Abstract
Polymicrobial biofilms provide a complex environment where co-infecting microorganisms can behave antagonistically, additively, or synergistically to alter the disease outcome compared to monomicrobial infections. Staphylococcus aureus skin and soft tissue infections (Sa-SSTIs) are frequently reported in healthcare and community settings, and they can also involve other bacterial and fungal microorganisms. This polymicrobial aetiology is usually found in chronic wounds, such as diabetic foot ulcers, pressure ulcers, and burn wounds, where the establishment of multi-species biofilms in chronic wounds has been extensively described. This review article explores the recent updates on the microorganisms commonly found together with S. aureus in SSTIs, such as Pseudomonas aeruginosa, Escherichia coli, Enterococcus spp., Acinetobacter baumannii, and Candida albicans, among others. The molecular mechanisms behind these polymicrobial interactions in the context of infected wounds and their impact on pathogenesis and antimicrobial susceptibility are also revised.
1. Introduction
Skin and soft tissue infections (SSTIs) comprise a group of infections that affect the skin and underlying subcutaneous tissue, fascia, or muscle. These infections can vary in severity, ranging from superficial infections of mild to moderate severity to deeper necrotizing infections [1,2]. SSTIs have significant global impact, increasing hospitalizations, length of stay, and mortality [3,4,5].
Several classifications can be adopted for SSTIs, depending on specific variables such as anatomical localization, etiological agent(s), skin extension, progression rate, clinical presentation, and severity [6,7,8,9]. The Infectious Diseases Society of America (IDSA) classification is based on three different distinctions: (i) skin extension, uncomplicated, typically superficial infections, and complicated infection, basing the latter definition for those reaching deep structures of the skin; (ii) rate of progression, acute wound infections (traumatic, bite-related, postoperative) and chronic wound infections (diabetic foot infections, venous stasis ulcers, pressure sores); (iii) tissue necrosis, necrotizing (fascitis, myonecrosis, gangrena) and non-necrotizing infections [7]. There is one last criterion that allows the differentiation of SSTIs as monomicrobial and polymicrobial [3]. Especially those infections with a long lasting or chronic course can be sustained by multiple microbial species [10].
All these classifications include patients with the following clinical entities: (i) cellulitis/erysipelas, defined as a skin infection characterised by spreading areas of redness, edema, or induration; (ii) wound infection, characterised by purulent drainage from a wound with surrounding redness, edema, or induration; and (iii) major cutaneous abscess characterised by a collection of pus within the dermis or deeper that is accompanied by redness, edema, or induration [3].
Staphylococcus aureus, an opportunistic Gram-positive pathogen, is a common cause of SSTIs, ranging from the benign (e.g., impetigo and uncomplicated cellulitis) to the immediately life-threatening [3,11]. Among S. aureus strains, methicillin-resistant S. aureus (MRSA) isolates are of particular concern because they can also exhibit concomitant resistance to many commonly used antibiotics. The specific multidrug-resistant pattern of MRSA can vary depending of the geographic location and includes resistance to macrolides (erythromycin and clarithromycin), lincosamides (clindamycin), aminoglycosides (gentamicin), tetracyclines (tetracycline and doxycycline), and fluoroquinolones (ciprofloxacin) [12,13,14].
S. aureus expresses several factors that facilitate skin colonization and infection. These include various toxins and immune evasion factors, and a large array of protein and non-protein factors that enable host colonisation during infection [15,16]. S. aureus avoids being eliminated by neutrophils on many levels that include: (i) the inhibition of neutrophil extravasation from the bloodstream into the tissues, neutrophil activation, and chemotaxis, (ii) inhibition of phagocytosis by aggregation, protective surface structures, and biofilm formation, (iii) inhibition of opsonisation, (iv) inhibition of neutrophil killing mechanisms, and (v) direct elimination of neutrophils by cytolytic toxins or triggering of apoptosis [16,17].
Furthermore, biofilm formation has been postulated as a common behaviour of S. aureus isolates from skin infections [18,19,20,21]. Biofilms pose a significant challenge in the treatment of these infections due to their unique characteristics and the protective environment they create. In this regard, biofilms show increased resistance to host immunity and increased tolerance to antibiotics compared to their planktonic counterparts [22,23].
Polymicrobial SSTIs involving S. aureus have been reported (Sa-SSTIs) [10,11,24,25,26]. Most mixed-species SSTIs are associated with chronicinfections such as diabetic foot infections (DFIs), pressure ulcers infection, and burn infection, among others [10]. These chronic wounds can commonly become infected with polymicrobial biofilms containing bacterial and fungal microorganisms [27,28]. Mixed biofilm communities provide a complex environment in which a variety of interactions may occur, ranging from cooperative interactions to antagonism [29,30]. The polymicrobial interactions in wounds may help the partner species to establish and infect the tissues. It has been reported that the high polymicrobial load in wounds delays the wound closure and favours the emergence of antibiotic-resistant strains compared to the single-species biofilms [31,32]. Understanding the microbial species involved, predisposing factors of the disease progression, and the polymicrobial interaction between microorganisms is essential for diagnosing and developing treatment strategies.
This review article explores the recent updates on the microorganisms commonly found together with S. aureus in SSTIs, such as Pseudomonas aeruginosa, Escherichia coli, Enterococcus spp., Acinetobacter baumannii, and Candida albicans. The molecular mechanisms behind these polymicrobial interactions and their impact on pathogenesis and antimicrobial susceptibility are also revised.
2. Occurrence of PolymicrobialSSTI Associated with S. aureus (Sa-SSTIs)
Infections with a long-lasting or chronic course are usually sustained by multiple microbial aetiologies [30,33,34,35]. In this regard, polymicrobial SSTIs are usually observed for diabetic foot ulcers, pressure ulcers, and burn wounds [3,10]. Microbiological assessment of polymicrobial SSTIs, performed by standard culturing techniques or molecular methods, can be challenging [3,36,37]. Culture-dependent techniques are biasedtoward those microorganisms that develop well under laboratory conditions, and might inadequately represent fungal and bacterial communities in chronic wounds [20,38]. On the other hand, culture-independent, amplicon-based sequencing methods (i.e., bacterial and fungal 16S rRNA gene sequencing) have the major limitation of failing to distinguish individual species [21,39]. Recently, exhaustive strain-level classification of microbial communities has been achieved by shotgun metagenomic sequencing [40]. Consequently, a combinationof metagenomic approach and culturing methods seems to be more adequate to identify the complex microbial communities formed in chronic wounds [41].
The microbiology of SSTIs shows that S. aureus is a frequent aetiology, with a high incidence of MRSA [2,8]. Cumulative data indicate that up to 70% of Sa-SSTIs are polymicrobial [2,10,25]. As mentioned early, mixed-species SSTIs are usually associated with chronic infections such as diabetic foot infections (DFIs), pressure ulcers infection, and burn infections [3].
2.1. Diabetic Foot Infections (DFIs)
Diabetic foot ulcers are a common complication of diabetic patients affecting their lower extremities; these wounds occur due to a combination of factors such as reduced blood flow and nerve damage (neuropathy) [42,43]. Although usually initially characterised as acute wounds, their inability to progress through the healing stages converts them into chronic wounds [11].
Diabetic foot ulcers are highly susceptible to infections due to several reasons: (i) diabetes can cause peripheral neuropathy, which damages the nerves in the feet, making it difficult for the patient to notice a foot ulcer infection developing; (ii) compromised blood circulation that impairs the immune cells to reach the wound efficiently; and (iii) prolonged healing due to the underlying complications mentioned before, which provides an extended window of opportunity for bacteria to multiply and establish an infection. Once an ulcer becomes infected, the bacteria can spread through the tissues, leading to cellulitis, abscess formation, osteomyelitis (infection of the bone), or systemic infection if left untreated. In severe cases, the infection can progress to a point where amputation becomes necessary [44].
Chronic diabetic foot ulcers usually become infected with bacterial biofilms, which constitute a significant factor contributing to the severity and delayed healing of diabetic foot infections (DFIs) [20]. Diabetic foot ulcers are typically colonised with skin commensal bacteria establishing biofilms that increase their microbial diversity over time and with progression of the ulcer [21,45]. Some common microorganisms associated with DFIs are Staphylococcus spp., Corynebacterium spp., and P. aeruginosa [26,46]; however, these infections involve a great diversity of microbes. The microorganisms reported to co-exist with S. aureus in polymicrobial DFIs are mainly gram negative bacteria: P. aeruginosa, Acinetobacter spp., Escherichia coli, Enterobacter spp., Citrobacter spp., Proteus spp., Klebsiella spp. In addition, Gram-positive Enterococcus spp. have also been reported to co-occur with S. aureus in DFIs (Table 1) [10,27].

Table 1.
Microbial species in polymicrobial Sa-SSTIs.
2.2. Pressure Ulcer Infections
Pressure ulcers (PUs) are injuries to the skin and underlying tissue resulting from ischemia caused by prolonged pressure on the skin [28]. PUs can affect any part of the body that is put under pressure. They are most common on bony parts of the body, such as the heels, elbows, hips, and base of the spine [53]. PUs are a significant health problem worldwide that commonly occurs among inpatients and elderly people with physical-motor limitations. The overall prevalence of pressure ulcers in hospitalised patients has been estimated to range from 5% to 15% but may be significantly higher in intensive care units and certain long-term care settings [54,55].
PUs are typically categorised into stages based on their severity: (i) Stage 1, the skin is intact, but there may be non-blanchable erythema; (ii) Stage 2, partial-thickness skin loss with exposed dermis; (iii) Stage 3, full-thickness skin loss: (iv) Stage 4, full-thickness skinand tissue loss; (v) unstageable pressure injury, obscured full-thickness skinand tissue loss; (vi) deep tissue pressure injury, persistent nonblanchable deep red, maroon, or purple discoloration [54,55].
These wounds are frequently exacerbated by the presence of bacteria and advanced stages of PUs are described to be polymicrobial and linked with biofilm-associated infections [28,29]. The most common organisms identified in PUs are S. aureus, Proteus mirabilis, P. aeruginosa, and E. faecalis [53]. In chronically infected PUs, S. aureus has been found together with P. aeruginosa, E. coli, P. mirabilis, Enterobacter cloacae, and E. faecalis (Table 1) [29,47,48].
2.3. Burn Wound Infections
Burn wounds refer to injuries that result from exposure to heat, chemicals, electricity, or radiation, and they are considered a public health issue all over the world, especially in low- or middle-income countries [56,57]. Burn wounds can vary in severity and are typically classified, based on the depth and extent of tissue damage, as follows: (i) first-degree burns, superficial burns, called erythema, that only affect the epidermis; (ii) second-degree burns, partial-thickness superficial burns where the epidermis and the dermis are damaged; (iii) third-degree burns, full-thickness deep burns that affect all layers of the skin, including the subcutaneous tissue and the muscle; (iv) fourth-degree burns, full-thickness burns including deeper lying tissues such as muscles, tendons, or bones [58].
Burn wounds are particularly susceptible to infections because the damaged skin provides an entry point for microbes, including bacteria and fungi. Microbial infections in burn wound patients are difficult to control; moreover, biofilm formation in burns is a major concern [49,59]. Some of the bacteria commonly found in chronic burn wound infections are P. aeruginosa, S. aureus, Streptococcus spp., Klebsiella spp., Enterococcus spp., and E. coli. In addition, the most prevalent fungi are Aspergillus niger and Candida spp. [60]. In particular, chronic burn wounds co-infected by S. aureus/P. aeruginosa and S. aureus/C. albicans have been widely reported (Table 1) [49,50,51,52].
3. Implications of Polymicrobial Interactionson Infection Outcome
Polymicrobial infections, which are being recognised with increasing frequency, can occur in various parts of the body including the oral cavity, respiratory tract, urinary tract, skin, and wounds [30,61,62]. The presence of multiple microbial species in a polymicrobial infection can lead to several challenges in diagnosis, treatment, and management [61,63,64]. This is partly because infectious polymicrobial communities are often found to be more resistant to antibiotics than their mono-culture counterparts [65,66].
A polymicrobial biofilm is a complex community of microorganisms (fungi, bacteria, and viruses) that adhere to a biotic or abiotic surface, and it is embedded in a self- and/or host-derived hydrated matrix, often consisting of polysaccharides, proteins, and extracellular DNA [30,67]. Biofilm formation involves a series of steps: aggregation or attachment of cells to a surface, growth of the cells into a sessile biofilm colony, and detachment of the cells from the colony into the surrounding medium [22,68]. Because of the large variety and concentration of microbes present in polymicrobial biofilms, each of these stages can be shaped by species-specific physical and chemical interactions, ranging from cooperative relationships to microbial competition [34,69].
3.1. Beneficial Interactions
In polymicrobial biofilms, the synergism and cooperation between microbial species are important to keep the coexistence of different microorganism, outcompeting possible mutual antagonistic effects [70,71].
A behaviour that helps to promote multispecies coexistence within a biofilm occurs when microbes initiate cohesion and coaggregation by producing several adhesion molecules that induce intercellular interactions. Coaggregation has been very well studied in the oral biofilm–dental plaque, and it can involve fimbriae, other surface proteins with adhesive properties, and extracellular polysaccharides; for example, the short fimbriae of Porphyromonasgingivalis play a role in coadhesion with Streptococcus gordonii [72,73].
Another important cooperative strategy is related to metabolic interactions, such as cross-feeding. This occurs when different strains have access to distinctive nutrient substrates, and the product of one strain’s metabolism can be utilised in the nutrition of another. An example is that of Aggregatibacter actinomycetemcomitans and Streptococcus gordonii, bacteria isolated from the human oral cavity. It has been shown that Streptococcus gordonii can secrete lactate as a metabolic byproduct, and this lactate is used as a preferred carbon source by Aggregatibacter actinomycetemcomitans, favouring its growth [74]. Cross-feeding is beneficial because it gives single or multispecies biofilm systems higher metabolic efficiency that can better support the growth of the microorganisms [71,75].
Quorum sensing, a type of cell signalling related to the ability to detect and respond to cell population density by gene regulation, is important in biofilm formation and interspecies communication. Quorum sensing acts through small diffusible signal molecules (autoinducers) that have been implicated in interspecies cooperation [71]. In this regard, Autoinducer 2 (AI-2) produced by Enterococcus faecalis promotes collective behaviours of Escherichia coli at lower cell densities, enhancing autoaggregation of E. coli but also leading to chemotaxis-dependent coaggregation between the two species [76].
In addition, within biofilms, different species of bacteria can use horizontal gene transfer to exchange antibiotic resistant genes, helping the entire community survive antibiotic exposure. The mechanisms of horizontal gene transfer include conjugation, transformation, transduction, membrane vesicles, and gene transfer agents [77,78,79]. It has been widely reported that horizontal gene transfer allows microbes to acquire new sources of antibiotic resistant genes [80]. For example, one study described that a plasmid harboring a carbapenemase resistance gene (blaNDM-1) can be transferred from E. coli to either P. aeruginosa or Acinetobacter baumannii via conjugation within dual-species biofilms [81].
Finally, microorganisms in polymicrobial biofilms may benefit each other by secreting certain beneficial molecules, such as enzymes that inactivate detrimental agents. In this context, studies of polymicrobial biofilms related to otitis media evidenced that beta-lactamase production by Moraxella catarrhalis provides passive protection to Streptococcus pneumoniae from beta-lactam antibiotic killing [82].
3.2. Competitive Interactions
Bacteria in mixed-species biofilms have to coexist and compete for limited space and nutrients. Competition between species appears to define the interactions that predominate in microbial communities [83]. Competition is categorized into two modes, exploitative and interference. Exploitative competition refers to indirect interactions between organisms, by which one organism prevents access to and/or limits the use of resources by another organism whereas interference competition is related to the production of antagonistic factors to impede competitors [84,85,86].
In biofilms, bacteria live under severe environmental conditions, characterized by low nutrient concentrations and low rates of gas renewal or exchanges [68]. Due to the requirements for limited nutrients, different bacterial species compete for nutrients to survive. Competition for iron has been widely observed and is related to the production of iron-chelating molecules (siderophores) by microorganisms [69]. For example, iron competition has been postulated to modulate bacterial composition of dual-species biofilms formed by uropathogenic Klebsiella pneumoniae and E. coli strains, promoting K. pneumoniae growth to the detriment of E. coli [87]. Moreover, oxygen competition has been described between aerobic microorganisms growing in polymicrobial biofilm pellicles at the air liquid interface. For instance, the facultative aerobe Pseudoxanthomonas outcompeted the obligate aerobe Brevibacillus in dual-species pellicles through severe competition for oxygen [88].
In addition, metabolic byproducts generated by one microorganism can be toxic for the surrounding organisms; this provides the first one a competitive advantage. In the upper respiratory tract, hydrogen peroxide is a byproduct of the Streptococcus pneumoniae metabolism that diminish cell viability of Neisseria meningitidis and Moraxella catarrhalis [89].
Bacterial competition can also be driven by the production of small antimicrobial compounds, such as colicins, microcins, and bacteriocins. For example, Streptococcus salivarius in the oral cavity secretes bacteriocins that inhibit several Gram-positive pathogens, such as Streptococcus pneumoniae [90].
Contact-dependent growth inhibition mediated by the type 6 secretion system (T6SS) is able to inject a toxic molecule into other competitor bacteria. In this regard, T6SS of Burkholderia thailandensis conferred an ecological advantage to this species in mixed biofilms because it protected B. thailandensis from invasion by other competitor species, for example, Pseudomonas putida [91].
Finally, interference competition may occur by alteration of biofilm development. Bacteria can use several biofilm-inhibiting strategies including: (i) quorum sensing inhibition as a result of degradation of quorum sensing molecules or by blocking its synthesis [92,93], (ii) inhibition of adhesion by modifying the surface with biosurfactants or by down-regulating adhesion molecules [94,95], (iii) matrix degradation caused by secreted enzymes [96,97], and (iv) the induction of biofilm dispersal on the competitor species by secreting specific messenger molecules [98].
4. Interactions between S. aureus and P. aeruginosa
S. aureus and P. aeruginosa are two common microorganisms colonising chronic wounds [50,99,100]. These two organisms, used as model organisms to study polymicrobial interactions, have been shown to display both cooperative and competitive interactions within the wound (Figure 1). The subtle balance between the competitive and cooperative behaviours of S. aureus and P. aeruginosa could be the key to understanding this interspecies relationship.

Figure 1.
Scheme of S. aureus-P. aeruginosa interactions. Coexistence has been observed, with each bacterial species occupying a discrete niche. Competitive interactions mediated by secreted P. aeruginosa molecules, such as the siderophores pioverdine and piochelin, have also been reported. Synergistic effects with increasing production of virulence factors also occur.
4.1. Interactions Observed In Vitro in Co-Cultivation Experiments
Several competitive interactions between S. aureus and P. aeruginosa have been observed by performing co-cultivation experiments under standard laboratory conditions. P. aeruginosa excretes several small respiratory toxins that kill or inhibit growth of S. aureus, including pyocyanin that permeates the cells where it produces reactive oxygen species [101,102]; the quorum sensing effector molecule 2-heptyl-4-hydroxyquinoline n-oxide (HQNO) [103]; the LasA protease (also known as staphylolysin) that cleaves the S. aureus peptidoglycan and induces its lysis [104]; rhamnolipids, which present antiadhesive and dispersing properties on S. aureus biofilms [105,106]; and the iron-chelating siderophores pyoverdine and pyochelin [107].
In response to this antagonistic attack, S. aureus reduces its metabolism, favouring small-colony variant selection as a survival strategy [108]. These S. aureus small-colony variants are well known for stable aminoglycoside resistance and persistence in chronic infections [109,110].
In vitro co-cultivation experiments using a wound-like medium demonstrated that the quorum sensing systems of P. aeruginosa are inhibited by the albumin present in the serum; consequently, the bacteria was unable to produce the virulence factors that kill S. aureus such as HQNO. This results in the survival of S. aureus in the presence of P. aeruginosa [111].
4.2. Interactions Observed in Wound Infection Models
In contrast to the reported antagonisms described above, the results obtained in wound infection models showed coexistence between S. aureus and P. aeruginosa. Studies during early stages of wound coinfection evidenced a predominance of S. aureus in non-attached bacterial aggregates and biofilm, favouring the subsequent attachment of P. aeruginosa to human keratinocytes [112]. Moreover, P. aeruginosa promoted S. aureus invasion to these cells. Co-infected keratinocytes showed an intermediate inflammatory response that is in agreement with the maintenance of low-level tissue damage and can be associated with chronicity, prolonged colonisation, and impaired wound repair [112].
In addition, P. aeruginosa showed a higher tolerance to gentamicin in S. aureus/P.aeruginosa polymicrobial infection when compared to mono-infection in a murine chronic wound infection model [113].On the other hand, the presence of P. aeruginosa induced the expression of S. aureus virulence factors alpha-toxin and Panton-Valentine leukocidin in a porcine wound model when compared to infection with S. aureus alone [50]. A recent report showed that S. aureus inactivated the P. aeruginosa-derived siderophore pyochelin via the methyltransferase Spm (staphylococcal pyochelin methyltransferase), increasing S. aureus survival during in vivo competition with P. aeruginosa in a murine wound co-infection model [114]. Furthermore, the secreted P. aeruginosa molecule HQNO induced the production of S. aureus membrane-bound pigment staphyloxanthin (STX), which consequently promotes resistance of both pathogens to innate immune effectors such as hydrogen peroxide [115].
Analysis of chronic wound biopsies suggests that S. aureus and P. aeruginosa occupy distinct niches, albeit separated by a few hundred micrometres [116]. In the same way, using a mouse chronic wound model, it has been observed that S. aureus and P. aeruginosa coexist at high cell densities in murine wounds, establishing a patchy distribution [117,118]. A precise microbial spatial distribution at both the macro (mm)- and micro (μm)-scales was mediated by P. aeruginosa production of the antimicrobial HQNO, while pyocyanin had no impact. This precise spatial structure enhances S. aureus tolerance to aminoglycoside antibiotics but not vancomycin [117]. Pougetet al. found that the percentages of biofilm formation were significantly higher in the mixed S. aureus/P. aeruginosa biofilm compared to those determined for the bacterial species alone and that S. aureus aggregates were located close to the wound surface, whereas P. aeruginosa was located deeper in the wound bed [118].
5. Interactions of S. aureus with Microorganisms other than P. aeruginosa
5.1. S. aureus and Enterococcus faecalis
S. aureus and E. faecalis have been implicated in biofilm-associated infections such as chronic wounds, among others [27,119]. The transfer of vancomycin resistance genes from E. faecalis to S. aureus has been observed in clinical settings [120,121]. Additionally, it has been reported that in combination, these two species act synergistically, producing augmented biofilm biomass (Figure 2) [122]. For this, heme cross-feeding has been reported, and it was postulated to involve gelatinase-mediated heme acquisition by E. faecalis from secreted S. aureus hemoproteins. Heme acquisition by E. faecalis facilitates its oxidative respiration [122].
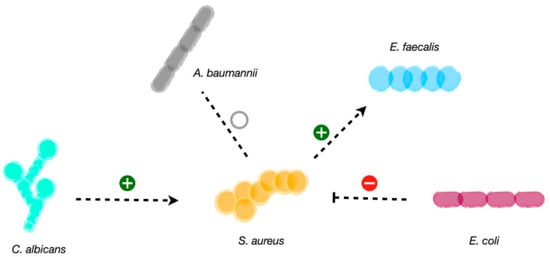
Figure 2.
Scheme of the studied microbial interactions of S. aureus. S. aureus can establish neutral interactions and co-exist with A. baumannii. Competitive interactions have been reported for E. coli on S. aureus trough the genotoxin colibactin. Synergistic interactions occur between S. aureus and E. faecalis, where heme cross-feeding facilitates oxidative respiration in E. faecalis. C. albicans also favors S. aureus proliferation, biofilm formation and virulence factors upregulation.
5.2. S. aureus and Escherichia coli
S. aureus and E. coli are among the most frequent cultured microorganisms from wound infections [12,27,123]. By using a mouse excisional wound model, E. coli was shown to antagonize the growth of S. aureus via the genotoxin colibactin (Figure 2) [124]. The prevalence of polyketide synthase island (pks) in E. coli isolated from human wound swabs was nearly 30% [124]. While the mechanism for colibactin release from E. coli or penetration into target cells is not known, it has been shown that the colibactin intermediate N-myristoyl-D-Asn (NMDA) is able to disrupt the S. aureus membrane [124]. Moreover, during interspecies competition, the E. coli BarA-UvrY two-component system senses S. aureus and responds by upregulating pks island gene expression [124]. Given that E. coli and S. aureus are co-isolated from wounds, it may be possible that these E. coli strains are unable to express the pks island. Another possibility is related to a spatial segregation within wound biofilms such that colibactin-producing E. coli resides far enough from S. aureus to not be able to affect its viability [125].
5.3. S. aureus and Acinetobacter baumannii
Wound co-infections with S. aureus and A. baumannii are found in clinical settings. It has been reported that clinical strains of S. aureus and A. baumannii that were recovered from the same site of infection (diabetic foot ulcer) exhibit a state of commensalism between the two when co-cultured in vitro, without an effect of one another, whether beneficial or detrimental (Figure 2) [126]. More recently, evidence was published that A. baumannii can sense and respond to molecules secreted by S. aureus, modulating virulence responses, such as motility and biofilm formation [127]. In addition, it has been shown that the fitness requirements of S. aureus in vivowere dramatically changed by co-infection with A. baumannii, with around 50% of the essential genes needed during mono-infection converted to non-essential during co-infection [128].
5.4. S. aureus and Candida Albicans
The mixed species of S. aureus and C. albicans can cause skin infections. An increase in S. aureus proliferation and biofilm formation was observed in S. aureus and C. albicans dual-species culture [129]. According to the transcriptome analysis of the dual-species culture, virulence factors of S. aureus were significantly upregulated. Moreover, the beta-lactams and vancomycin-resistant genes in S. aureus as well as azole-resistant genes in C. albicans were also significantly increased [129].
5.5. S. aureus and Commensal Skin Bacteria
It has been demonstrated that co-infection of S. aureus with commensal skin flora can increase S. aureus virulence. This effect, termed augmentation, has been observed in several infection models, including mouse soft-tissue infection [130]. A natural mix of mammalian skin microflora, as well as isolated Staphylococcus epidermidis or Micrococcus luteus strains, was able to augment S. aureus virulence. Moreover, pathogenesis augmentation could be mediated by particulate cell wall peptidoglycan from a range of Gram-positive species including Staphylococcus epidermidis, Curtobacterium flaccumfaciens, and Bacillus subtilis, reducing the S. aureus infectious dose by over 1000-fold [130]. More recently, in vitro and in vivo studies have evidenced that the molecular basis for augmentation is absorption of reactive oxygen species by augmenting material (peptidoglycan), shielding S. aureus from macrophage-mediated killing [131].
6. Conclusion and Perspectives
Polymicrobial human infections are of significant concern on human health. These infections have been reported to be more tolerant to antibiotics and to cause worse clinical outcomes compared to their single-species counterparts.
S. aureus in polymicrobial infections constitutes a greater medical problem than S. aureus in single-species infections. The complex network of microbial S. aureus partners and their interactions has the potential, through diversity in beneficial and/or competitive crosstalk, to accelerate, delay, or worsen wound healing. Microorganisms coexisting in the same site of infection can alter growth, gene expression, invasion ability, and antimicrobial sensitivity patterns.
Further investigations are required to better understand the multi-species interactions between S. aureus and co-infecting organisms to design appropriate treatment strategies and to improve the management of chronic polymicrobial skin and soft-tissue infections involving S. aureus.
Author Contributions
Conceptualization, F.M. and E.M.G.; original draft preparation, F.M. and E.M.G., review and editing, E.M.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
F.M. is a fellow and E.M.G. is a researcher member of CONICET, Argentina.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rajan, S. Skin and Soft-Tissue Infections: Classifying and Treating a Spectrum. Cleve. Clin. J. Med. 2012, 79, 57–66. [Google Scholar] [CrossRef]
- Esposito, S.; Pagliano, P.; De Simone, G.; Pan, A.; Brambilla, P.; Gattuso, G.; Mastroianni, C.; Kertusha, B.; Contini, C.; Massoli, L.; et al. Epidemiology, Aetiology and Treatment of Skin and Soft Tissue Infections: Final Report of a Prospective Multicentre National Registry. J. Chemother. 2022, 34, 524–533. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Ascione, T.; Pagliano, P. Management of Bacterial Skin and Skin Structure Infections with Polymicrobial Etiology. Expert Rev. Anti. Infect. Ther. 2019, 17, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Poulakou, G.; Lagou, S.; Tsiodras, S. What’s New in the Epidemiology of Skin and Soft Tissue Infections in 2018? Curr. Opin. Infect. Dis. 2019, 32, 77–86. [Google Scholar] [CrossRef]
- Yakupu, A.; Aimaier, R.; Yuan, B.; Chen, B.; Cheng, J.; Zhao, Y.; Peng, Y.; Dong, J.; Lu, S. The Burden of Skin and Subcutaneous Diseases: Findings from the Global Burden of Disease Study 2019. Front. Public Health 2023, 11, 1145513. [Google Scholar] [CrossRef] [PubMed]
- Bus, S.A.; Van Netten, J.J.; Hinchliffe, R.J.; Apelqvist, J.; Lipsky, B.A.; Schaper, N.C. Standards for the Development and Methodology of the 2019 International Working Group on the Diabetic Foot Guidelines. Diabetes. Metab. Res. Rev. 2020, 36 (Suppl. 1), e3267. [Google Scholar] [CrossRef]
- Stevens, D.L.; Bisno, A.L.; Chambers, H.F.; Dellinger, E.P.; Goldstein, E.J.C.; Gorbach, S.L.; Hirschmann, J.V.; Kaplan, S.L.; Montoya, J.G.; Wade, J.C. Practice Guidelines for the Diagnosis and Management of Skin and Soft Tissue Infections: 2014 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2014, 59, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Sartelli, M.; Coccolini, F.; Kluger, Y.; Agastra, E.; Abu-Zidan, F.M.; Abbas, A.E.S.; Ansaloni, L.; Adesunkanmi, A.K.; Augustin, G.; Bala, M.; et al. WSES/GAIS/WSIS/SIS-E/AAST Global Clinical Pathways for Patients with Skin and Soft Tissue Infections. World J. Emerg. Surg. 2022, 17, 3. [Google Scholar] [CrossRef]
- Sartelli, M.; Malangoni, M.A.; May, A.K.; Viale, P.; Kao, L.S.; Catena, F.; Ansaloni, L.; Moore, E.E.; Moore, F.A.; Peitzman, A.B.; et al. World Society of Emergency Surgery (WSES) Guidelines for Management of Skin and Soft Tissue Infections. World J. Emerg. Surg. 2014, 9, 57. [Google Scholar] [CrossRef]
- Shettigar, K.; Jain, S.; Bhat, D.V.; Acharya, R.; Ramachandra, L.; Satyamoorthy, K.; Murali, T.S. Virulence Determinants in Clinical Staphylococcus Aureus from Monomicrobial and Polymicrobial Infections of Diabetic Foot Ulcers. J. Med. Microbiol. 2016, 65, 1392–1404. [Google Scholar] [CrossRef]
- Afonso, A.C.; Oliveira, D.; Saavedra, M.J.; Borges, A.; Simões, M. Biofilms in Diabetic Foot Ulcers: Impact, Risk Factors and Control Strategies. Int. J. Mol. Sci. 2021, 22, 8278. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G.J. Staphylococcus Aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Vella, V.; Galgani, I.; Polito, L.; Arora, A.K.; Creech, C.B.; David, M.Z.; Lowy, F.D.; Macesic, N.; Ridgway, J.P.; Uhlemann, A.-C.; et al. Staphylococcus Aureus Skin and Soft Tissue Infection Recurrence Rates in Outpatients: A Retrospective Database Study at 3 US Medical Centers. Clin. Infect. Dis. 2021, 73, e1045–e1053. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.; Frees, D.; Ingmer, H. Antibiotic Resistance and the MRSA Problem. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Olaniyi, R.; Pozzi, C.; Grimaldi, L.; Bagnoli, F. Staphylococcus Aureus-Associated Skin and Soft Tissue Infections: Anatomical Localization, Epidemiology, Therapy and Potential Prophylaxis. Curr. Top. Microbiol. Immunol. 2017, 409, 199–227. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus Aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- de Jong, N.W.M.; van Kessel, K.P.M.; van Strijp, J.A.G. Immune Evasion by Staphylococcus Aureus. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Schilcher, K.; Horswill, A.R. Staphylococcal Biofilm Development: Structure, Regulation, and Treatment Strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Malone, M.; Bjarnsholt, T.; McBain, A.J.; James, G.A.; Stoodley, P.; Leaper, D.; Tachi, M.; Schultz, G.; Swanson, T.; Wolcott, R.D. The Prevalence of Biofilms in Chronic Wounds: A Systematic Review and Meta-Analysis of Published Data. J. Wound Care 2017, 26, 20–25. [Google Scholar] [CrossRef]
- Jnana, A.; Muthuraman, V.; Varghese, V.K.; Chakrabarty, S.; Murali, T.S.; Ramachandra, L.; Shenoy, K.R.; Rodrigues, G.S.; Prasad, S.S.; Dendukuri, D.; et al. Microbial Community Distribution and Core Microbiome in Successive Wound Grades of Individuals with Diabetic Foot Ulcers. Appl. Environ. Microbiol. 2020, 86, e02608-19. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef]
- Lazar, V.; Oprea, E.; Ditu, L.-M. Resistance, Tolerance, Virulence and Bacterial Pathogen Fitness-Current State and Envisioned Solutions for the Near Future. Pathogens 2023, 12, 746. [Google Scholar] [CrossRef]
- Nair, N.; Biswas, R.; Götz, F.; Biswas, L. Impact of Staphylococcus Aureus on Pathogenesis in Polymicrobial Infections. Infect. Immun. 2014, 82, 2162–2169. [Google Scholar] [CrossRef]
- Tanveer, F.; Bhargava, A.; Riederer, K.; Johnson, L.B.; Khatib, R. Low Frequency of Staphylococcus Aureus in Lower Extremity Skin and Soft Tissue Infections. Am. J. Med. Sci. 2018, 356, 528–530. [Google Scholar] [CrossRef]
- Mudrik-Zohar, H.; Carasso, S.; Gefen, T.; Zalmanovich, A.; Katzir, M.; Cohen, Y.; Paitan, Y.; Geva-Zatorsky, N.; Chowers, M. Microbiome Characterization of Infected Diabetic Foot Ulcers in Association With Clinical Outcomes: Traditional Cultures Versus Molecular Sequencing Methods. Front. Cell. Infect. Microbiol. 2022, 12, 836699. [Google Scholar] [CrossRef] [PubMed]
- Sloan, T.J.; Turton, J.C.; Tyson, J.; Musgrove, A.; Fleming, V.M.; Lister, M.M.; Loose, M.W.; Sockett, R.E.; Diggle, M.; Game, F.L.; et al. Examining Diabetic Heel Ulcers through an Ecological Lens: Microbial Community Dynamics Associated with Healing and Infection. J. Med. Microbiol. 2019, 68, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Braga, I.A.; Brito, C.S.; Filho, A.D.; Filho, P.P.G.; Ribas, R.M. Pressure Ulcer as a Reservoir of Multiresistant Gram-Negative Bacilli: Risk Factors for Colonization and Development of Bacteremia. Brazilian J. Infect. Dis. 2017, 21, 171–175. [Google Scholar] [CrossRef]
- Gomes, F.; Furtado, G.E.; Henriques, M.; Sousa, L.B.; Santos-Costa, P.; Bernardes, R.; Apóstolo, J.; Parreira, P.; Salgueiro-Oliveira, A. The Skin Microbiome of Infected Pressure Ulcers: A Review and Implications for Health Professionals. Eur. J. Clin. Investig. 2022, 52, e13688. [Google Scholar] [CrossRef]
- Anju, V.T.; Busi, S.; Imchen, M.; Kumavath, R.; Mohan, M.S.; Salim, S.A.; Subhaswaraj, P.; Dyavaiah, M. Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies. Antibiotics 2022, 11, 1713. [Google Scholar] [CrossRef]
- Metcalf, D.G.; Bowler, P.G. Biofilm Delays Wound Healing: A Review of the Evidence. Burn. Trauma 2013, 1, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Bessa, L.J.; Fazii, P.; Di Giulio, M.; Cellini, L. Bacterial Isolates from Infected Wounds and Their Antibiotic Susceptibility Pattern: Some Remarks about Wound Infection. Int. Wound J. 2015, 12, 47–52. [Google Scholar] [CrossRef]
- Kvich, L.; Burmølle, M.; Bjarnsholt, T.; Lichtenberg, M. Do Mixed-Species Biofilms Dominate in Chronic Infections?-Need for in Situ Visualization of Bacterial Organization. Front. Cell. Infect. Microbiol. 2020, 10, 396. [Google Scholar] [CrossRef] [PubMed]
- Burmølle, M.; Thomsen, T.R.; Fazli, M.; Dige, I.; Christensen, L.; Homøe, P.; Tvede, M.; Nyvad, B.; Tolker-Nielsen, T.; Givskov, M.; et al. Biofilms in Chronic Infections—A Matter of Opportunity—Monospecies Biofilms in Multispecies Infections. FEMS Immunol. Med. Microbiol. 2010, 59, 324–336. [Google Scholar] [CrossRef]
- Xu, Y.; Moser, C.; Al-Soud, W.A.; Sorensen, S.; Hoiby, N.; Nielsen, P.H.; Thomsen, T.R. Culture-Dependent and -Independent Investigations of Microbial Diversity on Urinary Catheters. J. Clin. Microbiol. 2012, 50, 3901–3908. [Google Scholar] [CrossRef] [PubMed]
- Tatum, O.L.; Dowd, S.E. Wound Healing Finally Enters the Age of Molecular Diagnostic Medicine. Adv. Wound Care 2012, 1, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Kalan, L.R.; Meisel, J.S.; Loesche, M.A.; Horwinski, J.; Soaita, I.; Chen, X.; Uberoi, A.; Gardner, S.E.; Grice, E.A. Strain- and Species-Level Variation in the Microbiome of Diabetic Wounds Is Associated with Clinical Outcomes and Therapeutic Efficacy. Cell Host Microbe 2019, 25, 641–655.e5. [Google Scholar] [CrossRef]
- Gardner, S.E.; Hillis, S.L.; Heilmann, K.; Segre, J.A.; Grice, E.A. The Neuropathic Diabetic Foot Ulcer Microbiome Is Associated with Clinical Factors. Diabetes 2013, 62, 923–930. [Google Scholar] [CrossRef]
- Loesche, M.; Gardner, S.E.; Kalan, L.; Horwinski, J.; Zheng, Q.; Hodkinson, B.P.; Tyldsley, A.S.; Franciscus, C.L.; Hillis, S.L.; Mehta, S.; et al. Temporal Stability in Chronic Wound Microbiota Is Associated With Poor Healing. J. Investig. Dermatol. 2017, 137, 237–244. [Google Scholar] [CrossRef]
- Kalan, L.R.; Brennan, M.B. The Role of the Microbiome in Nonhealing Diabetic Wounds. Ann. N. Y. Acad. Sci. USA 2019, 1435, 79–92. [Google Scholar] [CrossRef]
- Be, N.A.; Allen, J.E.; Brown, T.S.; Gardner, S.N.; McLoughlin, K.S.; Forsberg, J.A.; Kirkup, B.C.; Chromy, B.A.; Luciw, P.A.; Elster, E.A.; et al. Microbial Profiling of Combat Wound Infection through Detection Microarray and Next-Generation Sequencing. J. Clin. Microbiol. 2014, 52, 2583–2594. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.C.; Chhatbar, K.C.; Kashikar, A.; Mehndiratta, A. Diabetic Foot. BMJ 2017, 359, j5064. [Google Scholar] [CrossRef]
- Bandyk, D.F. The Diabetic Foot: Pathophysiology, Evaluation, and Treatment. Semin. Vasc. Surg. 2018, 31, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Boulton, A.J.M.; Armstrong, D.G.; Hardman, M.J.; Malone, M.; Embil, J.M.; Attinger, C.E.; Lipsky, B.A.; Aragón-Sánchez, J.; Li, H.K.; Schultz, G.; et al. Diagnosis and Management of Diabetic Foot Infections; American Diabetes Association: Arlington, VA, USA, 2020. [Google Scholar]
- Jneid, J.; Cassir, N.; Schuldiner, S.; Jourdan, N.; Sotto, A.; Lavigne, J.-P.; La Scola, B. Exploring the Microbiota of Diabetic Foot Infections With Culturomics. Front. Cell. Infect. Microbiol. 2018, 8, 282. [Google Scholar] [CrossRef]
- Mottola, C.; Mendes, J.J.; Cristino, J.M.; Cavaco-Silva, P.; Tavares, L.; Oliveira, M. Polymicrobial Biofilms by Diabetic Foot Clinical Isolates. Folia Microbiol. 2016, 61, 35–43. [Google Scholar] [CrossRef]
- Fayolle, M.; Morsli, M.; Gelis, A.; Chateauraynaud, M.; Yahiaoui-Martinez, A.; Sotto, A.; Lavigne, J.-P.; Dunyach-Remy, C. The Persistence of Staphylococcus Aureus in Pressure Ulcers: A Colonising Role. Genes 2021, 12, 1883. [Google Scholar] [CrossRef]
- Biglari, B.; vd Linden, P.H.; Simon, A.; Aytac, S.; Gerner, H.J.; Moghaddam, A. Use of Medihoney as a Non-Surgical Therapy for Chronic Pressure Ulcers in Patients with Spinal Cord Injury. Spinal Cord 2012, 50, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Maslova, E.; Eisaiankhongi, L.; Sjöberg, F.; McCarthy, R.R. Burns and Biofilms: Priority Pathogens and in Vivo Models. NPJ Biofilms Microbiomes 2021, 7, 73. [Google Scholar] [CrossRef] [PubMed]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of Methicillin Resistant Staphylococcus Aureus USA300 and Pseudomonas Aeruginosa in Polymicrobial Wound Infection. PLoS ONE 2013, 8, e56846. [Google Scholar] [CrossRef]
- Gupta, N.; Haque, A.; Mukhopadhyay, G.; Narayan, R.P.; Prasad, R. Interactions between Bacteria and Candida in the Burn Wound. Burns 2005, 31, 375–378. [Google Scholar] [CrossRef]
- Radlinski, L.; Rowe, S.E.; Kartchner, L.B.; Maile, R.; Cairns, B.A.; Vitko, N.P.; Gode, C.J.; Lachiewicz, A.M.; Wolfgang, M.C.; Conlon, B.P. Pseudomonas Aeruginosa Exoproducts Determine Antibiotic Efficacy against Staphylococcus Aureus. PLoS Biol. 2017, 15, e2003981. [Google Scholar] [CrossRef]
- Dana, A.N.; Bauman, W.A. Bacteriology of Pressure Ulcers in Individuals with Spinal Cord Injury: What We Know and What We Should Know. J. Spinal Cord Med. 2015, 38, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Mervis, J.S.; Phillips, T.J. Pressure Ulcers: Pathophysiology, Epidemiology, Risk Factors, and Presentation. J. Am. Acad. Dermatol. 2019, 81, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Edsberg, L.E.; Black, J.M.; Goldberg, M.; McNichol, L.; Moore, L.; Sieggreen, M. Revised National Pressure Ulcer Advisory Panel Pressure Injury Staging System: Revised Pressure Injury Staging System. J. Wound Ostomy Cont. Nurs. 2016, 43, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Peck, M.D. Epidemiology of Burns throughout the World. Part I: Distribution and Risk Factors. Burns 2011, 37, 1087–1100. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, D.G. Management of Burns. N. Engl. J. Med. 2019, 380, 2349–2359. [Google Scholar] [CrossRef] [PubMed]
- Markiewicz-Gospodarek, A.; Kozioł, M.; Tobiasz, M.; Baj, J.; Radzikowska-Büchner, E.; Przekora, A. Burn Wound Healing: Clinical Complications, Medical Care, Treatment, and Dressing Types: The Current State of Knowledge for Clinical Practice. Int. J. Environ. Res. Public Health 2022, 19, 1338. [Google Scholar] [CrossRef]
- Thomas, R.E.; Thomas, B.C. Reducing Biofilm Infections in Burn Patients’ Wounds and Biofilms on Surfaces in Hospitals, Medical Facilities and Medical Equipment to Improve Burn Care: A Systematic Review. Int. J. Environ. Res. Public Health 2021, 18, 13195. [Google Scholar] [CrossRef]
- Escandón-Vargas, K.; Tangua, A.R.; Medina, P.; Zorrilla-Vaca, A.; Briceño, E.; Clavijo-Martínez, T.; Tróchez, J.P. Healthcare-Associated Infections in Burn Patients: Timeline and Risk Factors. Burns 2020, 46, 1775–1786. [Google Scholar] [CrossRef]
- Peters, B.M.; Jabra-Rizk, M.A.; O’May, G.A.; Costerton, J.W.; Shirtliff, M.E. Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin. Microbiol. Rev. 2012, 25, 193–213. [Google Scholar] [CrossRef]
- Wolcott, R.; Costerton, J.W.; Raoult, D.; Cutler, S.J. The Polymicrobial Nature of Biofilm Infection. Clin. Microbiol. Infect. 2013, 19, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Rosch, J.W. Polymicrobial Interactions Operative during Pathogen Transmission. MBio 2021, 12, e01027-21. [Google Scholar] [CrossRef] [PubMed]
- Baishya, J.; Wakeman, C.A. Selective Pressures during Chronic Infection Drive Microbial Competition and Cooperation. NPJ Biofilms Microbiomes 2019, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.R.; Déziel, E.; D’Argenio, D.A.; Lépine, F.; Emerson, J.; McNamara, S.; Gibson, R.L.; Ramsey, B.W.; Miller, S.I. Selection for Staphylococcus Aureus Small-Colony Variants Due to Growth in the Presence of Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19890–19895. [Google Scholar] [CrossRef]
- Magalhães, A.P.; Lopes, S.P.; Pereira, M.O. Insights into Cystic Fibrosis Polymicrobial Consortia: The Role of Species Interactions in Biofilm Development, Phenotype, and Response to In-Use Antibiotics. Front. Microbiol. 2016, 7, 2146. [Google Scholar] [CrossRef] [PubMed]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The Biofilm Life Cycle: Expanding the Conceptual Model of Biofilm Formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Burmolle, M.; Ren, D.; Bjarnsholt, T.; Sorensen, S.J. Interactions in Multispecies Biofilms: Do They Actually Matter? Trends Microbiol. 2014, 22, 84–91. [Google Scholar] [CrossRef]
- Huang, R.; Li, M.; Gregory, R.L. Bacterial Interactions in Dental Biofilm. Virulence 2011, 2, 435–444. [Google Scholar] [CrossRef]
- Luo, A.; Wang, F.; Sun, D.; Liu, X.; Xin, B. Formation, Development, and Cross-Species Interactions in Biofilms. Front. Microbiol. 2021, 12, 757327. [Google Scholar] [CrossRef]
- Park, Y.; Simionato, M.R.; Sekiya, K.; Murakami, Y.; James, D.; Chen, W.; Hackett, M.; Yoshimura, F.; Demuth, D.R.; Lamont, R.J. Short Fimbriae of PorphyromonasGingivalis and Their Role in Coadhesion with Streptococcus Gordonii. Infect. Immun. 2005, 73, 3983–3989. [Google Scholar] [CrossRef]
- Kuboniwa, M.; Tribble, G.D.; James, C.E.; Kilic, A.O.; Tao, L.; Herzberg, M.C.; Shizukuishi, S.; Lamont, R.J. Streptococcus Gordonii Utilizes Several Distinct Gene Functions to Recruit PorphyromonasGingivalis into a Mixed Community. Mol. Microbiol. 2006, 60, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.M.; Rumbaugh, K.P.; Whiteley, M. Metabolite Cross-Feeding Enhances Virulence in a Model Polymicrobial Infection. PLoSPathog. 2011, 7, e1002012. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.V.; Gunawan, C.; Mann, R. We Are One: Multispecies Metabolism of a Biofilm Consortium and Their Treatment Strategies. Front. Microbiol. 2021, 12, 635432. [Google Scholar] [CrossRef] [PubMed]
- Laganenka, L.; Sourjik, V. Autoinducer 2-Dependent Escherichia Coli Biofilm Formation Is Enhanced in a Dual-Species Coculture. Appl. Environ. Microbiol. 2018, 84, e02638-17. [Google Scholar] [CrossRef] [PubMed]
- Molin, S.; Tolker-Nielsen, T. Gene Transfer Occurs with Enhanced Efficiency in Biofilms and Induces Enhanced Stabilisation of the Biofilm Structure. Curr. Opin. Biotechnol. 2003, 14, 255–261. [Google Scholar] [CrossRef]
- Toyofuku, M.; Nomura, N.; Eberl, L. Types and Origins of Bacterial Membrane Vesicles. Nat. Rev. Microbiol. 2019, 17, 13–24. [Google Scholar] [CrossRef]
- Bárdy, P.; Füzik, T.; Hrebík, D.; Pantůček, R.; Thomas Beatty, J.; Plevka, P. Structure and Mechanism of DNA Delivery of a Gene Transfer Agent. Nat. Commun. 2020, 11, 3034. [Google Scholar] [CrossRef]
- Orazi, G.; O’Toole, G.A. “It Takes a Village”: Mechanisms Underlying Antimicrobial Recalcitrance of Polymicrobial Biofilms. J. Bacteriol. 2019, 202, e00530-19. [Google Scholar] [CrossRef]
- Tanner, W.D.; Atkinson, R.M.; Goel, R.K.; Toleman, M.A.; Benson, L.S.; Porucznik, C.A.; VanDerslice, J.A. Horizontal Transfer of the BlaNDM-1 Gene to Pseudomonas Aeruginosa and Acinetobacter Baumannii in Biofilms. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Perez, A.C.; Pang, B.; King, L.B.; Tan, L.; Murrah, K.A.; Reimche, J.L.; Wren, J.T.; Richardson, S.H.; Ghandi, U.; Swords, W.E. Residence of Streptococcus Pneumoniae and Moraxella Catarrhalis within Polymicrobial Biofilm Promotes Antibiotic Resistance and Bacterial Persistence in Vivo. Pathog. Dis. 2014, 70, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.R.; Bell, T. Competition, Not Cooperation, Dominates Interactions among Culturable Microbial Species. Curr. Biol. 2012, 22, 1845–1850. [Google Scholar] [CrossRef] [PubMed]
- Hibbing, M.E.; Fuqua, C.; Parsek, M.R.; Peterson, S.B. Bacterial Competition: Surviving and Thriving in the Microbial Jungle. Nat. Rev. Microbiol. 2010, 8, 15–25. [Google Scholar] [CrossRef]
- Rendueles, O.; Ghigo, J.-M. Mechanisms of Competition in Biofilm Communities. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; Straight, P.D. Multifaceted Interfaces of Bacterial Competition. J. Bacteriol. 2016, 198, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Juarez, G.E.; Galvan, E.M. Role of Nutrient Limitation in the Competition between Uropathogenic Strains of Klebsiella Pneumoniae and Escherichia Coli in Mixed Biofilms. Biofouling 2018, 34, 287–298. [Google Scholar] [CrossRef]
- Yamamoto, K.; Haruta, S.; Kato, S.; Ishii, M.; Igarashi, Y. Determinative Factors of Competitive Advantage between Aerobic Bacteria for Niches at the Air-Liquid Interface. Microbes Environ. 2010, 25, 317–320. [Google Scholar] [CrossRef]
- Pericone, C.D.; Overweg, K.; Hermans, P.W.; Weiser, J.N. Inhibitory and Bactericidal Effects of Hydrogen Peroxide Production by Streptococcus Pneumoniae on Other Inhabitants of the Upper Respiratory Tract. Infect. Immun. 2000, 68, 3990–3997. [Google Scholar] [CrossRef] [PubMed]
- Santagati, M.; Scillato, M.; Patanè, F.; Aiello, C.; Stefani, S. Bacteriocin-Producing Oral Streptococci and Inhibition of Respiratory Pathogens. FEMS Immunol. Med. Microbiol. 2012, 65, 23–31. [Google Scholar] [CrossRef]
- Schwarz, S.; West, T.E.; Boyer, F.; Chiang, W.-C.; Carl, M.A.; Hood, R.D.; Rohmer, L.; Tolker-Nielsen, T.; Skerrett, S.J.; Mougous, J.D. Burkholderia Type VI Secretion Systems Have Distinct Roles in Eukaryotic and Bacterial Cell Interactions. PLoSPathog. 2010, 6, e1001068. [Google Scholar] [CrossRef]
- Augustine, N.; Kumar, P.; Thomas, S. Inhibition of Vibrio Cholerae Biofilm by AiiA Enzyme Produced from Bacillus Spp. Arch. Microbiol. 2010, 192, 1019–1022. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.; Zere, T.R.; Weber, M.M.; Wood, T.K.; Whiteley, M.; Hidalgo-Romano, B.; Valenzuela, E.J.; McLean, R.J.C. Indole Production Promotes Escherichia Coli Mixed-Culture Growth with Pseudomonas Aeruginosa by Inhibiting Quorum Signaling. Appl. Environ. Microbiol. 2012, 78, 411–419. [Google Scholar] [CrossRef]
- Rendueles, O.; Travier, L.; Latour-Lambert, P.; Fontaine, T.; Magnus, J.; Denamur, E.; Ghigo, J.-M. Screening of Escherichia Coli Species Biodiversity Reveals New Biofilm-Associated Antiadhesion Polysaccharides. MBio 2011, 2, e00043-11. [Google Scholar] [CrossRef] [PubMed]
- Christopher, A.B.; Arndt, A.; Cugini, C.; Davey, M.E. A Streptococcal Effector Protein That Inhibits PorphyromonasGingivalis Biofilm Development. Microbiology 2010, 156, 3469–3477. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Furukawa, S.; Fujita, S.; Mitobe, J.; Kawarai, T.; Narisawa, N.; Sekizuka, T.; Kuroda, M.; Ochiai, K.; Ogihara, H.; et al. Inhibition of Streptococcus Mutans Biofilm Formation by Streptococcus SalivariusFruA. Appl. Environ. Microbiol. 2011, 77, 1572–1580. [Google Scholar] [CrossRef]
- Sugimoto, S.; Iwamoto, T.; Takada, K.; Okuda, K.-I.; Tajima, A.; Iwase, T.; Mizunoe, Y. Staphylococcus Epidermidis Esp Degrades Specific Proteins Associated with Staphylococcus Aureus Biofilm Formation and Host-Pathogen Interaction. J. Bacteriol. 2013, 195, 1645–1655. [Google Scholar] [CrossRef]
- Davies, D.G.; Marques, C.N.H. A Fatty Acid Messenger Is Responsible for Inducing Dispersion in Microbial Biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef]
- Serra, R.; Grande, R.; Butrico, L.; Rossi, A.; Settimio, U.F.; Caroleo, B.; Amato, B.; Gallelli, L.; de Franciscis, S. Chronic Wound Infections: The Role of Pseudomonas Aeruginosa and Staphylococcus Aureus. Expert Rev. Anti. Infect. Ther. 2015, 13, 605–613. [Google Scholar] [CrossRef]
- Hotterbeekx, A.; Kumar-Singh, S.; Goossens, H.; Malhotra-Kumar, S. In Vivo and In Vitro Interactions between Pseudomonas Aeruginosa and Staphylococcus Spp. Front. Cell. Infect. Microbiol. 2017, 7, 106. [Google Scholar] [CrossRef]
- Kamer, A.M.A.; Abdelaziz, A.A.; Al-Monofy, K.B.; Al-Madboly, L.A. Antibacterial, Antibiofilm, and Anti-Quorum Sensing Activities of Pyocyanin against Methicillin-Resistant Staphylococcus Aureus: In Vitro and in Vivo Study. BMC Microbiol. 2023, 23, 116. [Google Scholar] [CrossRef]
- Gonçalves, T.; Vasconcelos, U. Colour Me Blue: The History and the Biotechnological Potential of Pyocyanin. Molecules 2021, 26, 927. [Google Scholar] [CrossRef]
- Machan, Z.A.; Taylor, G.W.; Pitt, T.L.; Cole, P.J.; Wilson, R. 2-Heptyl-4-Hydroxyquinoline N-Oxide, an Antistaphylococcal Agent Produced by Pseudomonas Aeruginosa. J. Antimicrob. Chemother. 1992, 30, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Kessler, E.; Safrin, M.; Olson, J.C.; Ohman, D.E. Secreted LasA of Pseudomonas Aeruginosa Is a Staphylolytic Protease. J. Biol. Chem. 1993, 268, 7503–7508. [Google Scholar] [CrossRef] [PubMed]
- Pamp, S.J.; Tolker-Nielsen, T. Multiple Roles of Biosurfactants in Structural Biofilm Development by Pseudomonas Aeruginosa. J. Bacteriol. 2007, 189, 2531–2539. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.S.E.; Carvalho, J.W.P.; Aires, C.P.; Nitschke, M. Disruption of Staphylococcus Aureus Biofilms Using Rhamnolipid Biosurfactants. J. Dairy Sci. 2017, 100, 7864–7873. [Google Scholar] [CrossRef]
- Cornelis, P. Iron Uptake and Metabolism in Pseudomonads. Appl. Microbiol. Biotechnol. 2010, 86, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Biswas, L.; Biswas, R.; Schlag, M.; Bertram, R.; Götz, F. Small-Colony Variant Selection as a Survival Strategy for Staphylococcus Aureus in the Presence of Pseudomonas Aeruginosa. Appl. Environ. Microbiol. 2009, 75, 6910–6912. [Google Scholar] [CrossRef] [PubMed]
- DeLeon, S.; Clinton, A.; Fowler, H.; Everett, J.; Horswill, A.R.; Rumbaugh, K.P. Synergistic Interactions of Pseudomonas Aeruginosa and Staphylococcus Aureus in an in Vitro Wound Model. Infect. Immun. 2014, 82, 4718–4728. [Google Scholar] [CrossRef] [PubMed]
- Massey, R.C.; Buckling, A.; Peacock, S.J. Phenotypic Switching of Antibiotic Resistance Circumvents Permanent Costs in Staphylococcus Aureus. Curr. Biol. 2001, 11, 1810–1814. [Google Scholar] [CrossRef]
- Smith, A.C.; Rice, A.; Sutton, B.; Gabrilska, R.; Wessel, A.K.; Whiteley, M.; Rumbaugh, K.P. Albumin Inhibits Pseudomonas Aeruginosa Quorum Sensing and Alters Polymicrobial Interactions. Infect. Immun. 2017, 85, 10–1128. [Google Scholar] [CrossRef]
- Alves, P.M.; Al-Badi, E.; Withycombe, C.; Jones, P.M.; Purdy, K.J.; Maddocks, S.E. Interaction between Staphylococcus Aureus and Pseudomonas Aeruginosa Is Beneficial for Colonisation and Pathogenicity in a Mixed Biofilm. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef] [PubMed]
- Dalton, T.; Dowd, S.E.; Wolcott, R.D.; Sun, Y.; Watters, C.; Griswold, J.A.; Rumbaugh, K.P. An in Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PLoS ONE 2011, 6, e27317. [Google Scholar] [CrossRef] [PubMed]
- Jenul, C.; Keim, K.C.; Jens, J.N.; Zeiler, M.J.; Schilcher, K.; Schurr, M.J.; Melander, C.; Phelan, V.V.; Horswill, A.R. Pyochelin Biotransformation by Staphylococcusaureus Shapes Bacterial Competition with Pseudomonas Aeruginosa in Polymicrobial Infections. Cell Rep. 2023, 42, 112540. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; McQuillen, E.A.; Rana, P.S.J.B.; Gloag, E.S.; Wozniak, D.J. Cross-Species Protection to Innate Immunity Mediated by A Bacterial Pigment. bioRxiv 2023. [Google Scholar] [CrossRef]
- Fazli, M.; Bjarnsholt, T.; Kirketerp-Møller, K.; Jørgensen, B.; Andersen, A.S.; Krogfelt, K.A.; Givskov, M.; Tolker-Nielsen, T. Nonrandom Distribution of Pseudomonas Aeruginosa and Staphylococcus Aureus in Chronic Wounds. J. Clin. Microbiol. 2009, 47, 4084–4089. [Google Scholar] [CrossRef]
- Ibberson, C.B.; Barraza, J.P.; Holmes, A.L.; Cao, P.; Whiteley, M. Precise Spatial Structure Impacts Antimicrobial Susceptibility of S. Aureus in Polymicrobial Wound Infections. Proc. Natl. Acad. Sci. USA 2022, 119, e2212340119. [Google Scholar] [CrossRef]
- Pouget, C.; Pantel, A.; Dunyach-Remy, C.; Magnan, C.; Sotto, A.; Lavigne, J.-P. Antimicrobial Activity of Antibiotics on Biofilm Formed by Staphylococcus Aureus and Pseudomonas Aeruginosa in an Open Microfluidic Model Mimicking the Diabetic Foot Environment. J. Antimicrob. Chemother. 2023, 78, 540–545. [Google Scholar] [CrossRef]
- Rajkumari, N.; Mathur, P.; Misra, M.C. Soft Tissue and Wound Infections Due to Enterococcus Spp. Among Hospitalized Trauma Patients in a Developing Country. J. Glob. Infect. Dis. 2014, 6, 189–193. [Google Scholar] [CrossRef]
- Weigel, L.M.; Donlan, R.M.; Shin, D.H.; Jensen, B.; Clark, N.C.; McDougal, L.K.; Zhu, W.; Musser, K.A.; Thompson, J.; Kohlerschmidt, D.; et al. High-Level Vancomycin-Resistant Staphylococcus Aureus Isolates Associated with a Polymicrobial Biofilm. Antimicrob. Agents Chemother. 2007, 51, 231–238. [Google Scholar] [CrossRef]
- Zhu, W.; Murray, P.R.; Huskins, W.C.; Jernigan, J.A.; McDonald, L.C.; Clark, N.C.; Anderson, K.F.; McDougal, L.K.; Hageman, J.C.; Olsen-Rasmussen, M.; et al. Dissemination of an Enterococcus Inc18-Like VanA Plasmid Associated with Vancomycin-Resistant Staphylococcus Aureus. Antimicrob. Agents Chemother. 2010, 54, 4314–4320. [Google Scholar] [CrossRef]
- Ch’ng, J.-H.; Muthu, M.; Chong, K.K.L.; Wong, J.J.; Tan, C.A.Z.; Koh, Z.J.S.; Lopez, D.; Matysik, A.; Nair, Z.J.; Barkham, T.; et al. Heme Cross-Feeding Can Augment Staphylococcus Aureus and Enterococcus Faecalis Dual Species Biofilms. ISME J. 2022, 16, 2015–2026. [Google Scholar] [CrossRef]
- Kaper, J.B. Pathogenic Escherichia coli. Int. J. Med. Microbiol. 2005, 295, 355–356. [Google Scholar] [CrossRef]
- Wong, J.J.; Ho, F.K.; Choo, P.Y.; Chong, K.K.L.; Ho, C.M.B.; Neelakandan, R.; Keogh, D.; Barkham, T.; Chen, J.; Liu, C.F.; et al. Escherichia Coli BarA-UvrY Regulates the Pks Island and Kills Staphylococci via the Genotoxin Colibactin during Interspecies Competition. PLoSPathog. 2022, 18, e1010766. [Google Scholar] [CrossRef]
- Nadell, C.D.; Drescher, K.; Foster, K.R. Spatial Structure, Cooperation and Competition in Biofilms. Nat. Rev. Microbiol. 2016, 14, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, N.; Nakanouchi, J.; Yüzen, D.I.; Fung, S.; Fernandez, J.S.; Barberis, C.; Tuchscherr, L.; Ramirez, M.S. A Study on Acinetobacter Baumannii and Staphylococcus Aureus Strains Recovered from the Same Infection Site of a Diabetic Patient. Curr. Microbiol. 2019, 76, 842–847. [Google Scholar] [CrossRef]
- Fernandez, J.S.; Tuttobene, M.R.; Montaña, S.; Subils, T.; Cantera, V.; Iriarte, A.; Tuchscherr, L.; Ramirez, M.S. Staphylococcus Aureus α-Toxin Effect on Acinetobacter Baumannii Behavior. Biology 2022, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Shen, W.; Gong, Y.; Li, M.; Rao, X.; Liu, Q.; Yu, Y.; Zhou, J.; Zhu, K.; Yuan, M.; et al. Essential Fitness Repertoire of Staphylococcus Aureus during Co-Infection with Acinetobacter Baumannii In Vivo. Msystems 2022, 7, e0033822. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Niu, Y.; Ye, X.; Zhu, C.; Tong, T.; Zhou, Y.; Zhou, X.; Cheng, L.; Ren, B. Staphylococcus Aureus Synergized with Candida Albicans to Increase the Pathogenesis and Drug Resistance in Cutaneous Abscess and Peritonitis Murine Models. Pathogens 2021, 10, 1036. [Google Scholar] [CrossRef] [PubMed]
- Boldock, E.; Surewaard, B.G.J.; Shamarina, D.; Na, M.; Fei, Y.; Ali, A.; Williams, A.; Pollitt, E.J.G.; Szkuta, P.; Morris, P.; et al. Human Skin Commensals Augment Staphylococcus Aureus Pathogenesis. Nat. Microbiol. 2018, 3, 881–890. [Google Scholar] [CrossRef]
- Gibson, J.F.; Pidwill, G.R.; Carnell, O.T.; Surewaard, B.G.J.; Shamarina, D.; Sutton, J.A.F.; Jeffery, C.; Derré-Bobillot, A.; Archambaud, C.; Siggins, M.K.; et al. Commensal Bacteria Augment Staphylococcus Aureus Infection by Inactivation of Phagocyte-Derived Reactive Oxygen Species. PLoS Pathog. 2021, 17, e1009880. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).