The Spatial–Temporal Effects of Bacterial Growth Substrates on Antibiotic Resistance Gene Spread in the Biofilm
Abstract
1. Introduction
2. Results and Discussion
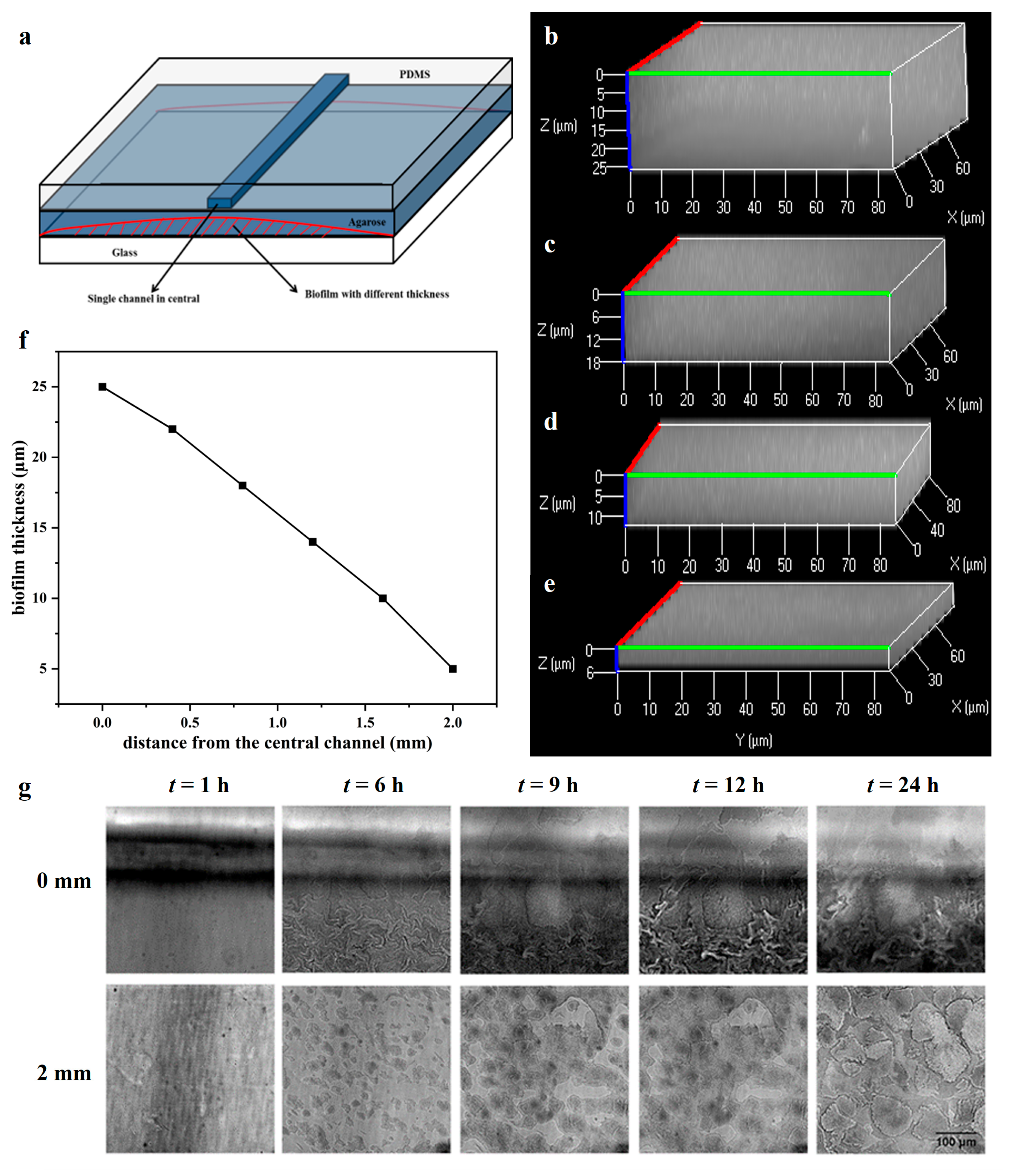
2.1. The Biofilm Formation with Different Thicknesses on the Microfluidic Chip
2.2. The Transfer Processes of Antibiotic Resistance Plasmid under Different Substrate Conditions on the Chip
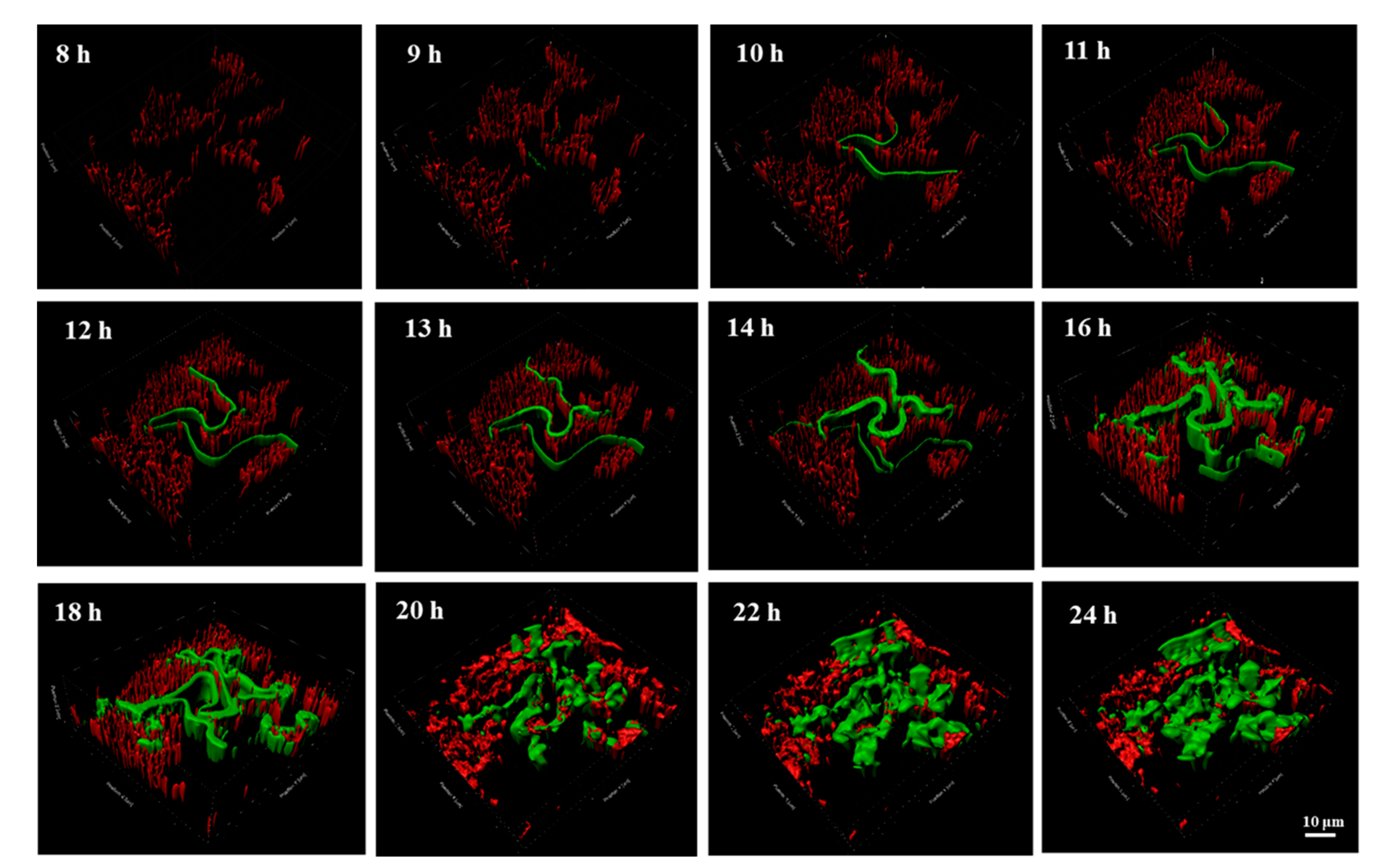
2.3. The Contribution of VGT to the ARG Spread Revealed by the 3D Distribution Analysis in the Biofilm
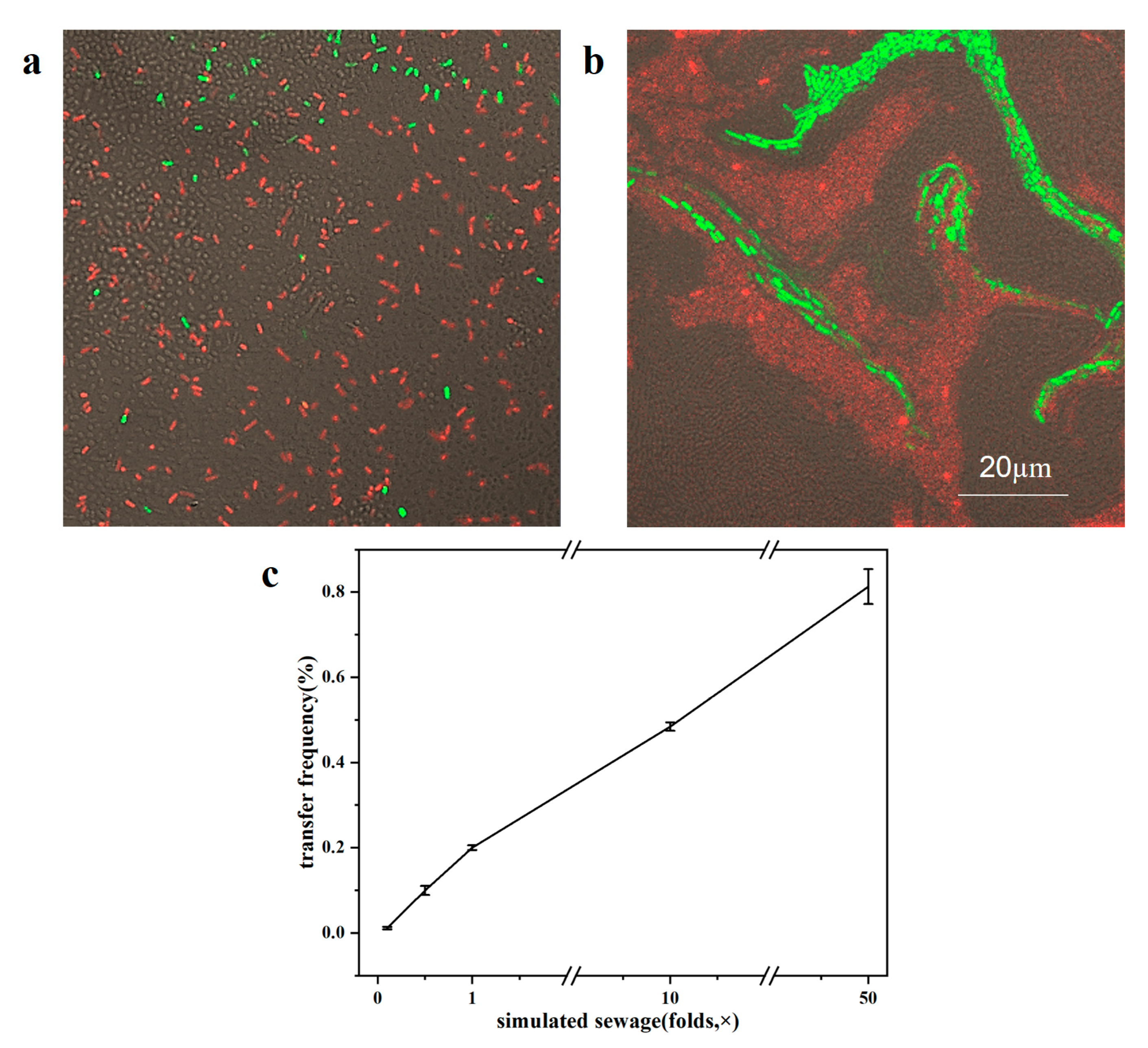
2.4. The Effect of Different Sewage Concentrations on the Transfer Frequency in Biofilms
3. Materials and Methods
3.1. Bacterial Growth Substrates
3.2. Bacterial Strains and Culture
3.3. Biofilm Formation and Mating Assays on Microfluidic Chips
3.4. Mating Assays in the Well Plate
3.5. Image Acquisition and Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Besemer, K. Biodiversity, community structure and function of biofilms in stream ecosystems. Res. Microbiol. 2015, 166, 774–781. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Balcázar, J.L.; Subirats, J.; Borrego, C.M. The role of biofilms as environmental reservoirs of antibiotic resistance. Front. Microbiol. 2015, 6, 1216. [Google Scholar] [CrossRef] [PubMed]
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 2020, 96, fiaa031. [Google Scholar] [CrossRef]
- Marti, E.; Huerta, B.; Rodríguez-Mozaz, S.; Barceló, D.; Jofre, J.; Balcázar, J.L. Characterization of ciprofloxacin-resistant isolates from a wastewater treatment plant and its receiving river. Water Res. 2014, 61, 67–76. [Google Scholar] [CrossRef]
- Nõlvak, H.; Truu, M.; Tiirik, K.; Oopkaup, K.; Sildvee, T.; Kaasik, A.; Mander, U.; Truu, J. Dynamics of antibiotic resistance genes and their relationships with system treatment efficiency in a horizontal subsurface flow constructed wetland. Sci. Total Environ. 2013, 461–462, 636–644. [Google Scholar] [CrossRef]
- Król, J.E.; Wojtowicz, A.J.; Rogers, L.M.; Heuer, H.; Smalla, K.; Krone, S.M.; Top, E.M. Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid 2013, 70, 110–119. [Google Scholar] [CrossRef]
- Anderson-Glenna, M.J.; Bakkestuen, V.; Clipson, N. Spatial and temporal variability in epilithic biofilm bacterial communities along an upland river gradient. FEMS Microbiol. Ecol. 2008, 64, 407–418. [Google Scholar] [CrossRef]
- Wilhelm, L.; Besemer, K.; Fasching, C.; Urich, T.; Singer, G.A.; Quince, C.; Battin, T.J. Rare but active taxa contribute to community dynamics of benthic biofilms in glacier-fed streams. Environ. Microbiol. 2014, 16, 2514–2524. [Google Scholar] [CrossRef]
- Hullar, M.; Kaplan, L.A.; Stahl, D.A. Recurring seasonal dynamics of microbial communities in stream habitats. Appl. Environ. Microbiol. 2006, 72, 713–722. [Google Scholar] [CrossRef]
- Heino, J.; Tolkkinen, M.; Pirttilä, A.M.; Aisala, H.; Mykrä, H. Microbial diversity and community-environment relationships in boreal streams. J. Biogeogr. 2014, 41, 2234–2244. [Google Scholar] [CrossRef]
- Cresson, R.; Carrère, H.; Delgenès, J.P.; Bernet, N. Biofilm formation during the start-up period of an anaerobic biofilm reactor—Impact of nutrient complementation. Biochem. Eng. J. 2006, 30, 55–62. [Google Scholar] [CrossRef]
- Liu, S.; Gunawan, C.; Barraud, N.; Rice, S.A.; Harry, E.J.; Amal, R. Understanding, Monitoring, and Controlling Biofilm Growth in Drinking Water Distribution Systems. Environ. Sci. Technol. 2016, 50, 8954–8976. [Google Scholar] [CrossRef]
- Jahid, I.K.; Mizan, M.F.R.; Ha, A.J.; Ha, S.D. Effect of salinity and incubation time of planktonic cells on biofilm formation, motility, exoprotease production, and quorum sensing of Aeromonas hydrophila. Food Microbiol. 2015, 49, 142–151. [Google Scholar] [CrossRef]
- Nesse, L.L.; Simm, R. Biofilm: A Hotspot for Emerging Bacterial Genotypes; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar] [CrossRef]
- Teal, T.K.; Lies, D.P.; Wold, B.J.; Newman, D.K. Spatiometabolic Stratification of Shewanella oneidensis Biofilms. Appl. Environ. Microbiol. 2006, 72, 7324–7330. [Google Scholar] [CrossRef]
- Werner, E.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Heydorn, A.; Molin, S.; Pitts, B.; Stewart, P.S. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2004, 70, 6188–6196. [Google Scholar] [CrossRef]
- Ping, Q.; Zhang, Z.P.; Ma, L.P.; Yan, T.T.; Wang, L.; Li, Y.M. The prevalence and removal of antibiotic resistance genes in full-scale wastewater treatment plants: Bacterial host, influencing factors and correlation with nitrogen metabolic pathway. Sci. Total Environ. 2022, 827, 154154. [Google Scholar] [CrossRef]
- Deng, W.W.; Zhang, A.Y.; Chen, S.J.; He, X.P.; Jin, L.; Yu, X.M.; Yang, S.Z.; Li, B.; Fan, L.Q.; Ji, L.; et al. Heavy metals, antibiotics and nutrients affect the bacterial community and resistance genes in chicken manure composting and fertilized soil. J. Environ. Manag. 2020, 257, 109980. [Google Scholar] [CrossRef]
- Deng, Y.Q.; Mao, C.; Lin, Z.Y.; Su, W.X.; Cheng, C.H.; Li, Y.; Gu, Q.H.; Gao, R.; Su, Y.L.; Feng, J. Nutrients, temperature, and oxygen mediate microbial antibiotic resistance in sea bass (Lateolabrax maculatus) ponds. Sci. Total Environ. 2022, 819, 153120. [Google Scholar] [CrossRef]
- Zhao, H.X.; Zhang, J.Y.; Chen, X.; Yang, S.; Huang, H.F.; Pan, L.H.; Huang, L.L.; Jiang, G.L.X.; Tang, J.L.; Xu, Q.S.; et al. Climate and nutrients regulate biographical patterns and health risks of antibiotic resistance genes in mangrove environment. Sci. Total Environ. 2023, 854, 158811. [Google Scholar] [CrossRef]
- Guliy, O.I.; Evstigneeva, S.S.; Bunin, V.D. Microfluidic bioanalytical system for biofilm formation indication. Talanta 2022, 247, 123541. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Han, S.; Jeon, J.S.; Yamamoto, K.; Zervantonakis, I.K.; Sudo, R.; Kamm, R.D.; Chung, S. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat. Protoc. 2012, 7, 1247–1259. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; Toh, Y.; Voldman, J.; Yu, H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip 2007, 7, 681. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zhang, J.; Li, B.; Wen, X.H.; Liang, P.; Huang, X. A novel microfluidic system enables visualization and analysis of antibiotic resistance gene transfer to activated sludge bacteria in biofilm. Sci. Total Environ. 2018, 642, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Torresi, E.; Polesel, F.; Bester, K.; Christensson, M.; Smets, B.F.; Trapp, S.; Andersen, H.R.; Plósz, B.G. Diffusion and sorption of organic micropollutants in biofilms with varying thicknesses. Water Res. 2017, 123, 388–400. [Google Scholar] [CrossRef]
- Heydorn, A.; Ersboll, B.; Kato, J.; Hentzer, M.; Parsek, M.R.; Tolker-Nielsen, T.; Givskov, M.; Molin, S. Statistical analysis of Pseudomonas aeruginosa biofilm development: Impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 2002, 68, 2008–2017. [Google Scholar] [CrossRef]
- Olapade, O.A.; Leff, L.G. Seasonal response of stream biofilm communities to dissolved organic matter and nutrient enrichments. Appl. Environ. Microbiol. 2005, 71, 2278–2287. [Google Scholar] [CrossRef]
- Rubin, M.A.; Leff, L.G. Nutrients and Other Abiotic Factors Affecting Bacterial Communities in an Ohio River (USA). Microb. Ecol. 2007, 54, 374–383. [Google Scholar] [CrossRef]
- Murugesan, N.; Dhar, P.; Panda, T.; Das, S.K. Interplay of chemical and thermal gradient on bacterial migration in a diffusive microfluidic device. Biomicrofluidics 2017, 11, 24108. [Google Scholar] [CrossRef]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef]
- Ehlers, L.J.; Bouwer, E.J. RP4 plasmid transfer among species of Pseudomonas in a biofilm reactor. Water Sci. Technol. 1999, 39, 163–171. [Google Scholar] [CrossRef]
- Jiang, Q.; Feng, M.B.; Ye, C.S.; Yu, X. Effects and relevant mechanisms of non-antibiotic factors on the horizontal transfer of antibiotic resistance genes in water environments: A review. Sci. Total Environ. 2022, 806, 150568. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Zhang, J.; Huang, X.; Shi, H.C.; Yin, H.B. Real-Time Study of Rapid Spread of Antibiotic Resistance Plasmid in Biofilm Using Microfluidics. Environ. Sci. Technol. 2018, 52, 11132–11141. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Song, Y.Q.; Lin, H.; Yin, H.B. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ. Int. 2019, 131, 105007. [Google Scholar] [CrossRef]
- Song, W.Z.; Wemheuer, B.; Steinberg, P.D.; Marzinelli, E.M.; Thomas, T. Contribution of horizontal gene transfer to the functionality of microbial biofilm on a macroalgae. ISME J. 2021, 15, 807–817. [Google Scholar] [CrossRef]
- Séveno, N.A.; Kallifidas, D.; Smalla, K.; van Elsas, J.D.; Collard, J.-M.; Karagouni, A.D.; Wellington, E.M.H. Occurrence and reservoirs of antibiotic resistance genes in the environment. Rev. Med. Microbiol. 2002, 13, 15–27. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Madsen, J.S.; Burmolle, M.; Hansen, L.H.; Sorensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef]
- Yang, D.; Wang, J.F.; Qiu, Z.G.; Jin, M.; Shen, Z.Q.; Chen, Z.L.; Wang, X.W.; Zhang, B.; Li, J.W. Horizontal transfer of antibiotic resistance genes in a membrane bioreactor. J. Biotechnol. 2013, 167, 441–447. [Google Scholar] [CrossRef]
- Musovic, S.; Klumper, U.; Dechesne, A.; Magid, J.; Smets, B.F. Long- term manure exposure increases soil bacterial community potential for plasmid uptake. Environ. Microbiol. Rep. 2014, 6, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Jacquiod, S.; Brejnrod, A.; Morberg, S.M.; Abu Al-Soud, W.; Sørensen, S.J.; Riber, L. Deciphering conjugative plasmid permissiveness in wastewater microbiomes. Mol. Ecol. 2017, 26, 3556–3571. [Google Scholar] [CrossRef] [PubMed]
- Neela, F.A.; Nonaka, L.; Rahman, M.H.; Suzuki, S. Transfer of the chromosomally encoded tetracycline resistance gene tet(M) from marine bacteria to Escherichia coli and Enterococcus faecalis. World J. Microbiol. Biotechnol. 2009, 25, 1095–1101. [Google Scholar] [CrossRef]
- Jiao, Y.N.; Chen, H.; Gao, R.X.; Zhu, Y.G.; Rensing, C. Organic compounds stimulate horizontal transfer of antibiotic resistance genes in mixed wastewater treatment systems. Chemosphere 2017, 184, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.T.; Yuan, Q.B.; Yang, J. Distinguishing Effects of Ultraviolet Exposure and Chlorination on the Horizontal Transfer of Antibiotic Resistance Genes in Municipal Wastewater. Environ. Sci. Technol. 2015, 49, 5771–5778. [Google Scholar] [CrossRef]
- Li, L.G.; Dechesne, A.; He, Z.M.; Madsen, J.S.; Nesme, J.; Sørensen, S.J.; Smets, B.F. Estimating the Transfer Range of Plasmids Encoding Antimicrobial Resistance in a Wastewater Treatment Plant Microbial Community. Environ. Sci. Technol. Lett. 2018, 5, 260–265. [Google Scholar] [CrossRef]
- Pallares-Vega, R.; Macedo, G.; Brouwer, M.S.M.; Leal, L.H.; van der Maas, P.; van Loosdrecht, M.C.M.; Weissbrodt, D.G.; Heederik, D.; Mevius, D.; Schmitt, H. Temperature and Nutrient Limitations Decrease Transfer of Conjugative IncP-1 Plasmid pKJK5 to Wild Escherichia coli Strains. Front. Microbiol. 2021, 12, 656250. [Google Scholar] [CrossRef]
- Shafieifini, M.; Sun, Y.P.; Staley, Z.R.; Riethoven, J.-J.; Li, X.; Elkins, C.A. Effects of Nutrient Level and Growth Rate on the Conjugation Process That Transfers Mobile Antibiotic Resistance Genes in Continuous Cultures. Appl. Environ. Microbiol. 2022, 88, e112122. [Google Scholar] [CrossRef]
- Gutiérrez, M.; Etxebarria, J.; de Las Fuentes, L. Evaluation of wastewater toxicity: Comparative study between Microtox® and activated sludge oxygen uptake inhibition. Water Res. 2002, 36, 919–924. [Google Scholar] [CrossRef]
- Li, B.; Qiu, Y.; Glidle, A.; McIlvenna, D.; Luo, Q.; Cooper, J.; Shi, H.C.; Yin, H.B. Gradient Microfluidics Enables Rapid Bacterial Growth Inhibition Testing. Anal. Chem. 2014, 86, 3131–3137. [Google Scholar] [CrossRef]
- Lin, H.; Jiang, L.T.; Li, B.; Dong, Y.B.; He, Y.H.; Qiu, Y. Screening and evaluation of heavy metals facilitating antibiotic resistance gene transfer in a sludge bacterial community. Sci. Total Environ. 2019, 695, 133862. [Google Scholar] [CrossRef]


| Donor/Recipient | Substrates | Culture Method | Transfer Frequency (Transconjugants/Recipients) | Ref. |
|---|---|---|---|---|
| E. coli K12 (ATCC 47076)/activated sludge bacteria | CODCr 194.3–315.9 mg/L | MBR reactor | 2.76 × 10−5 | [41] |
| Pseudomonas putida KT2442/soil bacteria | Soil extract and R2A medium | Filter mating | 8.24 × 10−5–4.56 × 10−4 | [42] |
| E. coli MG1655/influent, effluent | LB medium | Membrane filter | 5.1 × 10−2–7 × 10−1 | [43] |
| P. damselae/E. coli K12 | LB medium | Filter mating | (6.62 ± 1.61) × 10−3 | [44] |
| E. coli HB101/E. coli NK5449 | LB medium | Mixed cultivation | 10−5–10−3 | [45] |
| E. coli K12/E. coli NK5449 | Sterile wastewater (NaAc, α-lactose, glucose) | Mixed cultivation | 10−7–10−4 | [46] |
| E. coli MG1655, P. putida KT2440/activated sludge bacteria | Synthetic wastewater medium | Filter mating | 3.39 × 10−5–5.05 × 10−4 | [47] |
| E. coli MG1655/E. coli | Synthetic wastewater, soil extract | Filter mating | 3, 6 logs decrease respectively than LB medium | [48] |
| E. coli CV601/E. coli J53 | 1/10, 1/3-strength Mueller–Hinton broth(MHB) | Filter mating | 101–103, 102–104 CFU/mL transconjugants | [49] |
| E. coli MG1655/sludge bacteria | Simulated wastewater with different concentrations | Mixed cultivation | 10−4–10−3 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Liu, B.; Zhu, Y.; Qiu, Y.; Li, B. The Spatial–Temporal Effects of Bacterial Growth Substrates on Antibiotic Resistance Gene Spread in the Biofilm. Antibiotics 2023, 12, 1154. https://doi.org/10.3390/antibiotics12071154
Liu S, Liu B, Zhu Y, Qiu Y, Li B. The Spatial–Temporal Effects of Bacterial Growth Substrates on Antibiotic Resistance Gene Spread in the Biofilm. Antibiotics. 2023; 12(7):1154. https://doi.org/10.3390/antibiotics12071154
Chicago/Turabian StyleLiu, Shuzhen, Bingwen Liu, Yin Zhu, Yong Qiu, and Bing Li. 2023. "The Spatial–Temporal Effects of Bacterial Growth Substrates on Antibiotic Resistance Gene Spread in the Biofilm" Antibiotics 12, no. 7: 1154. https://doi.org/10.3390/antibiotics12071154
APA StyleLiu, S., Liu, B., Zhu, Y., Qiu, Y., & Li, B. (2023). The Spatial–Temporal Effects of Bacterial Growth Substrates on Antibiotic Resistance Gene Spread in the Biofilm. Antibiotics, 12(7), 1154. https://doi.org/10.3390/antibiotics12071154







