Multi-Host Pathogen Staphylococcus aureus—Epidemiology, Drug Resistance and Occurrence in Humans and Animals in Poland
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Evaluation of Drug Resistance
4.3. Detection of Resistance Genes
4.4. MLST
4.5. ADSRRS-Fingerprinting
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Garoy, E.Y.; Gebreab, Y.B.; Achila, O.O.; Tekeste, D.G.; Kesete, R.; Ghirmay, R.; Kiflay, R.; Tesfu, T. Methicillin-Resistant Staphylococcus aureus (MRSA): Prevalence and Antimicrobial Sensitivity Pattern among Patients-A Multicenter Study in Asmara, Eritrea. Can. J. Infect. Dis. Med. Microbiol. 2019, 2019, 8321834. [Google Scholar] [CrossRef] [PubMed]
- Nowakiewicz, A.; Ziółkowska, G.; Zięba, P.; Gnat, S.; Wojtanowicz-Markiewicz, K.; Trościańczyk, A. Coagulase-positive Staphylococcus isolated from wildlife: Identification, molecular characterization and evaluation of resistance profiles with focus on a methicillin-resistant strain. Comp. Immunol. Microbiol. Infect. Dis. 2016, 44, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2019, 21, 169–176. [Google Scholar] [CrossRef]
- Conly, J.M.; Johnston, B.L. VISA, hetero-VISA and VRSA: The end of the vancomycin era? Can. J. Infect. Dis. 2002, 13, 282–284. [Google Scholar] [CrossRef]
- Saunders, N.A.; Holmes, A. Multilocus sequence typing (MLST) of Staphylococcus aureus. Methods Mol. Biol. 2007, 391, 71–85. [Google Scholar] [CrossRef]
- Krawczyk, B.; Leibner, J.; Barańska-Rybak, W.; Samet, A.; Nowicki, R.; Kur, J. ADSRRS-fingerprinting and PCR MP techniques for studies of intraspecies genetic relatedness in Staphylococcus aureus. J. Microbiol. Methods. 2007, 71, 114–122. [Google Scholar] [CrossRef]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Pantosti, A. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front. Microbiol. 2012, 3, 127. [Google Scholar] [CrossRef]
- Available online: https://www.who.int/health-topics/one-health#tab=tab_1 (accessed on 22 June 2023).
- Matynia, B.; Młodzinska, E.; Hryniewicz, W. Antimicrobial susceptibility patterns of Staphylococcus aureus in Poland obtained by the National Quality Assurance Programme. Clin. Microbiol. Infect. 2005, 11, 379–385. [Google Scholar] [CrossRef]
- Mroczkowska, A.; Żmudzki, J.; Marszałek, N.; Orczykowska-Kotyna, M.; Komorowska, I.; Nowak, A.; Grzesiak, A.; Czyżewska-Dors, E.; Dors, A.; Pejsak, Z.; et al. Livestock-associated Staphylococcus aureus on Polish pig farms. PLoS ONE 2017, 12, e0170745. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Fan, P.; Liu, T.; Yang, A.; Boughton, R.K.; Pepin, K.M.; Miller, R.S.; Jeong, K.C. Transmission of antibiotic resistance at the wildlife-livestock interface. Commun. Biol. 2022, 5, 585. [Google Scholar] [CrossRef] [PubMed]
- Boerlin, P.; Reid-Smith, R. Antimicrobial resistance: Its emergence and transmission. Anim. Health Res. Rev. 2008, 9, 115–126. [Google Scholar] [CrossRef]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Nowakiewicz, A.; Zięba, P.; Ziółkowska, G.; Gnat, S.; Muszyńska, M.; Tomczuk, K.; Majer Dziedzic, B.; Ulbrych, Ł.; Trościańczyk, A. Free-Living species of carnivorous mammals in Poland: Red Fox, Beech Marten, and Raccoon as a potential reservoir of Salmonella, Yersinia, Listeria spp. and coagulase-positive Staphylococcus. PLoS ONE 2016, 11, e0155533. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Giulieri, S.G.; Wong Fok Lung, T.; Baines, S.L.; Sharkey, L.K.; Lee, J.Y.H.; Hachani, A.; Monk, I.R.; Stinear, T.P. Staphylococcus aureus host interactions and adaptation. Nat. Rev. Microbiol. 2023, 21, 380–395. [Google Scholar] [CrossRef]
- European Medicines Agency, European Surveillance of Veterinary Antimicrobial Consumption. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2021 (EMA/795956/2022). 2022. Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2021-trends-2010-2021-twelfth-esvac_en.pdf. (accessed on 22 June 2023).
- Sineke, N.; Asante, J.; Amoako, D.G.; Abia, A.L.K.; Perrett, K.; Bester, L.A.; Essack, S.Y. Staphylococcus aureus in intensive pig production in South Africa: Antibiotic resistance, virulence determinants, and clonality. Pathogens 2021, 10, 317. [Google Scholar] [CrossRef]
- Gan, T.; Shu, G.; Fu, H.; Yan, Q.; Zhang, W.; Tang, H.; Yin, L.; Zhao, L.; Lin, J. Antimicrobial resistance and genotyping of Staphylococcus aureus obtained from food animals in Sichuan Province, China. BMC Vet. Res. 2021, 17, 177. [Google Scholar] [CrossRef]
- Mama, O.M.; Ruiz-Ripa, L.; Fernández-Fernández, R.; González-Barrio, D.; Ruiz-Fons, F.; Torres, C. High frequency of coagulase-positive staphylococci carriage in healthy wild boar with detection of MRSA of lineage ST398-t011. FEMS Microbiol. Lett. 2019, 366, fny292. [Google Scholar] [CrossRef]
- Krukowski, H.; Bakuła, Z.; Iskra, M.; Olender, A.; Bis-Wencel, H.; Jagielski, T. The first outbreak of methicillin-resistant Staphylococcus aureus in dairy cattle in Poland with evidence of on-farm and intrahousehold transmission. J. Dairy Sci. 2020, 103, 10577–10584. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef] [PubMed]
- Młynarczyk, G.; Młynarczyk, A.; Łuczak, M. VISA i hetero-VISA wśród szczepów Staphylococcus aureus izolowanych z przypadków klinicznych. [VISA and hetero-VISA among Staphylococcus aureus strains isolated from clinical cases]. Med. Mikrobiol. 2002, 54, 9–20. [Google Scholar]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef]
- European Medicines Agency. Reflection Paper on Use of Aminoglycosides in Animals in the European Union: Development of Resistance and Impact on Human and Animal Health. 2018, EMA/CVMP/AWP/721118/2014. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-use-aminoglycosides-animals-european-union-development-resistance-impact-human_en.pdf (accessed on 22 June 2023).
- van Duijkeren, E.; Schwarz, C.; Bouchard, D.; Catry, B.; Pomba, C.; Baptiste, K.E.; Moreno, M.A.; Rantala, M.; Ružauskas, M.; Sanders, P.; et al. The use of aminoglycosides in animals within the EU: Development of resistance in animals and possible impact on human and animal health: A review. J. Antimicrob. Chemother. 2019, 74, 2480–2496. [Google Scholar] [CrossRef] [PubMed]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 8088. [Google Scholar] [CrossRef] [PubMed]
- Trzcinski, K.; Cooper, B.S.; Hryniewicz, W.; Dowson, C.G. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2000, 45, 763–770. [Google Scholar] [CrossRef]
- European Medicines Agency. Reflection Paper on the Use of Macrolides, Lincosamides and Streptogramins (MLS) in Food-Producing Animals in the European Union: Development of Resistance and Impact on Human and Animal Health. 2011, EMA/CVMP/SAGAM/741087/2009. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/reflection-paper-use-macrolides-lincosamides-streptogramins-mls-food-producing-animals-european_en-0.pdf (accessed on 22 June 2023).
- Schmitz, F.-J.; Sadurski, R.; Kray, A.; Boos, M.; Geisel, R.; Köhrer, K.; Verhoef, J.; Fluit, A.C. Prevalence of macrolide-resistance genes in Staphylococcus aureus and Enterococcus faecium isolates from 24 European university hospitals. J. Antimicrob. Chemother. 2000, 45, 891–894. [Google Scholar] [CrossRef]
- Leclercq, R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef]
- Ojo, K.K.; Striplin, M.J.; Ulep, C.C.; Close, N.S.; Zittle, J.; Luis, H.; Bernardo, M.; Leitao, J.; Roberts, M.C. Staphylococcus efflux msr(A) Gene characterized in Streptococcus, Enterococcus, Corynebacterium, and Pseudomonas isolates. Antimicrob. Agents Chemother. 2006, 50, 1089–1091. [Google Scholar] [CrossRef]
- Ghanbari, F.; Ghajavand, H.; Havaei, R.; Jami, M.S.; Khademi, F.; Heydari, L.; Shahin, M.; Havaei, S.A. Distribution of erm genes among Staphylococcus aureus isolates with inducible resistance to clindamycin in Isfahan, Iran. Adv. Biomed. Res. 2016, 5, 62. [Google Scholar] [CrossRef]
- Trościańczyk, A.; Nowakiewicz, A.; Gnat, S.; Łagowski, D.; Osińska, M.; Chudzik-Rząd, B. Comparative study of multidrug-resistant Enterococcus faecium obtained from different hosts. J. Med. Microbiol. 2021, 70, 001340. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Shen, J.; Kadlec, K.; Wang, Y.; Michael, G.B.; Feßler, A.T.; Vester, B. Lincosamides, Streptogramins, Phenicols, and Pleuromutilins: Mode of Action and Mechanisms of Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a027037. [Google Scholar] [CrossRef] [PubMed]
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in animals. Microbiol. Spectrum. 2019, 7, GPP3-0060-2019. [Google Scholar] [CrossRef] [PubMed]
- Guinane, C.M.; Zakour, N.L.B.; Tormo-Mas, M.A.; Weinert, L.A.; Lowder, B.V.; Cartwright, R.A.; Smyth, D.S.; Smyth, C.J.; Lindsay, J.A.; Gould, K.A.; et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2010, 2, 454–466. [Google Scholar] [CrossRef]
- Rabello, R.F.; Moreira, B.M.; Lopes, R.M.; Teixeira, L.M.; Riley, L.W.; Castro, A.C. Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 2007, 56, 1505–1511. [Google Scholar] [CrossRef]
- Garbacz, K.; Piechowicz, L.; Podkowik, M.; Mroczkowska, A.; Empel, J.; Bania, J. Emergence and spread of worldwide Staphylococcus aureus clones among cystic fibrosis patients. Infect. Drug. Resist. 2018, 11, 247–255. [Google Scholar] [CrossRef]
- Lozano, C.; Gómez-Sanz, E.; Benito, D.; Aspiroz, C.; Zarazaga, M.; Torres, C. Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int. J. Med. Microbiol. 2011, 301, 500–505. [Google Scholar] [CrossRef]
- Pirolo, M.; Visaggio, D.; Gioffrè, A.; Artuso, I.; Gherardi, M.; Pavia, G.; Samele, P.; Ciambrone, L.; Di Natale, R.; Spatari, G.; et al. Unidirectional animal-to-human transmission of methicillin-resistant Staphylococcus aureus ST398 in pig farming; evidence from a surveillance study in southern Italy. Antimicrob. Resist. Infect. Control. 2019, 8, 187. [Google Scholar] [CrossRef]
- Lu, H.; Zhao, L.; Si, Y.; Jian, Y.; Wang, Y.; Li, T.; Dai, Y.; Huang, Q.; Ma, X.; He, L.; et al. The surge of hypervirulent ST398 MRSA lineage with higher biofilm-forming ability is a critical threat to clinics. Front. Microbiol. 2021, 12, 636788. [Google Scholar] [CrossRef]
- Ekroth, A.K.E.; Gerth, M.; Stevens, E.J.; Ford, S.A.; King, K.C. Host genotype and genetic diversity shape the evolution of a novel bacterial infection. ISME J. 2021, 15, 2146–2157. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Kozajda, A.; Jeżak, K.; Kapsa, A. Airborne Staphylococcus aureus in different environments—A review. Environ. Sci. Pollut. Res. 2019, 26, 34741–34753. [Google Scholar] [CrossRef]
- Sasaki, T.; Tsubakishita, S.; Tanaka, Y.; Sakusabe, A.; Ohtsuka, M.; Hirotaki, S.; Kawakami, T.; Fukata, T.; Hiramatsu, K. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. 2010, 48, 765–769. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI supplement M100; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2023; ISBN 978-1-68440-171-0. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 6th ed.; CLSI document Vet01S; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- PubMLST Website. Available online: https://pubmlst.org/organisms/staphylococcus-aureus (accessed on 21 March 2023).
- Krawczyk, B.; Lewandowski, K.; Bronk, M.; Samet, A.; Myjak, P.; Kur, J. Evaluation of a novel method based on amplification of DNA fragments surrounding rare restriction sites (ADSRRS fingerprinting) for typing strains of vancomycin-resistant Enterococcus faecium. J. Microbiol. Methods. 2003, 52, 341–351. [Google Scholar] [CrossRef]
- Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Alvarez, L.; Holden, M.T.G.; Lindsay, H.; Webb, C.; Brown, D.F.J.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Methicillin resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Martineau, F.; Picard, F.J.; Lansac, N.; Menard, C.; Roy, P.H.; Ouellette, M.; Bergeron, M. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2000, 44, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Strommenger, B.; Kettlitz, C.; Werner, G.; Witte, W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 4089–4094. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Agersù, Y.; Ahrens, P.; Jùrgensen, J.C.; Madsen, M.; Jensen, L.B. Antimicrobial susceptibility and presence of resistance genes in staphylococci from poultry. Vet. Microbiol. 2000, 74, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Schnellmann, C.; Gerber, V.; Rossano, A.; Jaquier, V.; Panchaud, Y.; Doherr, M.G.; Thomann, A.; Straub, R.; Perreten, V. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp.isolated from the skin of horses before and after clinic admission. J. Clin. Microbiol. 2006, 44, 4444–4454. [Google Scholar] [CrossRef] [PubMed]
- van de Klundert, J.A.M.; Vliegenthart, J.S. PCR detection of genes for aminoglycoside-modifying enzymes. In Diagnostic Molecular Microbiology. Principles and Applications; Persing, D.H., Smith, T.F., Tenover, F.C., White, T.J., Eds.; American Society for Microbiology: Washington, DC, USA, 1993; pp. 547–552. [Google Scholar]
- Depardieu, F.; Perichon, B.; Courvalin, P. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 2004, 42, 5857–5860. [Google Scholar] [CrossRef] [PubMed]
| Host | A † (n = 35) | B (n = 19) | C (n = 30) | D (n = 10) | E (n = 29) | F (n = 26) | G (n = 31) | H (n = 5) | I (n = 22) | J (n = 30) | K (n = 25) | L (n = 1) | M (n = 25) | N (n = 32) | O (n = 32) | P (n = 36) | R (n = 27) | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pigs (n = 248) | 12/37.5 | 3/9 | 36/100 | 0 | 51/21 | |||||||||||||
| Cattle (n = 680) | 1/3 | 1/5 | 1/3 | 0 | 12/41 | 3/11 | 4/13 | 3/60 | 1/4 | 0 | 8/32 | 1/100 | 1/4 | 36/5 | ||||
| Human carriers ‡ (n = 15) | nt § | 1 | 2 | 2 | 0 | 0 | 1 | nt | 1 | 1 | nt | nt | nt | 0 | nt | nt | 2 | 10/67 |
| Host | Agent † | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | FOX * | VAN | GEN | TET * | ERY * | CLI * CLIind | QD | CIP | ENR | CHL | SXT | RIF | NIT | LZD | |
| ‡ H (140) | § 96/69 | 6/4 | 25/18 | 13/9 | 24/17 | 49/35 | 26/19 7/5 | 3/2 | 26/19 | 13/9 | 7/5 | 2/1 | 1/1 | 3/2 | 1/1 |
| P (51) | 49/96 | 39/76 | 0/0 | 4/8 | 48/94 | 44/86 | 51/100 0/0 | 9/18 | 0/0 | 0/0 | 0/0 | 2/4 | 0/0 | 0/0 | 0/0 |
| C (36) | 3/8 | 1/3 | 5/14 | 0/0 | 2/6 | 1/3 | 1/3 0/0 | 0/0 | 2/6 | 1/3 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| W (12) | 2/17 | 0/0 | 0/0 | 1/8 | 1/8 | 0/0 | 0/0 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| T (239) | 150/63 | 46/19 | 30/13 | 18/8 | 75/31 | 94/39 | 78/33 7/3 | 12/5 | 28/12 | 14/6 | 7/3 | 4/2 | 1/0.4 | 3/1 | 1/0.4 |
| Host | Resistance Profile † | Proportion (Number/Percentage) |
|---|---|---|
| human (n = 140) § ca (n = 10) ho (n = 130) | PEN | 4/40 (B, I, J, G); 24/18 |
| CLI | 2/1.5 | |
| TET | 1/0.8 | |
| CHL | 1/0.8 | |
| CIP | 2/1.5 | |
| VAN | 3/2 | |
| PEN, VAN | 1/10 (C), 11/8 | |
| PEN, CLI | 1/10(C) | |
| PEN, ERY | 7/5 | |
| PEN, TET | 3/2 | |
| PEN, SXT | 1/0.8 | |
| PEN, GEN | 1/0.8 | |
| PEN, CHL | 1/0.8 | |
| PEN, CIP | 2/1.5 | |
| CLI, ERY | 3/2 | |
| CIP, ENR | 2/1.5 | |
| TET, GEN | 1/0.8 | |
| TET, VAN | 1/0.8 | |
| ERY, CHL | 1/0.8 | |
| PEN, CLI, VAN | 1/0.8 | |
| PEN, CLI, ERY | 4/3 | |
| PEN, TET, ERY | 2/20 (R), 3/2 | |
| PEN, TET, CIP | 1/0.8 | |
| PEN, ERY, CIP | 2/1.5 | |
| PEN, CIP, VAN | 1/0.8 | |
| CLI, ERY, GEN | 1/0.8 | |
| CLI, ERY, VAN | 1/0.8 | |
| PEN, CLI, ERY, CIP | 2/1.5 | |
| PEN, CLI, ERY, GEN | 2/1.5 | |
| PEN, CLI, ERY, ENR | 1/0.8 | |
| PEN, CLI, TET, ERY | 1/0.8 | |
| PEN, FOX, CIP, ENR | 1/0.8 | |
| PEN, QD, TET, CIP | 1/0.8 | |
| PEN, ERY, CIP, VAN | 1/0.8 | |
| PEN, FOX, TET, CHL | 1/0.8 | |
| PEN, CLI, TET, ERY, VAN | 1/10 (D) | |
| PEN, QD, TET, NIT, CIP, | 1/0.8 | |
| PEN, CLI, ERY, CIP, ENR | 4/3 | |
| PEN, CLI, TET, ERY, GEN | 1/10 (D), 1/0.8 | |
| PEN, FOX, CLI, ERY, CIP | 1/0.8 | |
| CLI, TET, ERY, CHL, CIP, ENR | 1/0.8 | |
| PEN, FOX, CLI, ERY, CIP, ENR | 1/0.8 | |
| PEN, CLI, TET, ERY, GEN, VAN | 2/1.5 | |
| PEN, ERY, GEN, CHL, CIP, ENR | 1/0.8 | |
| PEN, FOX, ERY, GEN, CIP, ENR, VAN | 1/0.8 | |
| CLI, SXT, TET, ERY, GEN, NIT, CHL, CIP, ENR | 1/0.8 | |
| PEN, FOX, CLI, QD, TET, RIF, ERY, GEN, NIT, LZD, VAN | 1/0.8 | |
| S | 24/17 | |
| pigs (n = 51) | PEN, CLI | 1/2 (N) |
| PEN, CLI, TET | 2/4 (N) | |
| CLI, QD, ERY | 1/2 (N) | |
| PEN, CLI, TET, ERY | 1/2 (N) | |
| PEN, CLI, TET, GEN | 1/2 (N) | |
| PEN, CLI, QD, TET | 3/6 (N) | |
| CLI, QD, ERY, GEN | 1/2 (N) | |
| PEN, FOX, CLI, TET, ERY | 36/71 (P) | |
| PEN, CLI, QD, TET, ERY | 1/2 (N) | |
| PEN, FOX, CLI, QD, TET, ERY | 1/2 (O) | |
| PEN, FOX, CLI, SXT, TET, ERY | 1/2 (O) | |
| PEN, CLI, SXT, QD, TET, ERY, GEN | 1/2 (N) | |
| PEN, FOX, CLI, QD, TET, ERY, GEN | 1/2 (O) | |
| cattle (n = 36) | PEN | 2/6 (B, H) |
| TET | 1/3 (A) | |
| CIP | 1/3 (E) | |
| VAN | 5/14 (E (n = 2), F (n = 1), I (n = 1), M (n = 1) | |
| CLI, ERY | 1/3 (G) | |
| PEN, FOX, TET, CIP, ENR | 1/3 (L) | |
| S | 25/69 (C (n = 1), E (n = 9), F (n = 2), G (n = 3), H (n = 2), K (n = 8)) | |
| wildlife (n = 12) | PEN | 1/8 |
| GEN | 1/8 | |
| PEN, TET | 1/8 | |
| S | 9/75 |
| Host † | Number of Drugs to Which Resistance Was Found | |||||||
|---|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 9 | 11 | Total | |
| H ca (n = 10) | 2/20 | 2/20 | 4/40 | |||||
| H ho (n = 130) | 14/11 | 10/8 | 7/5 | 5/4 | 1/0.8 | 1/0.8 | 1/0.8 | 39/30 |
| P (n = 51) | 3/6 | 6/12 | 37/73 | 2/4 | 2/4 | 50/98 | ||
| C (n = 36) | 1/3 | 1/3 | ||||||
| W (n = 12) | ||||||||
| Total (n = 239) | 19/8 | 16/7 | 47/20 | 7/3 | 3/1 | 1/0.4 | 1/0.4 | 94/39 |
| Host † | Resistance Genes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaZ * | mecA * | mecC * | aac(6′)-Ie-aph(2″)-Ia | tetM * | tetK * | ermA * | ermB * | ermC | msrA | Cat (pC221) | Cat (pC223) | |
| † H (140) | 88/63 | 5/4 | 5/4 | 10/7 | 23/16 | 14/10 | 9/6 | 21/15 | 12/9 | 5/4 | 4/3 | 1/1 |
| P (51) | 49/96 | 39/76 | 39/76 | 1/2 | 47/92 | 39/76 | 36/71 | 39/76 | 3/6 | 0/0 | 0/0 | 0/0 |
| C (36) | 3/8 | 1/3 | 1/3 | 0/0 | 2/6 | 2/6 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| W (12) | 1/8 | 0/0 | 0/0 | 0/0 | 1/8 | 1/8 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 |
| T (239) | 141/59 | 45/19 | 45/19 | 11/5 | 73/31 | 56/23 | 45/19 | 60/25 | 15/6 | 5/2 | 4/2 | 1/0.4 |
| No. of Isolates Resistant to: | Number of Resistance Genes | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| penicillin (n = 150) | 8/5 | 97/65: blaZ | 1/0.7: mecA, mecC | 44/29: mecA, mecC, blaZ | |
| cefoxitin (methicillin) (n = 46) | 1/2 | 45/98: mecA, mecC | |||
| gentamicin (n = 18) | 7/39 | 11/61: aac(6′)-Ie-aph(2″)-Ia | |||
| tetracycline (n = 75) | 1/1 | 18/24: tetM | 56/75: tetM, tetK | ||
| erythromycin (n = 94) | 21/22 | 25/27: ermB (14/15) ermC (10/11) msrA (1/1) | 46/49: ermA, ermB (42/45) ermB, ermC (1/1) ermA, ermC (1/1) ermC, msrA (1/1) ermB, msrA (1/1) | 2/2: ermA, ermB, ermC, msrA | |
| clindamycin (n-85) | 18/21 | 21/25: ermB (10/12) ermC (11/13) | 44/52: ermA, ermB (42/49) ermB, ermC (1/1) ermA, ermC (1/1) | 2/2: ermA, ermB, ermC | |
| quinupristin-dalfopristin (n = 12) | 6/50 | 5/42: ermC (3/25) ermB (2/17) | 1/8: ermB, msrA | ||
| chloramphenicol (n = 7) | 2/29 | 5/71: cat (pC221) (4/57) cat (pC223) (1/14) | |||
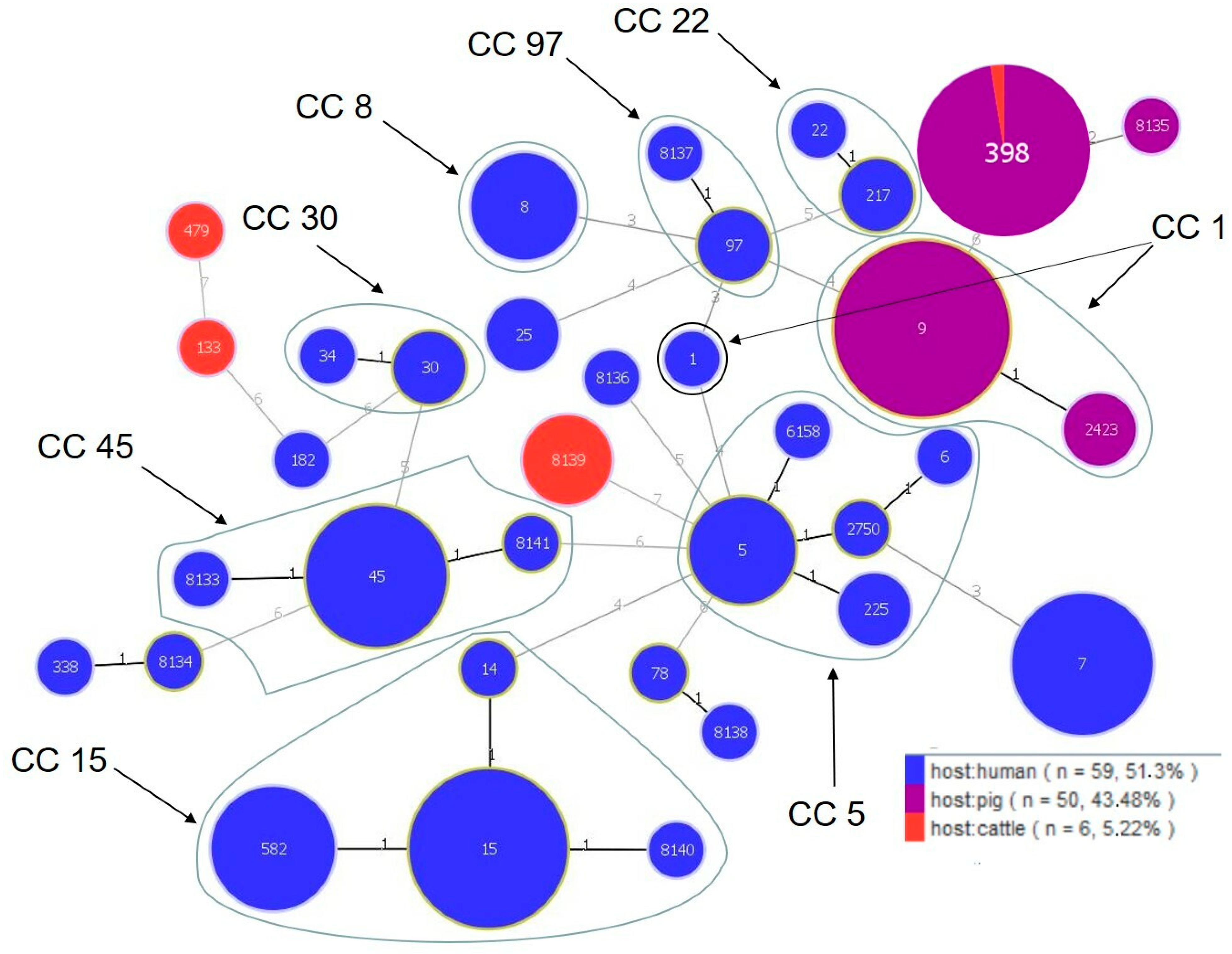
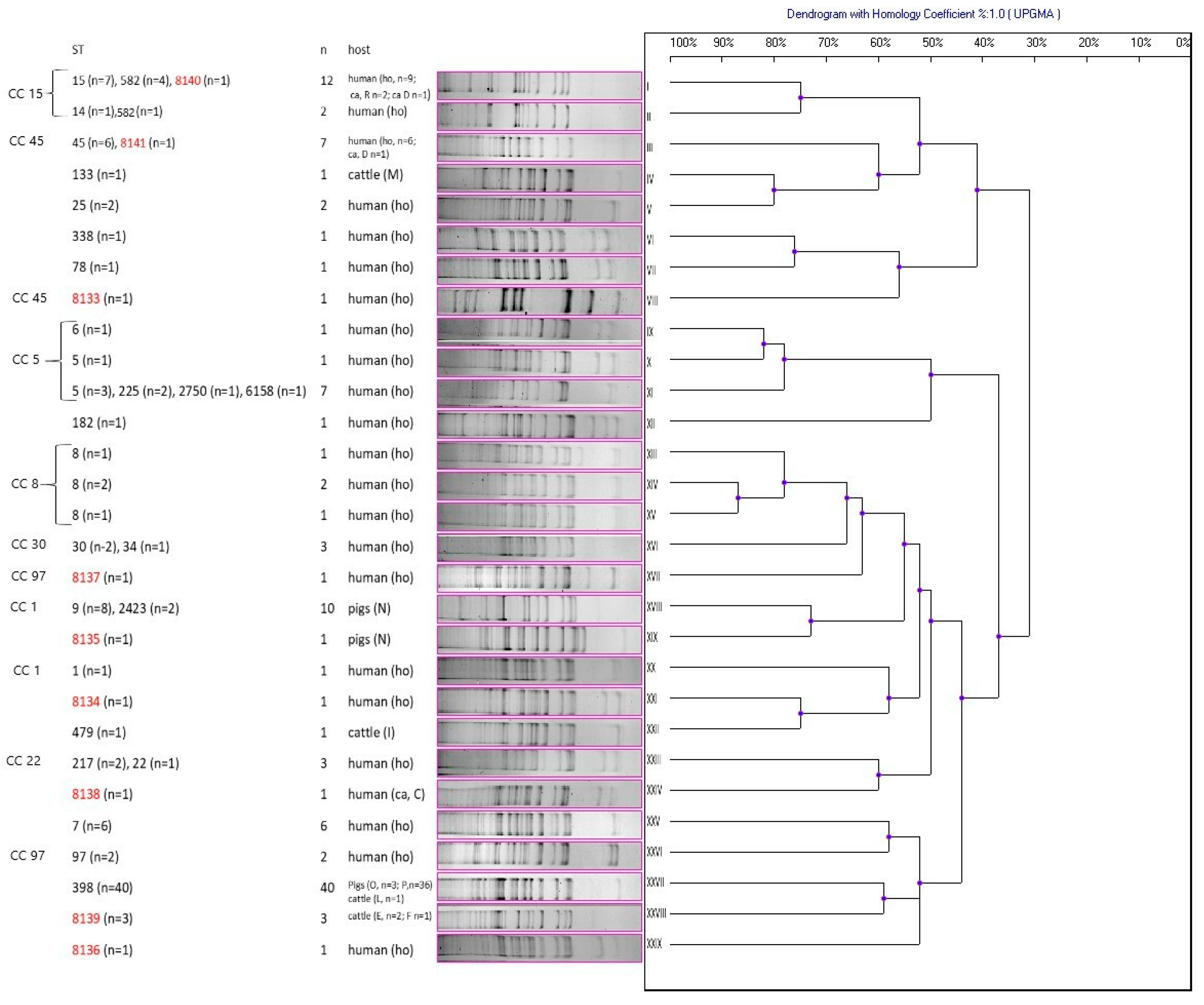
| CC a | ST | n | Host b | Genotypic Profile c | Resistance Profile d | Resistance Genes e | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaZ | mecA | mecC | aac | tetM | tetK | ermA | ermB | ermC | msrA | cat221 | cat223 | ||||||
| 1 | 1 | 1 | human (ho) | XX | PEN, CLI, ERY, CIP | + | + | ||||||||||
| 1 | 9 | 1 | pigs (N) | XVIII | PEN, CLI, TET | + | |||||||||||
| 1 | 9 | 3 | pigs (N) | XVIII | PEN, CLI, QD, TET | + | + | ||||||||||
| 1 | 9 | 1 | pigs (N) | XVIII | PEN, CLI, TET | + | + | ||||||||||
| 1 | 9 | 1 | pigs (N) | XVIII | PEN, CLI, TET, ERY | + | + | ||||||||||
| 1 | 9 | 1 | pigs (N) | XVIII | PEN, CLI, QD, TET, ERY | + | + | ||||||||||
| 1 | 9 | 1 | pigs (N) | XVIII | PEN, CLI, TET, GEN | + | + | ||||||||||
| 1 | 2423 | 1 | pigs (N) | XVIII | CLI, QD, ERY | + | |||||||||||
| 1 | 2423 | 1 | pigs (N) | XVIII | CLI, QD, ERY, GEN | + | |||||||||||
| 5 | 5 | 1 | human (ho) | XI | PEN, ERY, GEN, CIP, ENR, VAN | + | + | + | + | ||||||||
| 5 | 5 | 1 | human (ho) | X | PEN, CLI, ERY, CIP, ENR | + | |||||||||||
| 5 | 5 | 1 | human (ho) | XI | CLI, SXT, TET, ERY, GEN, NIT, CIP, CHL, ENR | + | + | + | + | ||||||||
| 5 | 5 | 1 | human (ho) | XI | VAN | ||||||||||||
| 5 | 6 | 1 | human (ho) | IX | PEN, ERY, CIP | + | |||||||||||
| 5 | 225 | 2 | human (ho) | XI | PEN, CLI, ERY, CIP, ENR | + | + | + | |||||||||
| 5 | 2750 | 1 | human (ho) | XI | PEN, TET, FOX, CHL | + | + | + | + | + | |||||||
| 5 | 6158 | 1 | human (ho) | XI | PEN, TET, ERY | + | + | + | |||||||||
| 8 | 8 | 1 | human (ho) | XIII | PEN, CLI, FOX, ERY, CIP, ENR | + | + | + | + | ||||||||
| 8 | 8 | 1 | human (ho) | XIV | PEN, TET, ERY, GEN, CIP, ENR, VAN | + | + | + | + | ||||||||
| 8 | 8 | 1 | human (ho) | XV | CLI, TET, ERY, CIP, CHL, ENR | + | + | + | + | + | + | + | |||||
| 8 | 8 | 1 | human (ho) | XIV | PEN, VAN | + | |||||||||||
| 15 | 14 | 1 | human (ho) | II | PEN, ERY, CIP | + | |||||||||||
| 15 | 15 | 1 | human (ho) | I | PEN, QD, TET, CIP | + | + | + | |||||||||
| 15 | 15 | 1 | human (ho) | I | PEN, QD, TET, NIT, CIP | + | + | + | |||||||||
| 15 | 15 | 1 | human (ho) | I | PEN, CLI, TET, ERY | + | + | + | + | ||||||||
| 15 | 15 | 2 | human (ho) | I | PEN, TET, ERY | + | + | + | + | ||||||||
| 15 | 15 | 1 | human (ho) | I | PEN, TET, CIP | + | + | + | |||||||||
| 15 | 15 | 1 | human (ho) | I | PEN, VAN | + | |||||||||||
| 15 | 582 | 1 | human (ca, D) | I | PEN, CLI, TET, ERY, GEN | + | + | + | + | ||||||||
| 15 | 582 | 2 | human (ca, R) | I | PEN, TET, ERY | + | + | ||||||||||
| 15 | 582 | 1 | human (ho) | II | PEN, VAN | + | |||||||||||
| 15 | 582 | 1 | human (ho) | I | PEN, CLI, VAN | + | |||||||||||
| 15 | 8140 | 1 | human (ho) | I | PEN, VAN | + | |||||||||||
| 22 | 22 | 1 | human (ho) | XXIII | PEN, FOX, CIP, ENR | + | + | + | |||||||||
| 22 | 217 | 1 | human (ho) | XXIII | PEN, CLI, ERY | + | |||||||||||
| 22 | 217 | 1 | human (ho) | XXIII | PEN, VAN | + | |||||||||||
| 30 | 30 | 1 | human (ho) | XVI | PEN, CLI, ERY | + | |||||||||||
| 30 | 30 | 1 | human (ho) | XVI | PEN, VAN | + | |||||||||||
| 30 | 34 | 1 | human (ho) | XVI | PEN, VAN | + | |||||||||||
| 45 | 45 | 1 | human (ca, D) | III | PEN, CLI, TET, ERY, VAN | + | |||||||||||
| 45 | 45 | 1 | human (ho) | III | CLI, ERY, GEN | + | |||||||||||
| 45 | 45 | 1 | human (ho) | III | PEN, CLI, ERY | + | |||||||||||
| 45 | 45 | 1 | human (ho) | III | PEN, VAN | + | |||||||||||
| 45 | 45 | 1 | human (ho) | III | CLI, ERY, VAN | + | + | ||||||||||
| 45 | 45 | 1 | human (ho) | III | PEN, VAN | + | |||||||||||
| 45 | 8133 | 1 | human (ho) | VIII | PEN, CLI, QD, TET, FOX, RIF, ERY, GEN, NIT, LZD, VAN | + | + | + | + | + | |||||||
| 45 | 8141 | 1 | human (ho) | III | VAN | ||||||||||||
| 97 | 97 | 1 | human (ho) | XXVI | PEN, CIP, VEN | + | |||||||||||
| 97 | 97 | 1 | human (ho) | XXVI | TET, VAN | + | + | ||||||||||
| 97 | 8137 | 1 | human (ho) | XVII | PEN, VAN | ||||||||||||
| 7 | 1 | human (ho) | XXV | PEN, CLI, TET, ERY, GEN, VAN | + | + | + | + | |||||||||
| 7 | 1 | human (ho) | XXV | PEN, CLI, TET, ERY, GEN | + | + | + | + | |||||||||
| 7 | 1 | human (ho) | XXV | PEN, CLI, ERY, GEN | + | + | + | ||||||||||
| 7 | 1 | human (ho) | XXV | PEN, CLI, TET, ERY, GEN, VAN | + | + | + | + | + | ||||||||
| 7 | 1 | human (ho) | XXV | PEN, CIP, ERY, GEN | + | + | |||||||||||
| 7 | 1 | human (ho) | XXV | VAN | |||||||||||||
| 25 | 1 | human (ho) | V | PEN, CLI, ERY, CIP | + | ||||||||||||
| 25 | 1 | human (ho) | V | PEN, CLI, ERY, CIP, ENR | + | ||||||||||||
| 78 | 1 | human (ho) | VII | PEN, ERY, CIP, VAN | |||||||||||||
| 133 | 1 | cattle (M) | IV | VAN | |||||||||||||
| 182 | 1 | human (ho) | XII | PEN, VAN | + | ||||||||||||
| 338 | 1 | human (ho) | VI | PEN, CLI, ERY | + | + | |||||||||||
| 479 | 1 | cattle (I) | XXII | VAN | |||||||||||||
| 398 | 1 | pigs (O) | XXVII | PEN, CLI, QD, TET, FOX, ERY | + | + | + | + | + | + | + | ||||||
| 398 | 1 | pigs (O) | XXVII | PEN, CLI, QD, TET, FOX, ERY, GEN | + | + | + | + | + | + | |||||||
| 398 | 1 | pigs (O) | XXVII | PEN, CLI, SXT, TET, FOX, ERY | + | + | + | + | + | + | |||||||
| 398 | 36 | pigs (P) | XXVII | PEN, CLI, TET, FOX, ERY | + | + | + | + | + | + | + | ||||||
| 398 | 1 | cattle (L) | XXVII | PEN, TET, FOX, CIP, ENR | + | + | + | + | + | ||||||||
| 8134 | 1 | human (ho) | XXI | PEN, CLI, FOX, ERY, CIP | + | + | + | ||||||||||
| 8135 | 1 | pigs (N) | XIX | PEN, CLI, SXT, QD, TET, ERT, GEN | + | + | + | ||||||||||
| 8136 | 1 | human (ho) | XXIX | PEN, CLI, ERY, ENR | + | ||||||||||||
| 8138 | 1 | human (ca, C) | XXIV | PEN, VAN | + | ||||||||||||
| 8139 | 1 | cattle (F) | XXVIII | VAN | + | ||||||||||||
| 8139 | 2 | cattle (E) | XXVIII | VAN | |||||||||||||
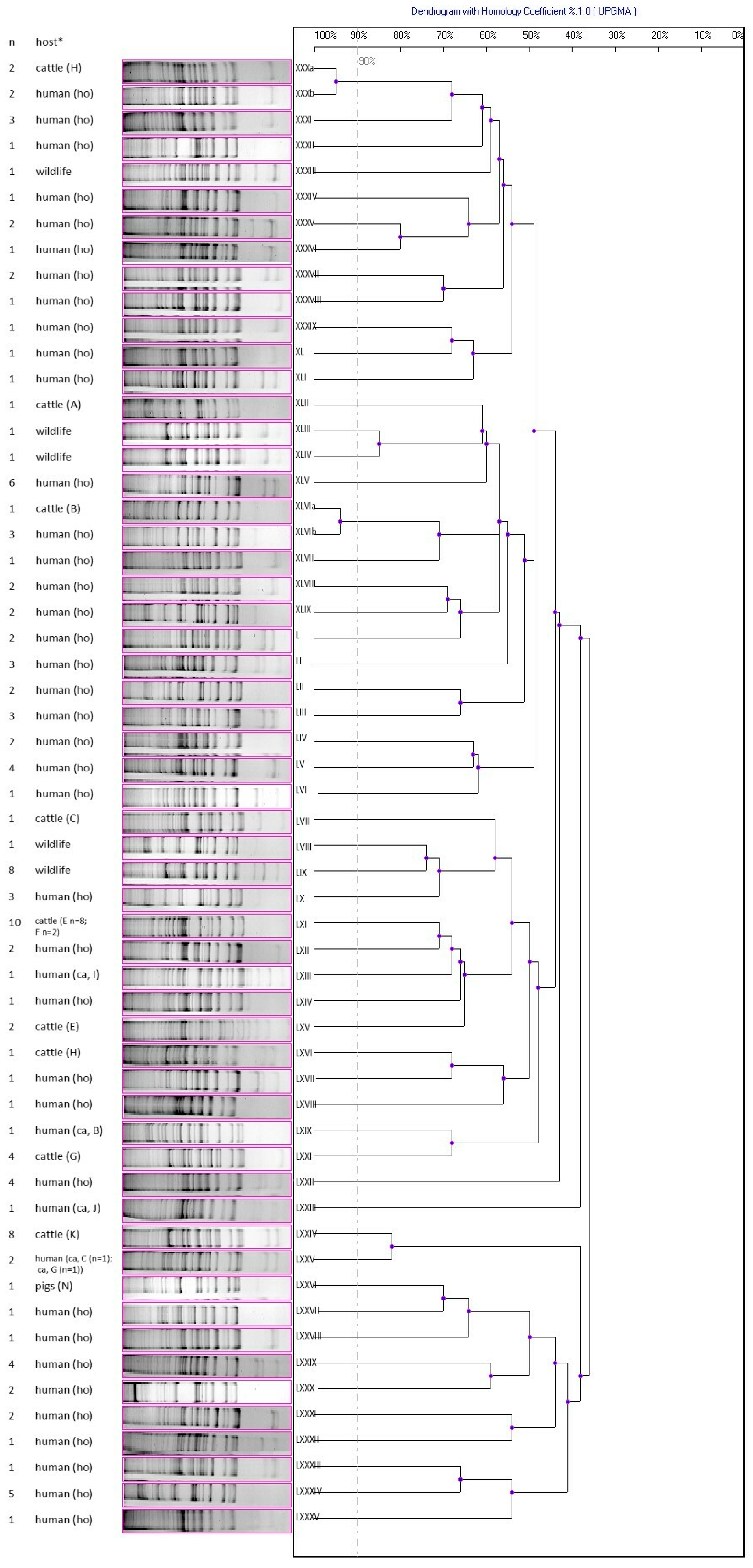
| Host | Source | Number | Animal Farm Marking |
|---|---|---|---|
| human ho † | S. aureus strains from skin and soft tissue infections (collection of the Department of Pharmaceutical Microbiology, Medical University of Lublin) | 130 | - |
| human ca (n = 15) | nasal swabs | 1 | N |
| 2 | R | ||
| 1 | B | ||
| 2 | C | ||
| 2 | D | ||
| 1 | E | ||
| 1 | F | ||
| 1 | G | ||
| 1 | I | ||
| 1 | J | ||
| 1 | AJ | ||
| 1 | AK | ||
| pigs (n = 248) | nasal swabs | 32 | N |
| 32 | O | ||
| 36 | P | ||
| 27 | R | ||
| 30 | S | ||
| 31 | T | ||
| 30 | U | ||
| 30 | W | ||
| cattle (n = 680) | nasal swabs | 35 | A |
| 19 | B | ||
| 30 | C | ||
| 10 | D | ||
| 29 | E | ||
| 26 | F | ||
| 31 | G | ||
| 5 | H | ||
| 22 | I | ||
| 30 | J | ||
| 25 | K | ||
| 1 | L | ||
| 25 | M | ||
| 27 | AA | ||
| 30 | AB | ||
| 37 | AC | ||
| 25 | AD | ||
| 40 | AF | ||
| 12 | AG | ||
| 22 | AH | ||
| 28 | AI | ||
| 25 | AJ | ||
| 67 | AK | ||
| 33 | AL | ||
| 46 | AM | ||
| wildlife (n = 113): Capreolus capreolus and Cervus elaphus | nasal swabs | ||
| 93 | - | ||
| 20 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trościańczyk, A.; Nowakiewicz, A.; Kasela, M.; Malm, A.; Tracz, A.M.; Hahaj-Siembida, A.; Osińska, M.; Gula, S.; Jankowiak, I. Multi-Host Pathogen Staphylococcus aureus—Epidemiology, Drug Resistance and Occurrence in Humans and Animals in Poland. Antibiotics 2023, 12, 1137. https://doi.org/10.3390/antibiotics12071137
Trościańczyk A, Nowakiewicz A, Kasela M, Malm A, Tracz AM, Hahaj-Siembida A, Osińska M, Gula S, Jankowiak I. Multi-Host Pathogen Staphylococcus aureus—Epidemiology, Drug Resistance and Occurrence in Humans and Animals in Poland. Antibiotics. 2023; 12(7):1137. https://doi.org/10.3390/antibiotics12071137
Chicago/Turabian StyleTrościańczyk, Aleksandra, Aneta Nowakiewicz, Martyna Kasela, Anna Malm, Anna Magdalena Tracz, Agata Hahaj-Siembida, Marcelina Osińska, Szczepan Gula, and Igor Jankowiak. 2023. "Multi-Host Pathogen Staphylococcus aureus—Epidemiology, Drug Resistance and Occurrence in Humans and Animals in Poland" Antibiotics 12, no. 7: 1137. https://doi.org/10.3390/antibiotics12071137
APA StyleTrościańczyk, A., Nowakiewicz, A., Kasela, M., Malm, A., Tracz, A. M., Hahaj-Siembida, A., Osińska, M., Gula, S., & Jankowiak, I. (2023). Multi-Host Pathogen Staphylococcus aureus—Epidemiology, Drug Resistance and Occurrence in Humans and Animals in Poland. Antibiotics, 12(7), 1137. https://doi.org/10.3390/antibiotics12071137









