Coralville, IA, USA). Reaction conditions are presented in Table 2. A melt curve and agarose gel electrophoresis were conducted to verify the specificity of the amplified products. Amplification efficiencies ranged from 93–103% and all r2 > 0.98. Gene abundance data are reported as copies mL−1 of river water.

Table 2.
Primers and reaction conditions for qPCR assays.
4.6. Data Analyses
All statistical tests and visualizations were performed using R (version 4.2.2) [75]. Non-parametric approaches were employed because ARG data were not normally distributed (Shapiro–Wilks tests, all p > 0.05). Alpha of 0.05 was used for all tests except correlations, which used a more conservative value of 0.01 to account for multiple comparisons.
Histograms were used to visualize the ARG data by plotting the fraction of samples (%) in each abundance interval (Figure 1). For simplicity and to facilitate statistical comparisons, data below the detection limit are interpreted as zeros. Mann–Whitney tests were performed to determine whether ARG abundances differed between the upstream site (HUG) and at the city center (CSO). Spearman correlation was then used to identify potential drivers of increased ARG abundance at the CSO site. We specifically considered covariation with bacterial abundance (16s rRNA copies mL−1), level of fecal contamination (E. coli CFU 100 mL−1), river discharge (m3 s−1), precipitation (cm), and nutrient concentrations. For precipitation, we evaluated two timeframes: “sampling-day precipitation” (cumulative rainfall from midnight on sampling day until sampling time (~12 h)) and “prior-day precipitation” (rainfall that fell between 12 and 36 h prior to sampling). For nutrients, we attributed increased concentrations of TN and TP at the CSO site to sewage overflow, and used the difference (Δ) as an indicator of the magnitude of the overflow event.
Author Contributions
Conceptualization, J.C.M. and R.B.F.; methodology, R.B.F.; software, J.C.M.; validation, J.C.M. and R.B.F.; formal analysis, J.C.M. and R.B.F.; investigation, J.C.M. and R.B.F.; resources, R.B.F.; data curation, J.C.M. and R.B.F.; writing—original draft preparation, J.C.M. and R.B.F.; writing—review and editing, J.C.M. and R.B.F.; visualization, J.C.M. and R.B.F.; supervision, R.B.F.; project administration, J.C.M. and R.B.F.; funding acquisition, J.C.M. and R.B.F. All authors have read and agreed to the published version of the manuscript.
Funding
Funding for this research was provided by a VCU Rice Center Student Research Grant, a VCU College of Humanities and Sciences Catalyst Award, and the VCU Breakthroughs Fund.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Raw data are available on request from the corresponding author.
Acknowledgments
We greatly appreciate David Hooper (Harvard University) for providing pTrcHis + qnrA, and Brian Badgley and Michael Strickland (Virginia Tech) for providing pCR®2.1-TOPO + tetW, pCR®2.1-TOPO + tetO, and pCR®2.1-TOPO + ampC for qPCR standards. We also want to thank Ella Balasa and Enjolie Levengood for assisting with qPCR assays, and Paul Bukaveckas and William “Mac” Lee for coordinating sample collection and nutrient data. This paper is contribution # 100 from the VCU Rice Rivers Center.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Zhang, Z.; Zhang, Q.; Wang, T.; Xu, N.; Lu, T.; Hong, W.; Penuelas, J.; Gillings, M.; Wang, M.; Gao, W.; et al. Assessment of Global Health Risk of Antibiotic Resistance Genes. Nat. Commun. 2022, 13, 1553. [Google Scholar] [CrossRef]
- Koch, N.; Islam, N.F.; Sonowal, S.; Prasad, R.; Sarma, H. Environmental Antibiotics and Resistance Genes as Emerging Contaminants: Methods of Detection and Bioremediation. Curr. Res. Microb. Sci. 2021, 2, 100027. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Proia, L.; Anzil, A.; Subirats, J.; Borrego, C.; Farrè, M.; Llorca, M.; Balcázar, J.L.; Servais, P. Antibiotic Resistance along an Urban River Impacted by Treated Wastewaters. Sci. Total Environ. 2018, 628–629, 453–466. [Google Scholar] [CrossRef]
- Thakali, O.; Malla, B.; Tandukar, S.; Sthapit, N.; Raya, S.; Furukawa, T.; Sei, K.; Sherchand, J.B.; Haramoto, E. Release of Antibiotic-Resistance Genes from Hospitals and a Wastewater Treatment Plant in the Kathmandu Valley, Nepal. Water 2021, 13, 2733. [Google Scholar] [CrossRef]
- Thai, P.K.; Ky, L.X.; Binh, V.N.; Nhung, P.H.; Nhan, P.T.; Hieu, N.Q.; Dang, N.T.T.; Tam, N.K.B.; Anh, N.T.K. Occurrence of Antibiotic Residues and Antibiotic-Resistant Bacteria in Effluents of Pharmaceutical Manufacturers and Other Sources around Hanoi, Vietnam. Sci. Total Environ. 2018, 645, 393–400. [Google Scholar] [CrossRef]
- Kotwani, A.; Joshi, J.; Kaloni, D. Pharmaceutical Effluent: A Critical Link in the Interconnected Ecosystem Promoting Antimicrobial Resistance. Environ. Sci. Pollut. Res. 2021, 28, 32111–32124. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. Npj Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Reddy, S.; Kaur, K.; Barathe, P.; Shriram, V.; Govarthanan, M.; Kumar, V. Antimicrobial Resistance in Urban River Ecosystems. Microbiol. Res. 2022, 263, 127135. [Google Scholar] [CrossRef]
- Sowah, R.A.; Molina, M.; Georgacopoulos, O.; Snyder, B.; Cyterski, M. Sources and Drivers of ARGs in Urban Streams in Atlanta, Georgia, USA. Microorganisms 2022, 10, 1804. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcazar, J.L. The Role of Aquatic Ecosystems as Reservoirs of Antibiotic Resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Balasa, G.; Levengood, E.S.; Battistelli, J.M.; Franklin, R.B. Diversity of Multidrug-Resistant Bacteria in an Urbanized River: A Case Study of the Potential Risks from Combined Sewage Overflows. Water 2021, 13, 2122. [Google Scholar] [CrossRef]
- Matsui, K.; Miki, T. Microbial Community Composition and Function in an Urban Waterway with Combined Sewer Overflows before and after Implementation of a Stormwater Storage Pipe. PeerJ 2023, 11, e14684. [Google Scholar] [CrossRef] [PubMed]
- Honda, R.; Tachi, C.; Yasuda, K.; Hirata, T.; Noguchi, M.; Hara-Yamamura, H.; Yamamoto-Ikemoto, R.; Watanabe, T. Estimated Discharge of Antibiotic-Resistant Bacteria from Combined Sewer Overflows of Urban Sewage System. Npj Clean Water 2020, 3, 15. [Google Scholar] [CrossRef]
- Collignon, P.; Beggs, J.J.; Walsh, T.R.; Gandra, S.; Laxminarayan, R. Anthropological and Socioeconomic Factors Contributing to Global Antimicrobial Resistance: A Univariate and Multivariable Analysis. Lancet Planet. Health 2018, 2, e398–e405. [Google Scholar] [CrossRef]
- Di Cesare, A.; Eckert, E.M.; Rogora, M.; Corno, G. Rainfall Increases the Abundance of Antibiotic Resistance Genes within a Riverine Microbial Community. Environ. Pollut. 2017, 226, 473–478. [Google Scholar] [CrossRef]
- Ouyang, W.-Y.; Huang, F.-Y.; Zhao, Y.; Li, H.; Su, J.-Q. Increased Levels of Antibiotic Resistance in Urban Stream of Jiulongjiang River, China. Appl. Microbiol. Biotechnol. 2015, 99, 5697–5707. [Google Scholar] [CrossRef]
- Xu, Y.; Guo, C.; Luo, Y.; Lv, J.; Zhang, Y.; Lin, H.; Wang, L.; Xu, J. Occurrence and Distribution of Antibiotics, Antibiotic Resistance Genes in the Urban Rivers in Beijing, China. Environ. Pollut. 2016, 213, 833–840. [Google Scholar] [CrossRef]
- Zhou, Z.-C.; Zheng, J.; Wei, Y.-Y.; Chen, T.; Dahlgren, R.A.; Shang, X.; Chen, H. Antibiotic Resistance Genes in an Urban River as Impacted by Bacterial Community and Physicochemical Parameters. Environ. Sci. Pollut. Res. 2017, 24, 23753–23762. [Google Scholar] [CrossRef]
- Pan, X.; Chen, L.; Zhang, L.; Zuo, J. Characteristics of Antibiotic Resistance Gene Distribution in Rainfall Runoff and Combined Sewer Overflow. Environ. Sci. Pollut. Res. 2022, 30, 30766–30778. [Google Scholar] [CrossRef]
- Petrie, B. A Review of Combined Sewer Overflows as a Source of Wastewater-Derived Emerging Contaminants in the Environment and Their Management. Environ. Sci. Pollut. Res. 2021, 28, 32095–32110. [Google Scholar] [CrossRef] [PubMed]
- United States Environmental Protection Agency. EPA Report to Congress on Impacts and Control of Combined Sewer Overflows and Sanitary Sewer Overflows; United States Environmental Protection Agency: Washington, DC, USA, 2004.
- Abdellatif, M.; Atherton, W.; Alkhaddar, R.M.; Osman, Y.Z. Quantitative Assessment of Sewer Overflow Performance with Climate Change in Northwest England. Hydrol. Sci. J. 2015, 60, 636–650. [Google Scholar] [CrossRef]
- Li, J.; Li, X.; Liu, H.; Gao, L.; Wang, W.; Wang, Z.; Zhou, T.; Wang, Q. Climate Change Impacts on Wastewater Infrastructure: A Systematic Review and Typological Adaptation Strategy. Water Res. 2023, 120282. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal Pollution Can Explain Antibiotic Resistance Gene Abundances in Anthropogenically Impacted Environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef]
- Zhang, X.; Zhi, X.; Chen, L.; Shen, Z. Spatiotemporal Variability and Key Influencing Factors of River Fecal Coliform within a Typical Complex Watershed. Water Res. 2020, 178, 115835. [Google Scholar] [CrossRef]
- Vitro, K.A.; BenDor, T.K.; Jordanova, T.V.; Miles, B. A Geospatial Analysis of Land Use and Stormwater Management on Fecal Coliform Contamination in North Carolina Streams. Sci. Total Environ. 2017, 603–604, 709–727. [Google Scholar] [CrossRef] [PubMed]
- Weyrauch, P.; Matzinger, A.; Pawlowsky-Reusing, E.; Plume, S.; Von Seggern, D.; Heinzmann, B.; Schroeder, K.; Rouault, P. Contribution of Combined Sewer Overflows to Trace Contaminant Loads in Urban Streams. Water Res. 2010, 44, 4451–4462. [Google Scholar] [CrossRef]
- Wang, J. Combined Sewer Overflows (CSOs) Impact on Water Quality and Environmental Ecosystem in the Harlem River. JEP 2014, 5, 1373–1389. [Google Scholar] [CrossRef]
- Diaz-Fierros, T.F.; Puerta, J.; Suarez, J.; Diaz-Fierros, V.F. Contaminant Loads of CSOs at the Wastewater Treatment Plant of a City in NW Spain. Urban Water 2002, 4, 291–299. [Google Scholar] [CrossRef]
- Damashek, J.; Westrich, J.R.; McDonald, J.M.B.; Teachey, M.E.; Jackson, C.R.; Frye, J.G.; Lipp, E.K.; Capps, K.A.; Ottesen, E.A. Non-Point Source Fecal Contamination from Aging Wastewater Infrastructure Is a Primary Driver of Antibiotic Resistance in Surface Waters. Water Res. 2022, 222, 118853. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Zhang, T.; Fang, H.H.P. Antibiotic Resistance Genes in Water Environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef]
- Patterson, A.J.; Rincon, M.T.; Flint, H.J.; Scott, K.P. Mosaic Tetracycline Resistance Genes Are Widespread in Human and Animal Fecal Samples. Antimicrob. Agents Chemother. 2007, 51, 1115–1118. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline Resistance Genes in Activated Sludge Wastewater Treatment Plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef]
- Pruden, A.; Larsson, D.G.J.; Amézquita, A.; Collignon, P.; Brandt, K.K.; Graham, D.W.; Lazorchak, J.M.; Suzuki, S.; Silley, P.; Snape, J.R.; et al. Management Options for Reducing the Release of Antibiotics and Antibiotic Resistance Genes to the Environment. Environ. Health Perspect. 2013, 121, 878–885. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, M.; Laird, E.; Gentry, T.J.; Brooks, J.P.; Karthikeyan, R. Increased Antimicrobial and Multidrug Resistance Downstream of Wastewater Treatment Plants in an Urban Watershed. Front. Microbiol. 2021, 12, 657353. [Google Scholar] [CrossRef] [PubMed]
- Keely, S.P.; Brinkman, N.E.; Wheaton, E.A.; Jahne, M.A.; Siefring, S.D.; Varma, M.; Hill, R.A.; Leibowitz, S.G.; Martin, R.W.; Garland, J.L.; et al. Geospatial Patterns of Antimicrobial Resistance Genes in the US EPA National Rivers and Streams Assessment Survey. Environ. Sci. Technol. 2022, 56, 14960–14971. [Google Scholar] [CrossRef]
- Thakali, O.; Tandukar, S.; Brooks, J.; Sherchan, S.; Sherchand, J.; Haramoto, E. The Occurrence of Antibiotic Resistance Genes in an Urban River in Nepal. Water 2020, 12, 450. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Wang, S.; Gao, X.; Gao, Y.; Li, Y.; Cao, M.; Xi, Z.; Zhao, L.; Feng, Z. Tetracycline Resistance Genes Identified from Distinct Soil Environments in China by Functional Metagenomics. Front. Microbiol. 2017, 8, 1406. [Google Scholar] [CrossRef]
- Daghrir, R.; Drogui, P. Tetracycline Antibiotics in the Environment: A Review. Environ. Chem. Lett. 2013, 11, 209–227. [Google Scholar] [CrossRef]
- Barr, B.S.; Waldridge, B.M.; Morresey, P.R.; Reed, S.M.; Clark, C.; Belgrave, R.; Donecker, J.M.; Weigel, D.J. Antimicrobial-Associated Diarrhoea in Three Equine Referral Practices: Antimicrobial-Associated Diarrhoea in Three Equine Referral Practices. Equine Vet. J. 2013, 45, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Rule, E.K.; Boyle, A.G.; Redding, L.E. Antimicrobial Prescribing Patterns in Equine Ambulatory Practice. Prev. Vet. Med. 2021, 193, 105411. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Cao, J.; Zhu, Y.; Chen, Q.; Shen, F.; Wu, Y.; Xu, S.; Fan, H.; Da, G.; Huang, R.; et al. Global Survey of Antibiotic Resistance Genes in Air. Environ. Sci. Technol. 2018, 52, 10975–10984. [Google Scholar] [CrossRef]
- Bajaj, P.; Singh, N.S.; Kanaujia, P.K.; Virdi, J.S. Distribution and Molecular Characterization of Genes Encoding CTX-M and AmpC β-Lactamases in Escherichia Coli Isolated from an Indian Urban Aquatic Environment. Sci. Total Environ. 2015, 505, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Lachmayr, K.L.; Kerkhof, L.J.; DiRienzo, A.G.; Cavanaugh, C.M.; Ford, T.E. Quantifying Nonspecific TEM β-Lactamase (BlaTEM) Genes in a Wastewater Stream. Appl. Environ. Microbiol. 2009, 75, 203–211. [Google Scholar] [CrossRef]
- Coertze, R.D.; Bezuidenhout, C.C. Detection and Quantification of Clinically Relevant Plasmid-Mediated AmpC Beta-Lactamase Genes in Aquatic Systems. Water Supply 2020, 20, 1745–1756. [Google Scholar] [CrossRef]
- Coertze, R.D.; Bezuidenhout, C.C. Relating the Prevalence of Plasmid-Mediated AmpC Beta-Lactamase Genes to Aquatic Environmental Factors. Sci. Total Environ. 2021, 763, 144119. [Google Scholar] [CrossRef]
- Martínez, J.L.; Coque, T.M.; Baquero, F. What Is a Resistance Gene? Ranking Risk in Resistomes. Nat. Rev. Microbiol. 2015, 13, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Coertze, R.D.; Bezuidenhout, C.C. Global Distribution and Current Research of AmpC Beta-Lactamase Genes in Aquatic Environments: A Systematic Review. Environ. Pollut. 2019, 252, 1633–1642. [Google Scholar] [CrossRef]
- Song, M.; Song, D.; Jiang, L.; Zhang, D.; Sun, Y.; Chen, G.; Xu, H.; Mei, W.; Li, Y.; Luo, C.; et al. Large-Scale Biogeographical Patterns of Antibiotic Resistome in the Forest Soils across China. J. Hazard. Mater. 2021, 403, 123990. [Google Scholar] [CrossRef]
- Scott, L.C.; Lee, N.; Aw, T.G. Antibiotic Resistance in Minimally Human-Impacted Environments. Int. J. Environ. Res. Public Health 2020, 17, 3939. [Google Scholar] [CrossRef] [PubMed]
- Adachi, F.; Yamamoto, A.; Takakura, K.-I.; Kawahara, R. Occurrence of Fluoroquinolones and Fluoroquinolone-Resistance Genes in the Aquatic Environment. Sci. Total Environ. 2013, 444, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Golovko, O.; Kumar, V.; Fedorova, G.; Randak, T.; Grabic, R. Seasonal Changes in Antibiotics, Antidepressants/Psychiatric Drugs, Antihistamines and Lipid Regulators in a Wastewater Treatment Plant. Chemosphere 2014, 111, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Shi, Y.; Li, W.; Niu, H.; Liu, J.; Cai, Y. Occurrence of Antibiotics in Eight Sewage Treatment Plants in Beijing, China. Chemosphere 2012, 86, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Janecko, N.; Pokludova, L.; Blahova, J.; Svobodova, Z.; Literak, I. Implications of Fluoroquinolone Contamination for the Aquatic Environment-A Review: Fluoroquinolone in the Aquatic Ecosystem-A Review. Environ. Toxicol. Chem. 2016, 35, 2647–2656. [Google Scholar] [CrossRef]
- Virginia Geographic Information Network. 2016 Download: Land Cover Dataset Download Application. Available online: https://www.arcgis.com/home/item.html?id=d3d51bb5431a4d26a313f586c7c2c848 (accessed on 1 July 2023).
- Maddox, T.W.; Clegg, P.D.; Williams, N.J.; Pinchbeck, G.L. Antimicrobial Resistance in Bacteria from Horses: Epidemiology of Antimicrobial Resistance. Equine Vet. J. 2015, 47, 756–765. [Google Scholar] [CrossRef]
- Dolejska, M.; Duskova, E.; Rybarikova, J.; Janoszowska, D.; Roubalova, E.; Dibdakova, K.; Maceckova, G.; Kohoutova, L.; Literak, I.; Smola, J.; et al. Plasmids Carrying BlaCTX-M-1 and Qnr Genes in Escherichia Coli Isolates from an Equine Clinic and a Horseback Riding Centre. J. Antimicrob. Chemother. 2011, 66, 757–764. [Google Scholar] [CrossRef]
- Lupo, A.; Haenni, M.; Saras, E.; Gradin, J.; Madec, J.-Y.; Börjesson, S. Is Bla CTX-M-1 Riding the Same Plasmid Among Horses in Sweden and France? Microb. Drug Resist. 2018, 24, 1580–1586. [Google Scholar] [CrossRef]
- Schmiedel, J.; Falgenhauer, L.; Domann, E.; Bauerfeind, R.; Prenger-Berninghoff, E.; Imirzalioglu, C.; Chakraborty, T. Multiresistant Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae from Humans, Companion Animals and Horses in Central Hesse, Germany. BMC Microbiol. 2014, 14, 187. [Google Scholar] [CrossRef]
- Li, J.; Wang, T.; Shao, B.; Shen, J.; Wang, S.; Wu, Y. Plasmid-Mediated Quinolone Resistance Genes and Antibiotic Residues in Wastewater and Soil Adjacent to Swine Feedlots: Potential Transfer to Agricultural Lands. Environ. Health Perspect. 2012, 120, 1144–1149. [Google Scholar] [CrossRef]
- FDA. Antimicrobials Sold or Distributed for Use in Food-Producing Animals; U.S. Food & Drug Administration: Center for Veterinary Medicine: Silver Spring, MD, USA, 2021.
- Stanton, I.C.; Murray, A.K.; Zhang, L.; Snape, J.; Gaze, W.H. Evolution of Antibiotic Resistance at Low Antibiotic Concentrations Including Selection below the Minimal Selective Concentration. Commun. Biol. 2020, 3, 467. [Google Scholar] [CrossRef] [PubMed]
- Turiel, E.; Martín-Esteban, A.; Bordin, G.; Rodríguez, A.R. Stability of Fluoroquinolone Antibiotics in River Water Samples and in Octadecyl Silica Solid-Phase Extraction Cartridges. Anal. Bioanal. Chem. 2004, 380. [Google Scholar] [CrossRef]
- EPA. Method 1603: Escherichia Coli (E. Coli) in Water by Membrane Filtration Using Modified Membrane-Thermotolerant Escherichia Coli Agar (Modified MTEC); United States Environmental Protection Agency: Washington, DC, USA, 2009.
- Bukaveckas, P.A.; Barry, L.E.; Beckwith, M.J.; David, V.; Lederer, B. Factors Determining the Location of the Chlorophyll Maximum and the Fate of Algal Production within the Tidal Freshwater James River. Estuaries Coasts 2011, 34, 569–582. [Google Scholar] [CrossRef]
- Morina, J.C.; Morrissey, E.M.; Franklin, R.B. Vegetation Effects on Bacteria and Denitrifier Abundance in the Soils of Two Tidal Freshwater Wetlands in Virginia. Appl. Environ. Soil. Sci. 2018, 2018, 1–10. [Google Scholar] [CrossRef]
- CDC. Outpatient Antibiotic Prescriptions—United States, 2015; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2015. [Google Scholar]
- Thames, C.H.; Pruden, A.; James, R.E.; Ray, P.P.; Knowlton, K.F. Excretion of Antibiotic Resistance Genes by Dairy Calves Fed Milk Replacers with Varying Doses of Antibiotics. Front. Microbio. 2012, 3, 139. [Google Scholar] [CrossRef]
- Aminov, R.I.; Garrigues-Jeanjean, N.; Mackie, R.I. Molecular Ecology of Tetracycline Resistance: Development and Validation of Primers for Detection of Tetracycline Resistance Genes Encoding Ribosomal Protection Proteins. Appl. Environ. Microbiol. 2001, 67, 22–32. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcázar, J.L. Bacteriophages as a Reservoir of Extended-Spectrum β -Lactamase and Fluoroquinolone Resistance Genes in the Environment. Clin. Microbiol. Infect. 2014, 20, O456–O459. [Google Scholar] [CrossRef]
- Shi, P.; Jia, S.; Zhang, X.-X.; Zhang, T.; Cheng, S.; Li, A. Metagenomic Insights into Chlorination Effects on Microbial Antibiotic Resistance in Drinking Water. Water Res. 2013, 47, 111–120. [Google Scholar] [CrossRef]
- Marti, E.; Balcázar, J.L. Real-Time PCR Assays for Quantification of Qnr Genes in Environmental Water Samples and Chicken Feces. Appl. Environ. Microbiol. 2013, 79, 1743–1745. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 3 March 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).