Acinetobacter baumannii Global Clone-Specific Resistomes Explored in Clinical Isolates Recovered from Egypt
Abstract
1. Introduction
2. Results
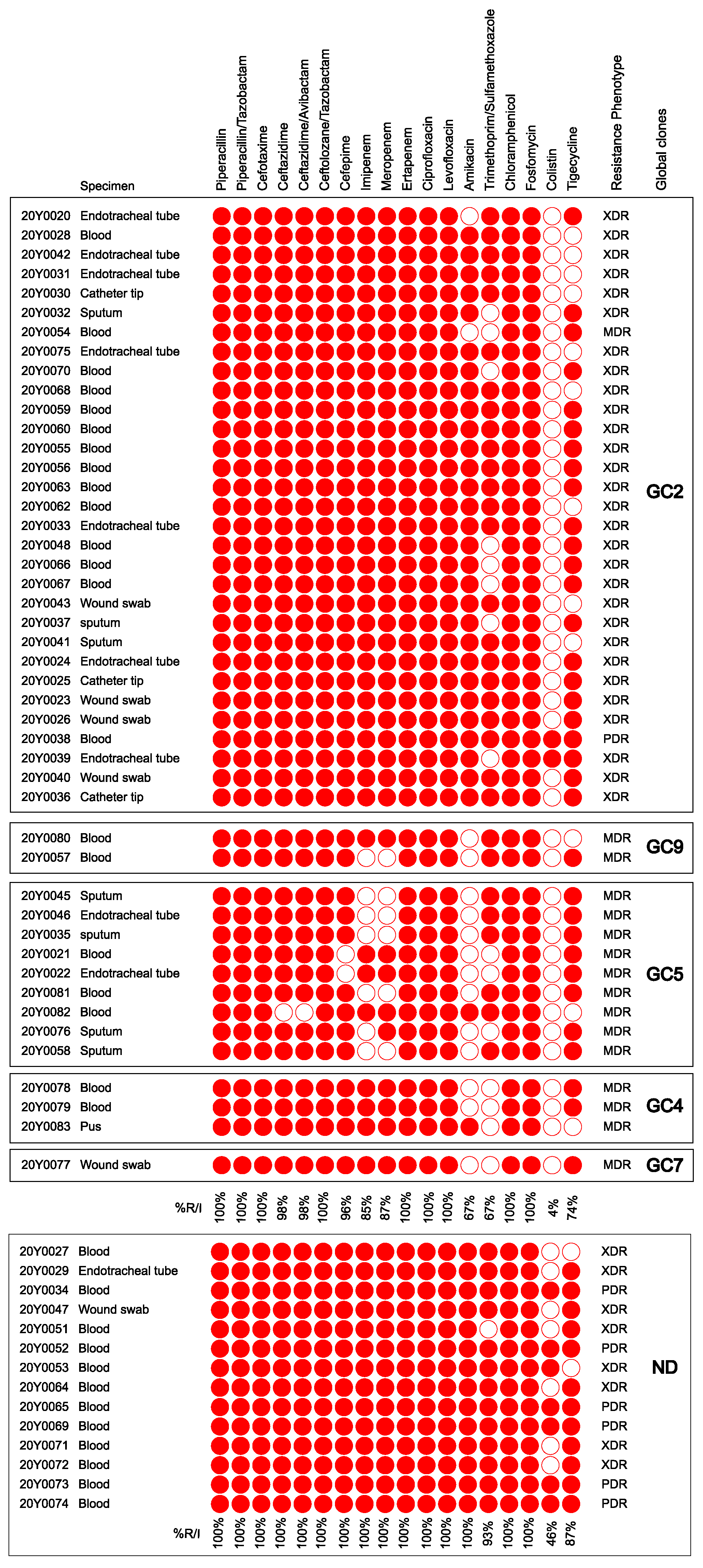
2.1. Bacterial Isolates and Their Antimicrobial Susceptibility Profiles
2.2. Draft Genomes and Annotation Features
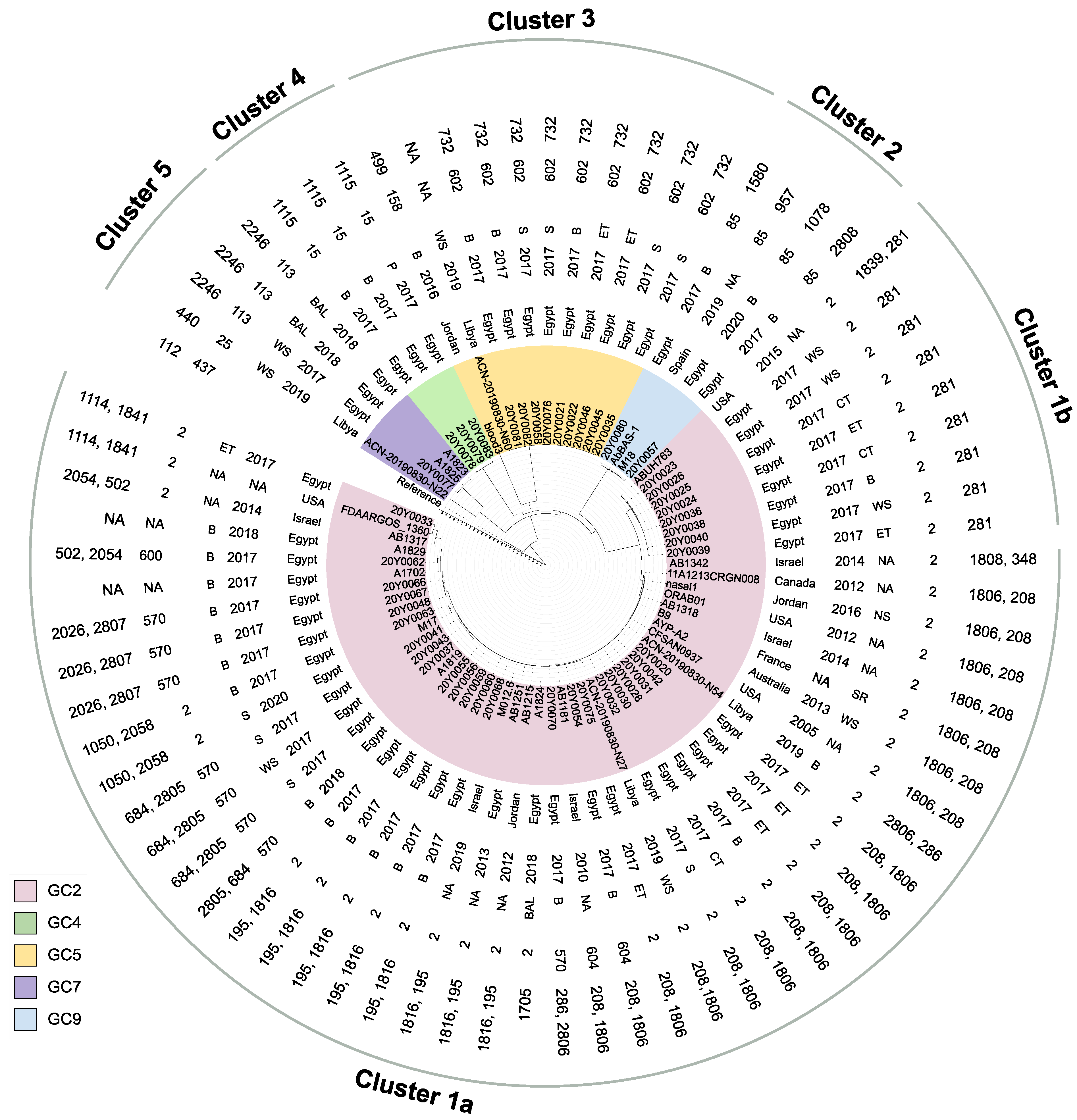
2.3. Phylogeny and Sequence Types
2.4. Correlation between Antimicrobial Resistance and Global Clones
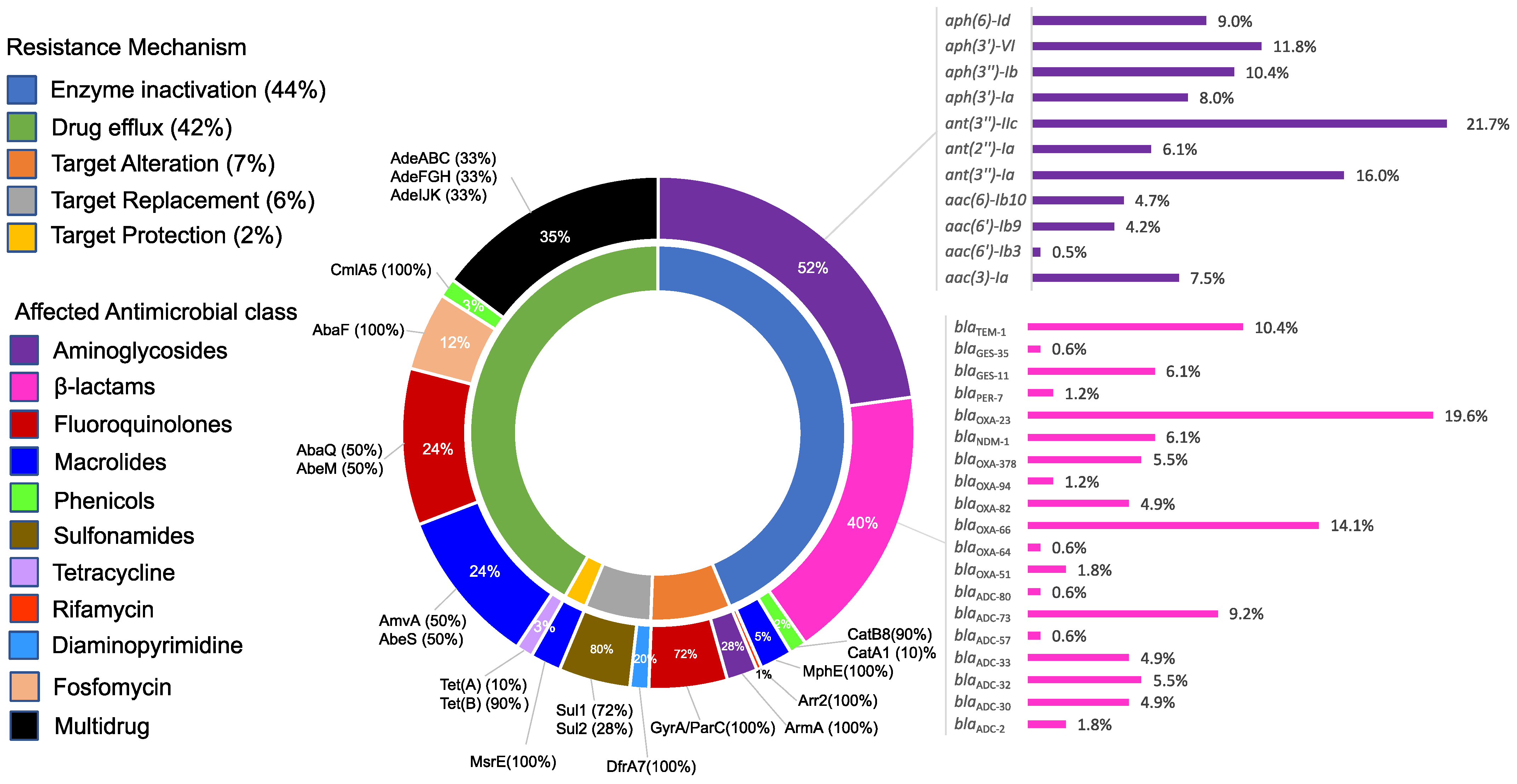
2.5. Resistome Structure and Its Correlation to the Resistance Phenotypes
2.5.1. Carbapenem Resistance
2.5.2. Amikacin Resistance
2.5.3. Colistin Resistance
2.5.4. Tigecycline Resistance
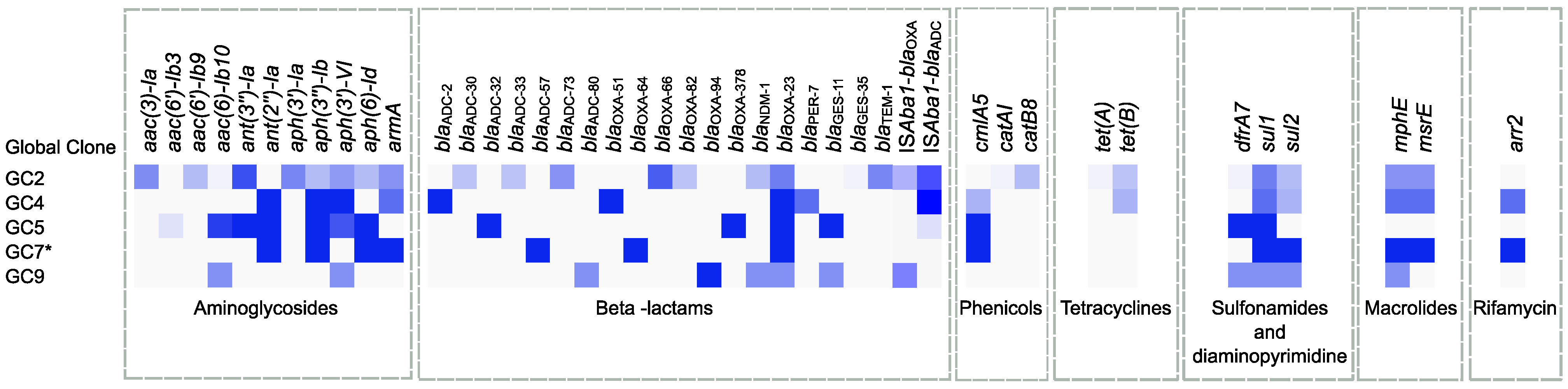
2.6. Global Clone-Specific Resistomes
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolates
4.2. Antimicrobial Susceptibility Testing
4.3. DNA Extraction, Library Preparation, and Whole-Genome Sequencing
4.4. Genome Assembly and Annotation
4.5. Multi-Locus Sequence Typing and Phylogenetic Analysis
4.6. Screening of Antimicrobial Resistance Determinants
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shelenkov, A.; Petrova, L.; Zamyatin, M.; Mikhaylova, Y.; Akimkin, V. Diversity of International High-Risk Clones of Acinetobacter baumannii Revealed in a Russian Multidisciplinary Medical Center during 2017–2019. Antibiotics 2021, 10, 1009. [Google Scholar] [CrossRef]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Garnacho-Montero, J.; Timsit, J.F. Managing Acinetobacter baumannii infections. Curr. Opin. Infect. Dis. 2019, 32, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Elkhatib, W.F.; Khalil, M.A.F.; Ashour, H.M. Integrons and Antiseptic Resistance Genes Mediate Resistance of Acinetobacter baumannii and Pseudomonas aeruginosa Isolates from Intensive Care Unit Patients with Wound Infections. Curr. Mol. Med. 2019, 19, 286–293. [Google Scholar] [CrossRef]
- Chen, C.T.; Wang, Y.C.; Kuo, S.C.; Shih, F.H.; Chen, T.L.; How, C.K.; Yang, Y.S.; Lee, Y.T. Community-acquired bloodstream infections caused by Acinetobacter baumannii: A matched case-control study. J. Microbiol. Immunol. Infect. 2018, 51, 629–635. [Google Scholar] [CrossRef]
- Leung, W.S.; Chu, C.M.; Tsang, K.Y.; Lo, F.H.; Lo, K.F.; Ho, P.L. Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest 2006, 129, 102–109. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Kritsotakis, E.I.; Gikas, A. Pandrug-resistant Gram-negative bacteria: A systematic review of current epidemiology, prognosis and treatment options. J. Antimicrob. Chemother. 2020, 75, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Villoria, A.M.; Valverde-Garduno, V. Antibiotic-Resistant Acinetobacter baumannii Increasing Success Remains a Challenge as a Nosocomial Pathogen. J. Pathog. 2016, 2016, 7318075. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, P.R.; Teng, L.J.; Chen, C.Y.; Chen, W.H.; Yu, C.J.; Ho, S.W.; Luh, K.T. Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg. Infect. Dis. 2002, 8, 827–832. [Google Scholar] [CrossRef]
- Papathanakos, G.; Andrianopoulos, I.; Papathanasiou, A.; Priavali, E.; Koulenti, D.; Koulouras, V. Colistin-Resistant Acinetobacter Baumannii Bacteremia: A Serious Threat for Critically Ill Patients. Microorganisms 2020, 8, 287. [Google Scholar] [CrossRef]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017.
- Jalal, D.; Elzayat, M.G.; Diab, A.A.; El-Shqanqery, H.E.; Samir, O.; Bakry, U.; Hassan, R.; Elanany, M.; Shalaby, L.; Sayed, A.A. Deciphering Multidrug-Resistant Acinetobacter baumannii from a Pediatric Cancer Hospital in Egypt. mSphere 2021, 6, e0072521. [Google Scholar] [CrossRef] [PubMed]
- Al-Agamy, M.H.; Khalaf, N.G.; Tawfick, M.M.; Shibl, A.M.; El Kholy, A. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. Int. J. Infect. Dis. 2014, 22, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Elwakil, W.H.; Rizk, S.S.; El-Halawany, A.M.; Rateb, M.E.; Attia, A.S. Multidrug-Resistant Acinetobacter baumannii Infections in the United Kingdom versus Egypt: Trends and Potential Natural Products Solutions. Antibiotics 2023, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.M.; Hussein, A.F.A.; Al-Agamy, M.H.; Radwan, H.H.; Zafer, M.M. Genetic Configuration of Genomic Resistance Islands in Acinetobacter baumannii Clinical Isolates From Egypt. Front. Microbiol. 2022, 13, 878912. [Google Scholar] [CrossRef] [PubMed]
- Zafer, M.M.; Hussein, A.F.A.; Al-Agamy, M.H.; Radwan, H.H.; Hamed, S.M. Genomic Characterization of Extensively Drug-Resistant NDM-Producing Acinetobacter baumannii Clinical Isolates With the Emergence of Novel bla ADC-257. Front. Microbiol. 2021, 12, 736982. [Google Scholar] [CrossRef] [PubMed]
- Hrabak, J.; Stolbova, M.; Studentova, V.; Fridrichova, M.; Chudackova, E.; Zemlickova, H. NDM-1 producing Acinetobacter baumannii isolated from a patient repatriated to the Czech Republic from Egypt, July 2011. Eurosurveillance 2012, 17, 20085. [Google Scholar] [CrossRef]
- Zarrilli, R.; Pournaras, S.; Giannouli, M.; Tsakris, A. Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 2013, 41, 11–19. [Google Scholar] [CrossRef]
- Müller, C.; Stefanik, D.; Wille, J.; Hackel, M.; Higgins, P.G.; Siefert, H. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii clinical isolates and identification of the novel international clone IC9: Results from a worldwide surveillance study (2012–2016). In Proceedings of the ECCMID 2019: Proceeding of the 29th European Congress of Clinical Microbiology & Infectious Diseases, Amsterdam, The Netherlands, 13–16 April 2019. [Google Scholar]
- Al-Hassan, L.; Elbadawi, H.; Osman, E.; Ali, S.; Elhag, K.; Cantillon, D.; Wille, J.; Seifert, H.; Higgins, P.G. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii From Khartoum State, Sudan. Front. Microbiol. 2021, 12, 628736. [Google Scholar] [CrossRef]
- Holt, K.; Kenyon, J.J.; Hamidian, M.; Schultz, M.B.; Pickard, D.J.; Dougan, G.; Hall, R. Five decades of genome evolution in the globally distributed, extensively antibiotic-resistant Acinetobacter baumannii global clone 1. Microb. Genom. 2016, 2, e000052. [Google Scholar] [CrossRef]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef]
- Salloum, T.; Tannous, E.; Alousi, S.; Arabaghian, H.; Rafei, R.; Hamze, M.; Tokajian, S. Genomic mapping of ST85 blaNDM-1 and blaOXA-94 producing Acinetobacter baumannii isolates from Syrian Civil War Victims. Int. J. Infect. Dis. 2018, 74, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cuenca, F.; Perez-Palacios, P.; Galan-Sanchez, F.; Lopez-Cerero, L.; Lopez-Hernandez, I.; Lopez Rojas, R.; Arca-Suarez, J.; Diaz-de Alba, P.; Rodriguez Iglesias, M.; Pascual, A. First identification of bla(NDM-1) carbapenemase in bla(OXA-94)-producing Acinetobacter baumannii ST85 in Spain. Enferm. Infecc. Y Microbiol. Clin. 2020, 38, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Karah, N.; Sundsfjord, A.; Towner, K.; Samuelsen, O. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updates 2012, 15, 237–247. [Google Scholar] [CrossRef]
- Cerezales, M.; Xanthopoulou, K.; Wille, J.; Bustamante, Z.; Seifert, H.; Gallego, L.; Higgins, P.G. Acinetobacter baumannii analysis by core genome multi-locus sequence typing in two hospitals in Bolivia: Endemicity of international clone 7 isolates (CC25). Int. J. Antimicrob. Agents 2019, 53, 844–849. [Google Scholar] [CrossRef]
- Zhang, G.; Leclercq, S.O.; Tian, J.; Wang, C.; Yahara, K.; Ai, G.; Liu, S.; Feng, J. A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3″)-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 2017, 13, e1006602. [Google Scholar] [CrossRef]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Nigro, S.J.; Hall, R.M. Amikacin resistance plasmids in extensively antibiotic-resistant GC2 Acinetobacter baumannii from two Australian hospitals. J. Antimicrob. Chemother. 2014, 69, 3435–3437. [Google Scholar] [CrossRef]
- Hammerstrom, T.G.; Beabout, K.; Clements, T.P.; Saxer, G.; Shamoo, Y. Acinetobacter baumannii Repeatedly Evolves a Hypermutator Phenotype in Response to Tigecycline That Effectively Surveys Evolutionary Trajectories to Resistance. PLoS ONE 2015, 10, e0140489. [Google Scholar] [CrossRef] [PubMed]
- Beabout, K.; Hammerstrom, T.G.; Perez, A.M.; Magalhaes, B.F.; Prater, A.G.; Clements, T.P.; Arias, C.A.; Saxer, G.; Shamoo, Y. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 2015, 59, 5561–5566. [Google Scholar] [CrossRef]
- Hornsey, M.; Ellington, M.J.; Doumith, M.; Thomas, C.P.; Gordon, N.C.; Wareham, D.W.; Quinn, J.; Lolans, K.; Livermore, D.M.; Woodford, N. AdeABC-mediated efflux and tigecycline MICs for epidemic clones of Acinetobacter baumannii. J. Antimicrob. Chemother. 2010, 65, 1589–1593. [Google Scholar] [CrossRef]
- Lin, M.F.; Lin, Y.Y.; Yeh, H.W.; Lan, C.Y. Role of the BaeSR two-component system in the regulation of Acinetobacter baumannii adeAB genes and its correlation with tigecycline susceptibility. BMC Microbiol. 2014, 14, 119. [Google Scholar] [CrossRef] [PubMed]
- Ghaith, D.M.; Zafer, M.M.; Al-Agamy, M.H.; Alyamani, E.J.; Booq, R.Y.; Almoazzamy, O. The emergence of a novel sequence type of MDR Acinetobacter baumannii from the intensive care unit of an Egyptian tertiary care hospital. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 34. [Google Scholar] [CrossRef] [PubMed]
- Sherif, M.M.; Elkhatib, W.F.; Khalaf, W.S.; Elleboudy, N.S.; Abdelaziz, N.A. Multidrug Resistant Acinetobacter baumannii Biofilms: Evaluation of Phenotypic-Genotypic Association and Susceptibility to Cinnamic and Gallic Acids. Front. Microbiol. 2021, 12, 716627. [Google Scholar] [CrossRef] [PubMed]
- Attia, N.M.; Elbaradei, A. Fluoroquinolone resistance conferred by gyrA, parC mutations, and AbaQ efflux pump among Acinetobacter baumannii clinical isolates causing ventilator-associated pneumonia. Acta Microbiològica Immunol. Hung. 2019, 67, 234–238. [Google Scholar] [CrossRef]
- Abdulzahra, A.T.; Khalil, M.A.F.; Elkhatib, W.F. First report of colistin resistance among carbapenem-resistant Acinetobacter baumannii isolates recovered from hospitalized patients in Egypt. New Microbes New Infect. 2018, 26, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Fam, N.S.; Gamal, D.; Mohamed, S.H.; Wasfy, R.M.; Soliman, M.S.; El-Kholy, A.A.; Higgins, P.G. Molecular Characterization of Carbapenem/Colistin-Resistant Acinetobacter baumannii Clinical Isolates from Egypt by Whole-Genome Sequencing. Infect. Drug Resist. 2020, 13, 4487–4493. [Google Scholar] [CrossRef]
- Nepka, M.; Perivolioti, E.; Kraniotaki, E.; Politi, L.; Tsakris, A.; Pournaras, S. In Vitro Bactericidal Activity of Trimethoprim-Sulfamethoxazole Alone and in Combination with Colistin against Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates. Antimicrob. Agents Chemother. 2016, 60, 6903–6906. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef]
- Kim, D.H.; Choi, J.Y.; Kim, H.W.; Kim, S.H.; Chung, D.R.; Peck, K.R.; Thamlikitkul, V.; So, T.M.; Yasin, R.M.; Hsueh, P.R.; et al. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob. Agents Chemother. 2013, 57, 5239–5246. [Google Scholar] [CrossRef]
- Wareth, G.; Linde, J.; Nguyen, N.H.; Nguyen, T.N.M.; Sprague, L.D.; Pletz, M.W.; Neubauer, H. WGS-Based Analysis of Carbapenem-Resistant Acinetobacter baumannii in Vietnam and Molecular Characterization of Antimicrobial Determinants and MLST in Southeast Asia. Antibiotics 2021, 10, 563. [Google Scholar] [CrossRef]
- Hujer, K.M.; Hamza, N.S.; Hujer, A.M.; Perez, F.; Helfand, M.S.; Bethel, C.R.; Thomson, J.M.; Anderson, V.E.; Barlow, M.; Rice, L.B.; et al. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: Defining a unique family of class C enzymes. Antimicrob. Agents Chemother. 2005, 49, 2941–2948. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.S.; Al-Agamy, M.H.; Ismail, M.A.; Shibl, A.M.; Al-Qahtani, A.A.; Al-Ahdal, M.N.; Forbes, K.J. The transferability of blaOXA-23 gene in multidrug-resistant Acinetobacter baumannii isolates from Saudi Arabia and Egypt. Int. J. Med. Microbiol. 2015, 305, 581–588. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed-Ahmed, M.A.; Amin, M.A.; Tawakol, W.M.; Loucif, L.; Bakour, S.; Rolain, J.M. High prevalence of bla(NDM-1) carbapenemase-encoding gene and 16S rRNA armA methyltransferase gene among Acinetobacter baumannii clinical Isolates in Egypt. Antimicrob. Agents Chemother. 2015, 59, 3602–3605. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Nicolau, D.P. Carbapenems: A potent class of antibiotics. Expert Opin. Pharmacother. 2008, 9, 23–37. [Google Scholar] [CrossRef]
- Yoon, Y.K.; Yang, K.S.; Lee, S.E.; Kim, H.J.; Sohn, J.W.; Kim, M.J. Effects of Group 1 versus Group 2 carbapenems on the susceptibility of Acinetobacter baumannii to carbapenems: A before and after intervention study of carbapenem-use stewardship. PLoS ONE 2014, 9, e99101. [Google Scholar] [CrossRef]
- Hassan, R.; Mukhtar, A.; Hasanin, A.; Ghaith, D. Role of insertion sequence Aba-1 and AdeS in reduced tigecycline susceptibility in MDR-Acinetobacter baumannii clinical isolates from Cairo, Egypt. J. Chemother. 2018, 30, 89–94. [Google Scholar] [CrossRef]
- Leelasupasri, S.; Santimaleeworagun, W.; Jitwasinkul, T. Antimicrobial Susceptibility among Colistin, Sulbactam, and Fosfomycin and a Synergism Study of Colistin in Combination with Sulbactam or Fosfomycin against Clinical Isolates of Carbapenem-Resistant Acinetobacter baumannii. J. Pathog. 2018, 2018, 3893492. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.J.; Yang, H.F. Synergy against extensively drug-resistant Acinetobacter baumannii in vitro by two old antibiotics: Colistin and chloramphenicol. Int. J. Antimicrob. Agents 2017, 49, 321–326. [Google Scholar] [CrossRef]
- Paranos, P.; Vourli, S.; Pournaras, S.; Meletiadis, J. Assessing Clinical Potential of Old Antibiotics against Severe Infections by Multi-Drug-Resistant Gram-Negative Bacteria Using In Silico Modelling. Pharmaceuticals 2022, 15, 1501. [Google Scholar] [CrossRef]
- Abouelfetouh, A.; Mattock, J.; Turner, D.; Li, E.; Evans, B.A. Diversity of carbapenem-resistant Acinetobacter baumannii and bacteriophage-mediated spread of the Oxa23 carbapenemase. Microb. Genom. 2022, 8, 000752. [Google Scholar] [CrossRef] [PubMed]
- Al-Hassan, L.; Zafer, M.M.; El-Mahallawy, H. Multiple sequence types responsible for healthcare-associated Acinetobacter baumannii dissemination in a single centre in Egypt. BMC Infect. Dis. 2019, 19, 829. [Google Scholar] [CrossRef] [PubMed]
- Khuntayaporn, P.; Kanathum, P.; Houngsaitong, J.; Montakantikul, P.; Thirapanmethee, K.; Chomnawang, M.T. Predominance of international clone 2 multidrug-resistant Acinetobacter baumannii clinical isolates in Thailand: A nationwide study. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, F.; Awan, F.; Jiang, H.; Zeng, Z.; Lv, W. Molecular Epidemiology and Clone Transmission of Carbapenem-Resistant Acinetobacter baumannii in ICU Rooms. Front. Cell. Infect. Microbiol. 2021, 11, 633817. [Google Scholar] [CrossRef]
- Levy-Blitchtein, S.; Roca, I.; Plasencia-Rebata, S.; Vicente-Taboada, W.; Velasquez-Pomar, J.; Munoz, L.; Moreno-Morales, J.; Pons, M.J.; Del Valle-Mendoza, J.; Vila, J. Emergence and spread of carbapenem-resistant Acinetobacter baumannii international clones II and III in Lima, Peru. Emerg. Microbes Infect. 2018, 7, 119. [Google Scholar] [CrossRef]
- Mathlouthi, N.; El Salabi, A.A.; Ben Jomaa-Jemili, M.; Bakour, S.; Al-Bayssari, C.; Zorgani, A.A.; Kraiema, A.; Elahmer, O.; Okdah, L.; Rolain, J.M.; et al. Early detection of metallo-beta-lactamase NDM-1- and OXA-23 carbapenemase-producing Acinetobacter baumannii in Libyan hospitals. Int. J. Antimicrob. Agents 2016, 48, 46–50. [Google Scholar] [CrossRef]
- Mathlouthi, N.; Ben Lamine, Y.; Somai, R.; Bouhalila-Besbes, S.; Bakour, S.; Rolain, J.M.; Chouchani, C. Incidence of OXA-23 and OXA-58 Carbapenemases Coexpressed in Clinical Isolates of Acinetobacter baumannii in Tunisia. Microb. Drug Resist. 2018, 24, 136–141. [Google Scholar] [CrossRef]
- Ababneh, Q.; Al-Rousan, E.; Jaradat, Z. Fresh produce as a potential vehicle for transmission of Acinetobacter baumannii. Int. J. Food Contam. 2022, 9, 5. [Google Scholar] [CrossRef]
- Jacobmeyer, L.; Semmler, T.; Stamm, I.; Ewers, C. Genomic Analysis of Acinetobacter baumannii Isolates Carrying OXA-23 and OXA-58 Genes from Animals Reveals ST1 and ST25 as Major Clonal Lineages. Antibiotics 2022, 11, 1045. [Google Scholar] [CrossRef]
- Hamed, S.M.; Elkhatib, W.F.; El-Mahallawy, H.A.; Helmy, M.M.; Ashour, M.S.; Aboshanab, K.M.A. Multiple mechanisms contributing to ciprofloxacin resistance among Gram negative bacteria causing infections to cancer patients. Sci. Rep. 2018, 8, 12268. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Lee, Y.S.; Park, Y.K.; Kim, B.S. Mutations in the gyrA and parC genes in ciprofloxacin-resistant clinical isolates of Acinetobacter baumannii in Korea. Microbiol. Immunol. 2005, 49, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.A.; Salim, M.T.A.; Anwer, B.E.; Aboshanab, K.M.; Aboulwafa, M.M. Impact of target site mutations and plasmid associated resistance genes acquisition on resistance of Acinetobacter baumannii to fluoroquinolones. Sci. Rep. 2021, 11, 20136. [Google Scholar] [CrossRef]
- Roy, S.; Chatterjee, S.; Bhattacharjee, A.; Chattopadhyay, P.; Saha, B.; Dutta, S.; Basu, S. Overexpression of Efflux Pumps, Mutations in the Pumps’ Regulators, Chromosomal Mutations, and AAC(6′)-Ib-cr Are Associated With Fluoroquinolone Resistance in Diverse Sequence Types of Neonatal Septicaemic Acinetobacter baumannii: A 7-Year Single Center Study. Front. Microbiol. 2021, 12, 602724. [Google Scholar] [CrossRef]
- Coyne, S.; Courvalin, P.; Perichon, B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011, 55, 947–953. [Google Scholar] [CrossRef]
- Tal-Jasper, R.; Katz, D.E.; Amrami, N.; Ravid, D.; Avivi, D.; Zaidenstein, R.; Lazarovitch, T.; Dadon, M.; Kaye, K.S.; Marchaim, D. Clinical and Epidemiological Significance of Carbapenem Resistance in Acinetobacter baumannii Infections. Antimicrob. Agents Chemother. 2016, 60, 3127–3131. [Google Scholar] [CrossRef]
- Nguyen, M.; Joshi, S.G. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: A scientific review. J. Appl. Microbiol. 2021, 131, 2715–2738. [Google Scholar] [CrossRef]
- Lima, W.G.; Silva Alves, G.C.; Sanches, C.; Fernandes, S.O.A.; de Paiva, M.C. Carbapenem-resistant Acinetobacter baumannii in patients with burn injury: A systematic review and meta-analysis. Burns 2019, 45, 1495–1508. [Google Scholar] [CrossRef]
- Donald, H.M.; Scaife, W.; Amyes, S.G.; Young, H.K. Sequence analysis of ARI-1, a novel OXA beta-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 2000, 44, 196–199. [Google Scholar] [CrossRef]
- Higgins, P.G.; Dammhayn, C.; Hackel, M.; Seifert, H. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 2010, 65, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Hassan, R.M.; Salem, S.T.; Hassan, S.I.M.; Hegab, A.S.; Elkholy, Y.S. Molecular characterization of carbapenem-resistant Acinetobacter baumannii clinical isolates from Egyptian patients. PLoS ONE 2021, 16, e0251508. [Google Scholar] [CrossRef] [PubMed]
- El Bannah, A.M.S.; Nawar, N.N.; Hassan, R.M.M.; Salem, S.T.B. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii in a Tertiary Care Hospital in Egypt: Clonal Spread of blaOXA-23. Microb. Drug Resist. 2018, 24, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, R.A.; Gebriel, M.G.; Kadry, H.M.; Mosallem, A. Carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa: Characterization of carbapenemase genes and E-test evaluation of colistin-based combinations. Infect. Drug Resist. 2018, 11, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Lopes, B.S.; Amyes, S.G.B. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J. Med. Microbiol. 2012, 61 Pt 8, 1103–1108. [Google Scholar] [CrossRef]
- Zander, E.; Chmielarczyk, A.; Heczko, P.; Seifert, H.; Higgins, P.G. Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J. Antimicrob. Chemother. 2013, 68, 308–311. [Google Scholar] [CrossRef]
- Jaidane, N.; Naas, T.; Oueslati, S.; Bernabeu, S.; Boujaafar, N.; Bouallegue, O.; Bonnin, R.A. Whole-genome sequencing of NDM-1-producing ST85 Acinetobacter baumannii isolates from Tunisia. Int. J. Antimicrob. Agents 2018, 52, 916–921. [Google Scholar] [CrossRef]
- Charfi-Kessis, K.; Mansour, W.; Khalifa, A.B.H.; Mastouri, M.; Nordmann, P.; Aouni, M.; Poirel, L. Multidrug-resistant Acinetobacter baumannii strains carrying the bla(OxA-23) and the bla(GES-11) genes in a neonatology center in Tunisia. Microb. Pathog. 2014, 74, 20–24. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Wu, X.; Schweppe, D.K.; Chavez, J.D.; Mathay, M.; Eng, J.K.; Keller, A.; Bruce, J.E. In Vivo Cross-Linking MS Reveals Conservation in OmpA Linkage to Different Classes of beta-Lactamase Enzymes. J. Am. Soc. Mass Spectrom. 2020, 31, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Wasfi, R.; Rasslan, F.; Hassan, S.S.; Ashour, H.M.; Abd El-Rahman, O.A. Co-Existence of Carbapenemase-Encoding Genes in Acinetobacter baumannii from Cancer Patients. Infect. Dis. Ther. 2021, 10, 291–305. [Google Scholar] [CrossRef]
- Mussi, M.A.; Limansky, A.S.; Viale, A.M. Acquisition of resistance to carbapenems in multidrug-resistant clinical strains of Acinetobacter baumannii: Natural insertional inactivation of a gene encoding a member of a novel family of beta-barrel outer membrane proteins. Antimicrob. Agents Chemother. 2005, 49, 1432–1440. [Google Scholar] [CrossRef]
- Pajand, O.; Rezaee, M.A.; Nahaei, M.R.; Mahdian, R.; Aghazadeh, M.; Soroush, M.H.; Tabrizi, M.S.; Hojabri, Z. Study of the carbapenem resistance mechanisms in clinical isolates of Acinetobacter baumannii: Comparison of burn and non-burn strains. Burns 2013, 39, 1414–1419. [Google Scholar] [CrossRef]
- Abbasi, E.; Goudarzi, H.; Hashemi, A.; Chirani, A.S.; Ardebili, A.; Goudarzi, M.; Sharahi, J.Y.; Davoudabadi, S.; Talebi, G.; Bostanghadiri, N. Decreased carO gene expression and OXA-type carbapenemases among extensively drug-resistant Acinetobacter baumannii strains isolated from burn patients in Tehran, Iran. Acta Microbiol. Immunol. Hung. 2020, 68, 48–54. [Google Scholar] [CrossRef]
- Wareth, G.; Linde, J.; Hammer, P.; Nguyen, N.H.; Nguyen, T.N.M.; Splettstoesser, W.D.; Makarewicz, O.; Neubauer, H.; Sprague, L.D.; Pletz, M.W. Phenotypic and WGS-derived antimicrobial resistance profiles of clinical and non-clinical Acinetobacter baumannii isolates from Germany and Vietnam. Int. J. Antimicrob. Agents 2020, 56, 106127. [Google Scholar] [CrossRef]
- Yoon, E.J.; Goussard, S.; Touchon, M.; Krizova, L.; Cerqueira, G.; Murphy, C.; Lambert, T.; Grillot-Courvalin, C.; Nemec, A.; Courvalin, P. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3′)-VI. mBio 2014, 5, e01972-14. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.M.; Hussein, A.F.A.; Al-Agamy, M.H.; Radwan, H.H.; Zafer, M.M. Tn7382, a novel composite transposon harboring bla(NDM-1) and aphA6 in Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2022, 30, 414–417. [Google Scholar] [CrossRef]
- Xanthopoulou, K.; Urrutikoetxea-Gutierrez, M.; Vidal-Garcia, M.; Diaz de Tuesta Del Arco, J.L.; Sanchez-Urtaza, S.; Wille, J.; Seifert, H.; Higgins, P.G.; Gallego, L. First Report of New Delhi Metallo-beta-Lactamase-6 (NDM-6) in a Clinical Acinetobacter baumannii Isolate From Northern Spain. Front. Microbiol. 2020, 11, 589253. [Google Scholar] [CrossRef]
- Vazquez-Lopez, R.; Solano-Galvez, S.G.; Juarez Vignon-Whaley, J.J.; Abello Vaamonde, J.A.; Padro Alonzo, L.A.; Rivera Resendiz, A.; Muleiro Alvarez, M.; Vega Lopez, E.N.; Franyuti-Kelly, G.; Alvarez-Hernandez, D.A.; et al. Acinetobacter baumannii Resistance: A Real Challenge for Clinicians. Antibiotics 2020, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; St Michael, F.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, L.A.; Herrera, C.M.; Fernandez, L.; Hankins, J.V.; Trent, M.S.; Hancock, R.E. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob. Agents Chemother. 2011, 55, 3743–3751. [Google Scholar] [CrossRef] [PubMed]
- Jenner, L.; Starosta, A.L.; Terry, D.S.; Mikolajka, A.; Filonava, L.; Yusupov, M.; Blanchard, S.C.; Wilson, D.N.; Yusupova, G. Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3812–3816. [Google Scholar] [CrossRef]
- Livermore, D.M. Tigecycline: What is it, and where should it be used? J. Antimicrob. Chemother. 2005, 56, 611–614. [Google Scholar] [CrossRef]
- Xu, C.; Bilya, S.R.; Xu, W. adeABC efflux gene in Acinetobacter baumannii. New Microbes New Infect. 2019, 30, 100549. [Google Scholar] [CrossRef]
- Moore, I.F.; Hughes, D.W.; Wright, G.D. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 2005, 44, 11829–11835. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, C.; Cui, C.Y.; Zhang, Y.; Liu, X.; Cui, Z.H.; Ma, X.Y.; Feng, Y.; Fang, L.X.; Lian, X.L.; et al. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat. Microbiol. 2019, 4, 1457–1464. [Google Scholar] [CrossRef]
- Zheng, W.; Huang, Y.; Wu, W.; Zhu, J.; Zhang, T. Analysis of Efflux Pump System and Other Drug Resistance Related Gene Mutations in Tigecycline-Resistant Acinetobacter baumannii. Comput. Math. Methods Med. 2023, 2023, 8611542. [Google Scholar] [CrossRef]
- Yoon, E.J.; Courvalin, P.; Grillot-Courvalin, C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 2013, 57, 2989–2995. [Google Scholar] [CrossRef]
- Sawant, A.R.; Pagal, S.; Amar, A.K.; Panda, L.; Devi, C.S.; Shashikala, P.; Kanungo, R.; Prashanth, K. Coexistence of blaNDM-1, blaOXA-51, blaOXA-23, and armA in conjunction with novel mutations detected in RND efflux pump regulators in tigecycline resistant clinical isolates of Acinetobacter baumannii. Pathog. Dis. 2022, 80, ftac020. [Google Scholar] [CrossRef] [PubMed]
- Foong, W.E.; Wilhelm, J.; Tam, H.K.; Pos, K.M. Tigecycline efflux in Acinetobacter baumannii is mediated by TetA in synergy with RND-type efflux transporters. J. Antimicrob. Chemother. 2020, 75, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Hagen, R.M.; Kreikemeyer, B.; Warnke, P.; Podbielski, A.; Frickmann, H.; Loderstadt, U. Molecular Epidemiology of Carbapenem-Resistant Acinetobacter baumannii Isolates from Northern Africa and the Middle East. Antibiotics 2021, 10, 291. [Google Scholar] [CrossRef]
- Bouvet, P.; Grimont, P. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov. Acinetobacter haemolyticus sp. nov. Acinetobacter johnsonii sp. nov. and Acinetobacter junii sp. nov. and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter Lwofii. Int. J. Syst. Evol. Microbiol. 1986, 36, 228–240. [Google Scholar]
- Wareth, G.; Linde, J.; Hammer, P.; Splettstoesser, W.D.; Pletz, M.W.; Neubauer, H.; Sprague, L.D. Molecular Characterization of German Acinetobacter baumannii Isolates and Multilocus Sequence Typing (MLST) Analysis Based on WGS Reveals Novel STs. Pathogens 2021, 10, 690. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Turton, J.F.; Livermore, D.M. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 2011, 35, 736–755. [Google Scholar] [CrossRef] [PubMed]
- Camargo, C.H.; Yamada, A.Y.; Nagamori, F.O.; de Souza, A.R.; Tiba-Casas, M.R.; de Moraes Franca, F.A.; Porto, M.; de Lima Garzon, M.L.; Higgins, P.; Madalosso, G.; et al. Clonal spread of ArmA- and OXA-23-coproducing Acinetobacter baumannii International Clone 2 in Brazil during the first wave of the COVID-19 pandemic. J. Med. Microbiol. 2022, 71, 001509. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro-Goncalves, B.; Francisco, A.P.; Vaz, C.; Ramirez, M.; Carrico, J.A. PHYLOViZ Online: Web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res. 2016, 44, W246–W251. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]

| Carbapenem Resistance Determinants | No. of Isolates | GC | IMPR | MERR |
|---|---|---|---|---|
| None | 2 | 2 | 100.0% | 100.0% |
| ISAba1-preceded blaOXA-51-like only | 5 | 2 | 100.0% | 100.0% |
| ISAba1-preceded blaADC only | 4 | 2 | 100.0% | 100.0% |
| blaOXA-23 only | 1 | 7 | 100.0% | 100.0% |
| ISAba1-preceded blaOXA-51-like + ISAba1-preceded blaADC | 2 | 2 | 100.0% | 100.0% |
| blaOXA-23 + ISAba1-preceded blaOXA-51-like | 1 | 2 | 100.0% | 100.0% |
| blaOXA-23 + ISAba1-preceded blaADC | 9 | 2 and 4 | 100.0% | 100.0% |
| blaNDM-1 + ISAba1-preceded blaOXA-51-like | 1 | 9 | 0.0% | 0.0% |
| blaOXA-23 + blaNDM-1 | 1 | 2 | 100.0% | 100.0% |
| blaOXA-23 + blaGES-11 | 9 | 5 and 9 | 44.4% | 55.6% |
| blaOXA-23 + ISAba1-preceded blaOXA-51-like + ISAba1-preceded blaADC | 1 | 2 | 100.0% | 100.0% |
| blaOXA-23 + blaNDM-1 + ISAba1-preceded blaADC | 8 | 2 | 100.0% | 100.0% |
| blaOXA-23 + blaGES-11 + ISAba1-preceded blaADC | 1 | 5 | 0.0% | 0.0% |
| blaOXA-23 + blaGES-35 + ISAba1-preceded blaADC | 1 | 2 | 100.0% | 100.0% |
| Aminoglycoside Resistance Determinants | No. of Isolates | GC | AMKR |
|---|---|---|---|
| None | 15 | 2 and 9 | 86.7% |
| aph(3′)-VI only | 3 | 2 and 4 | 100.0% |
| aac(6′)-Ib10 only | 2 | 5 | 50.0% |
| armA only | 2 | 2 and 7 | 50.0% |
| armA + aac(6′)-Ib9 | 2 | 2 | 100.0% |
| armA + aph(3′)-VI | 6 | 2 and 4 | 66.7% |
| aph(3′)-VI + aac(6′)-Ib10 | 7 | 5 and 9 | 0.0% |
| aph(3′)-VI + aac(6′)-Ib3 | 1 | 5 | 0.0% |
| aph(3′)-VI + aac(6′)-Ib10 + armA | 1 | 2 | 0.0% |
| aph(3′)-VI + aac(6′)-Ib9 + armA | 7 | 2 | 100.0% |
| GC | Unique | Fully Conserved | Not Fully Conserved |
|---|---|---|---|
| blaOXA-23/blaTEM-1-positive GC2 | aph(3′)-Ia/blaTEM-1/blaADC-73/blaGES-35/catA1/catB8/tetA | aph(3′)-Ia/blaTEM-1/blaOXA-66/blaOXA-23/ISAba1-blaADC | aac(3)-Ia/aac(6′)-Ib/ant(3″)-Ia/aph(3″)-Ib/aph(3′)-VI/aph(6)-Id/armA/blaADC-30 */blaADC-73/blaNDM-1/blaGES-35/ISAba1-blaOXA-51-like/catA1/catB8/mphE/msrE/tetA/tetB/sul1/sul2/dfrA7 |
| blaOXA-23/blaTEM-1-negative GC2 | blaADC-33/blaOXA-82 | aac(3)-Ia/ant(3″)-Ia/aph(3′)-VI/blaADC-30 */blaADC-33/blaOXA-66/blaOXA-82/blaOXA-23/ISAba1-blaOXA-51-like/ISAba1-blaADC/sul1/sul2 | |
| GC4 | blaADC-2/blaOXA-51/blaPER-7 | ant(2″)-Ia/aph(3″)-Ib/aph(3′)-VI/blaADC-2/blaOXA-51/blaOXA-23/ISAba1-blaADC | armA/blaPER-7/cmlA5/mphE/msrE/tetB/sul1/sul2/arr2 |
| GC5 | blaADC-32/blaOXA-378 | aac(6′)-Ib/ant(3″)-Ia/ant(2″)-Ia/aph(3″)-Ib/aph(6)-Id/blaADC-32/blaOXA-378/blaOXA-23/blaGES-11/cmlA5/sul1/dfrA7 | aph(3′)-VI/ISAba1-blaADC |
| GC9 | blaADC-32/blaOXA-94 | blaOXA-94 | aac(6′)-Ib/aph(3′)-VI/blaADC-32/blaNDM-1/blaOXA-23/blaGES-11/ISAba1-blaOXA-51-like/mphE/sul1/sul2/dfrA7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamed, S.M.; Elkhatib, W.F.; Brangsch, H.; Gesraha, A.S.; Moustafa, S.; Khater, D.F.; Pletz, M.W.; Sprague, L.D.; Neubauer, H.; Wareth, G. Acinetobacter baumannii Global Clone-Specific Resistomes Explored in Clinical Isolates Recovered from Egypt. Antibiotics 2023, 12, 1149. https://doi.org/10.3390/antibiotics12071149
Hamed SM, Elkhatib WF, Brangsch H, Gesraha AS, Moustafa S, Khater DF, Pletz MW, Sprague LD, Neubauer H, Wareth G. Acinetobacter baumannii Global Clone-Specific Resistomes Explored in Clinical Isolates Recovered from Egypt. Antibiotics. 2023; 12(7):1149. https://doi.org/10.3390/antibiotics12071149
Chicago/Turabian StyleHamed, Samira M., Walid F. Elkhatib, Hanka Brangsch, Ahmed S. Gesraha, Shawky Moustafa, Dalia F. Khater, Mathias W. Pletz, Lisa D. Sprague, Heinrich Neubauer, and Gamal Wareth. 2023. "Acinetobacter baumannii Global Clone-Specific Resistomes Explored in Clinical Isolates Recovered from Egypt" Antibiotics 12, no. 7: 1149. https://doi.org/10.3390/antibiotics12071149
APA StyleHamed, S. M., Elkhatib, W. F., Brangsch, H., Gesraha, A. S., Moustafa, S., Khater, D. F., Pletz, M. W., Sprague, L. D., Neubauer, H., & Wareth, G. (2023). Acinetobacter baumannii Global Clone-Specific Resistomes Explored in Clinical Isolates Recovered from Egypt. Antibiotics, 12(7), 1149. https://doi.org/10.3390/antibiotics12071149









