Phylogeny, Virulence, and Antimicrobial Resistance Gene Profiles of Enterococcus faecium Isolated from Australian Feedlot Cattle and Their Significance to Public and Environmental Health
Abstract
1. Introduction
2. Results
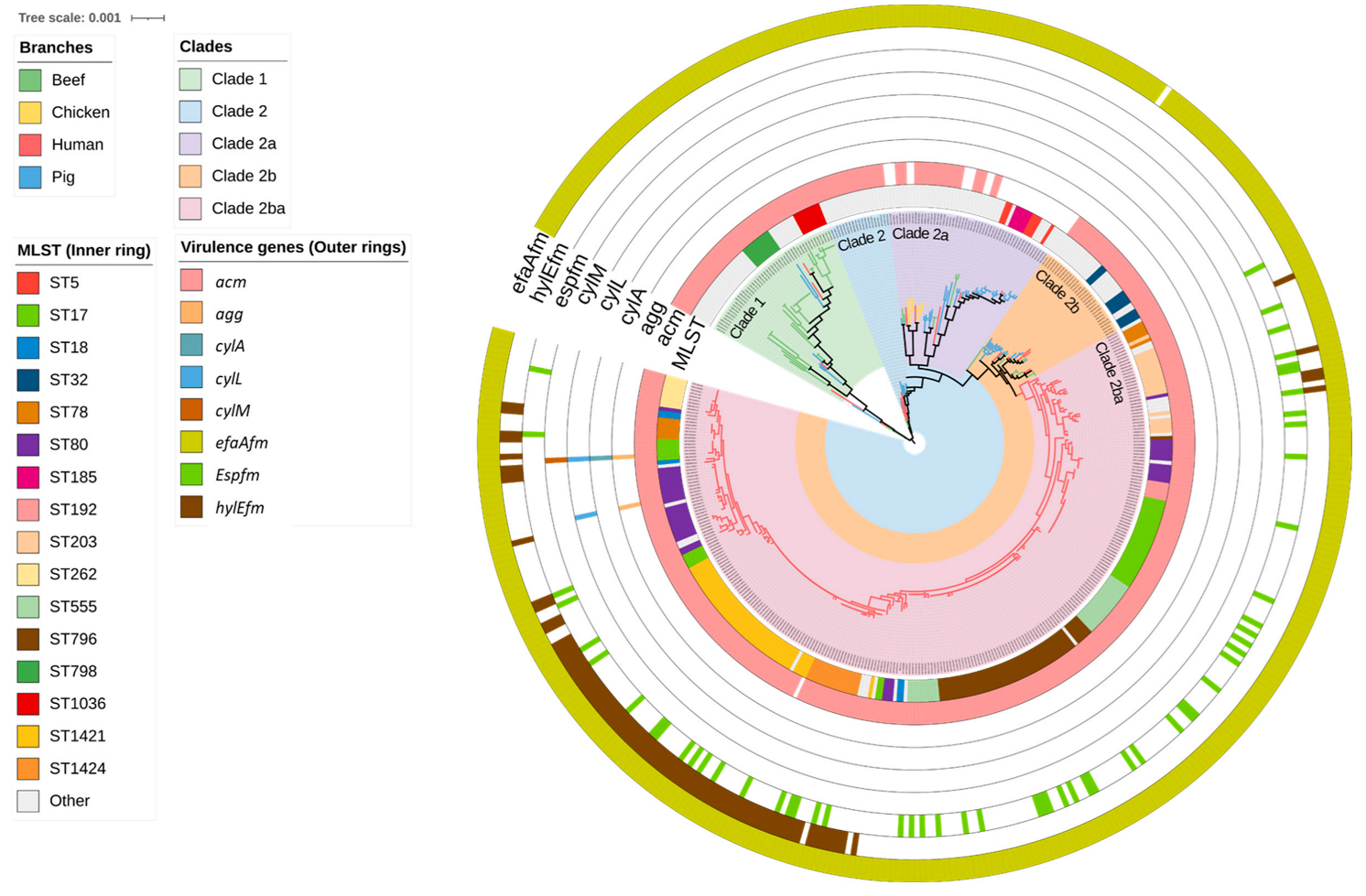
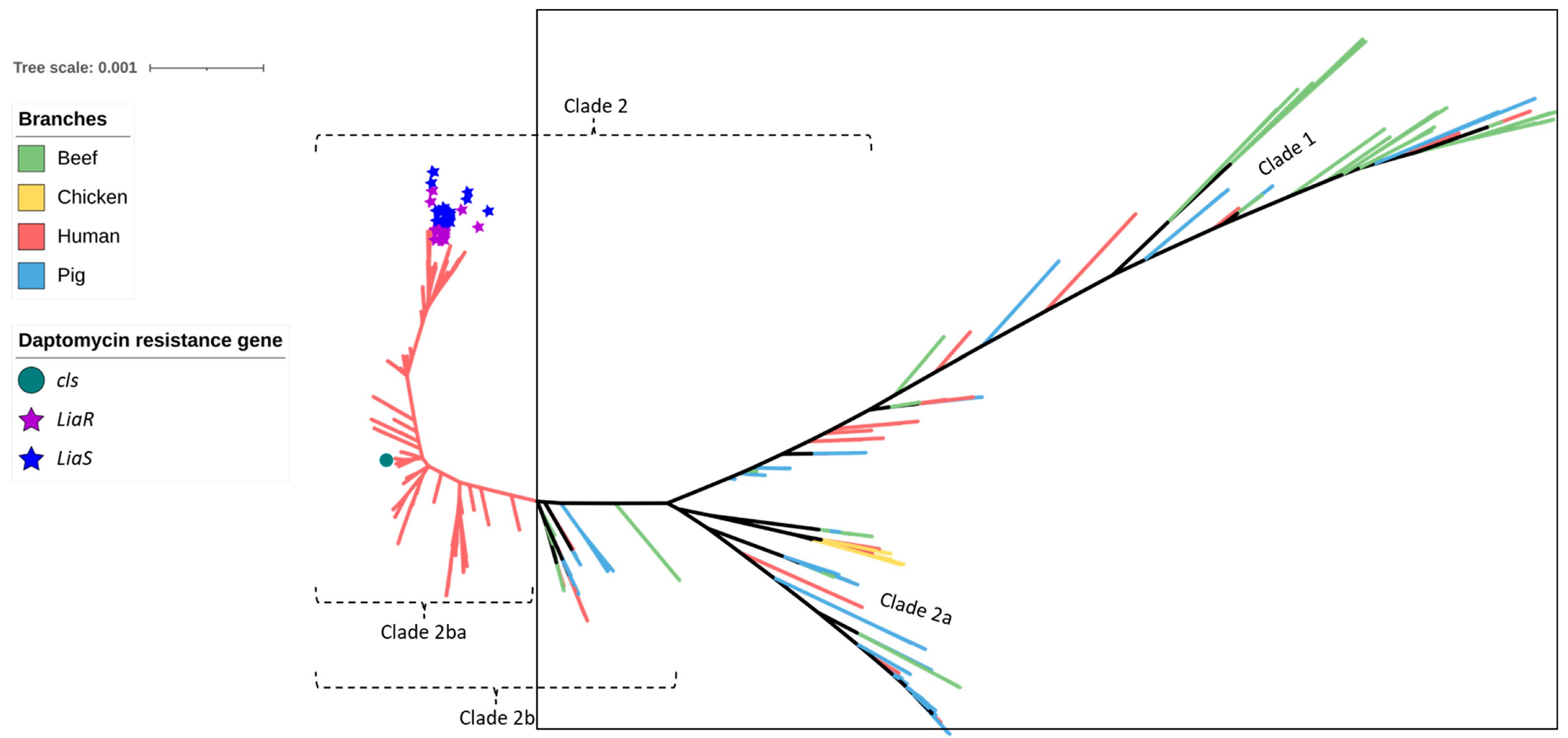
2.1. Distribution of MLST Genotypes and Virulence Factors
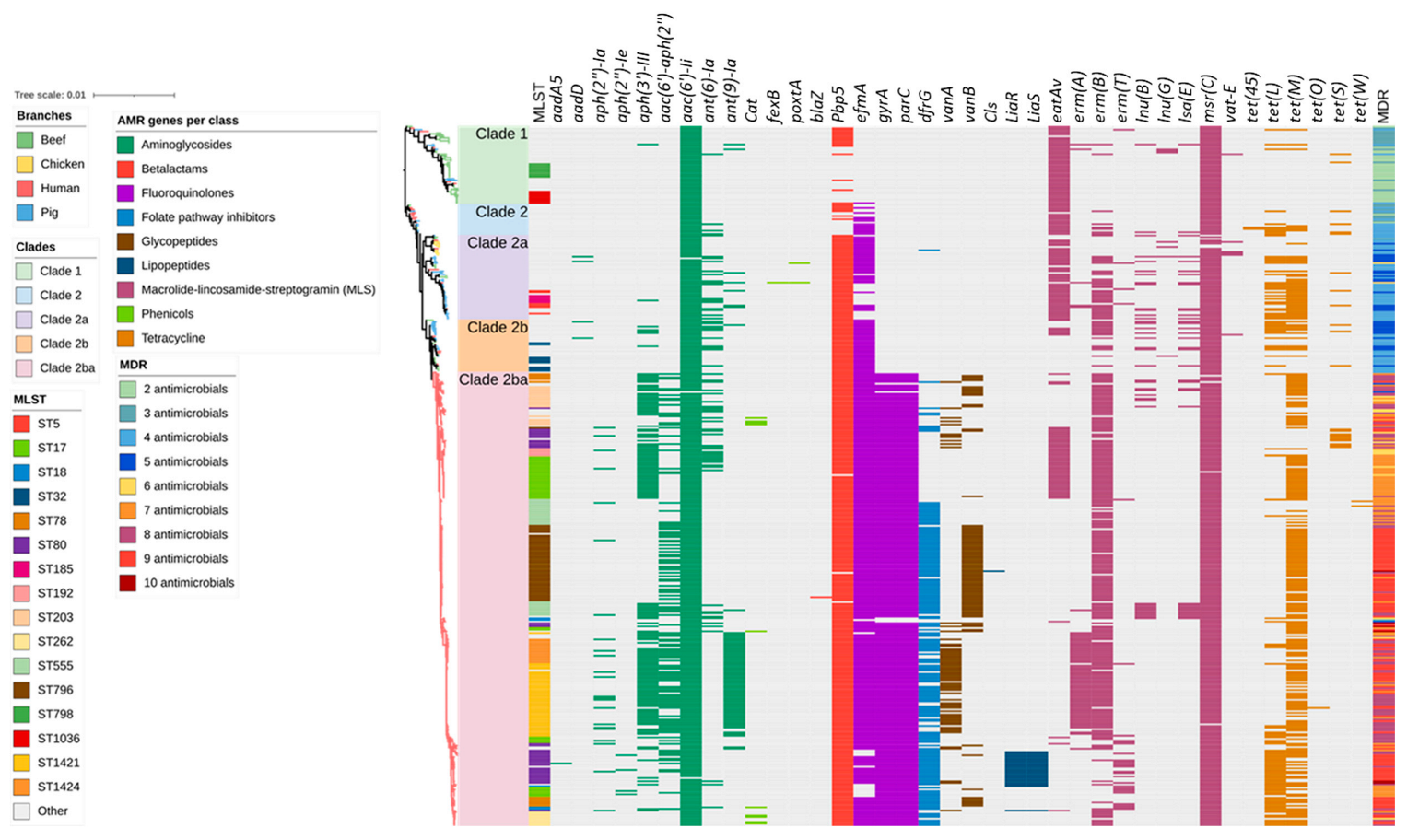
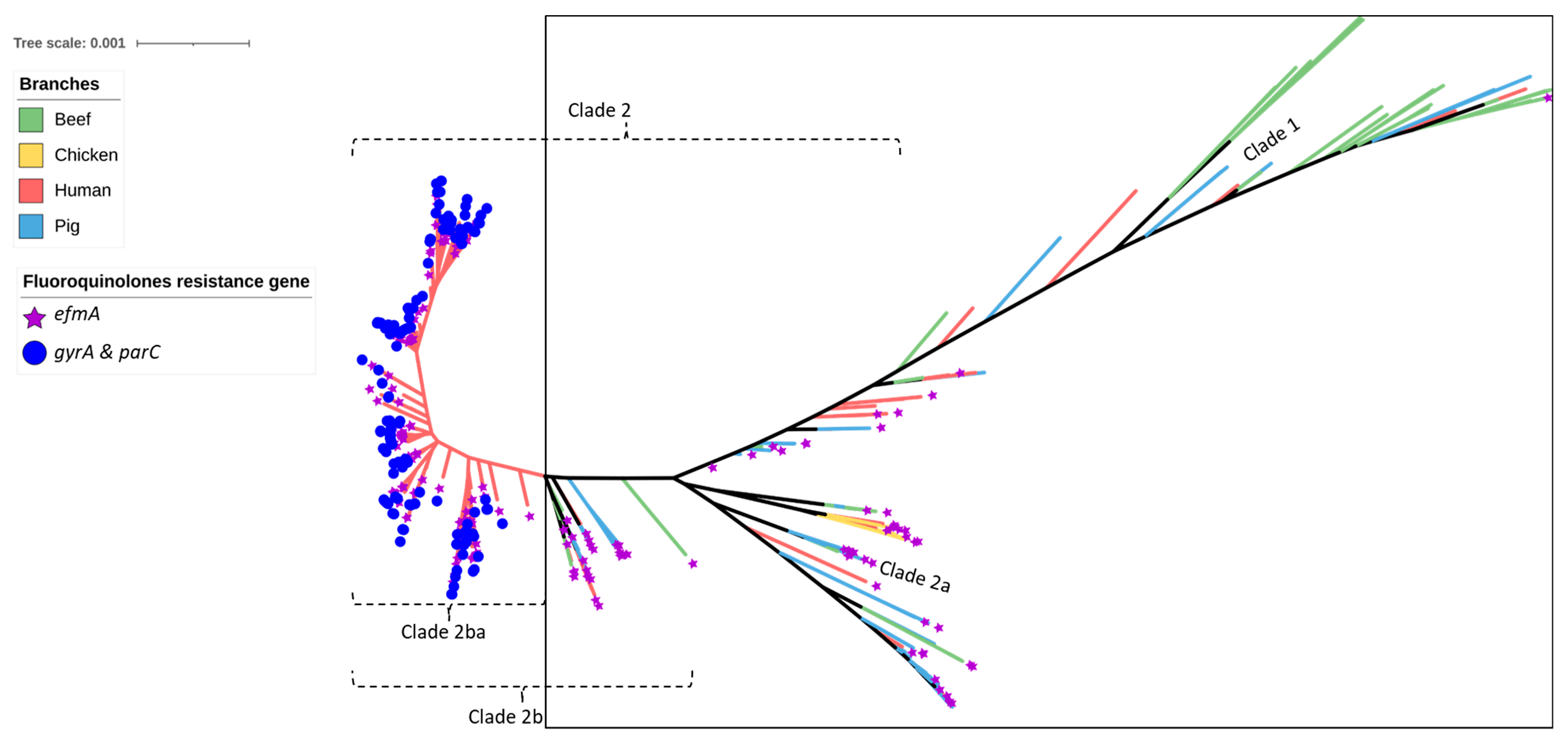
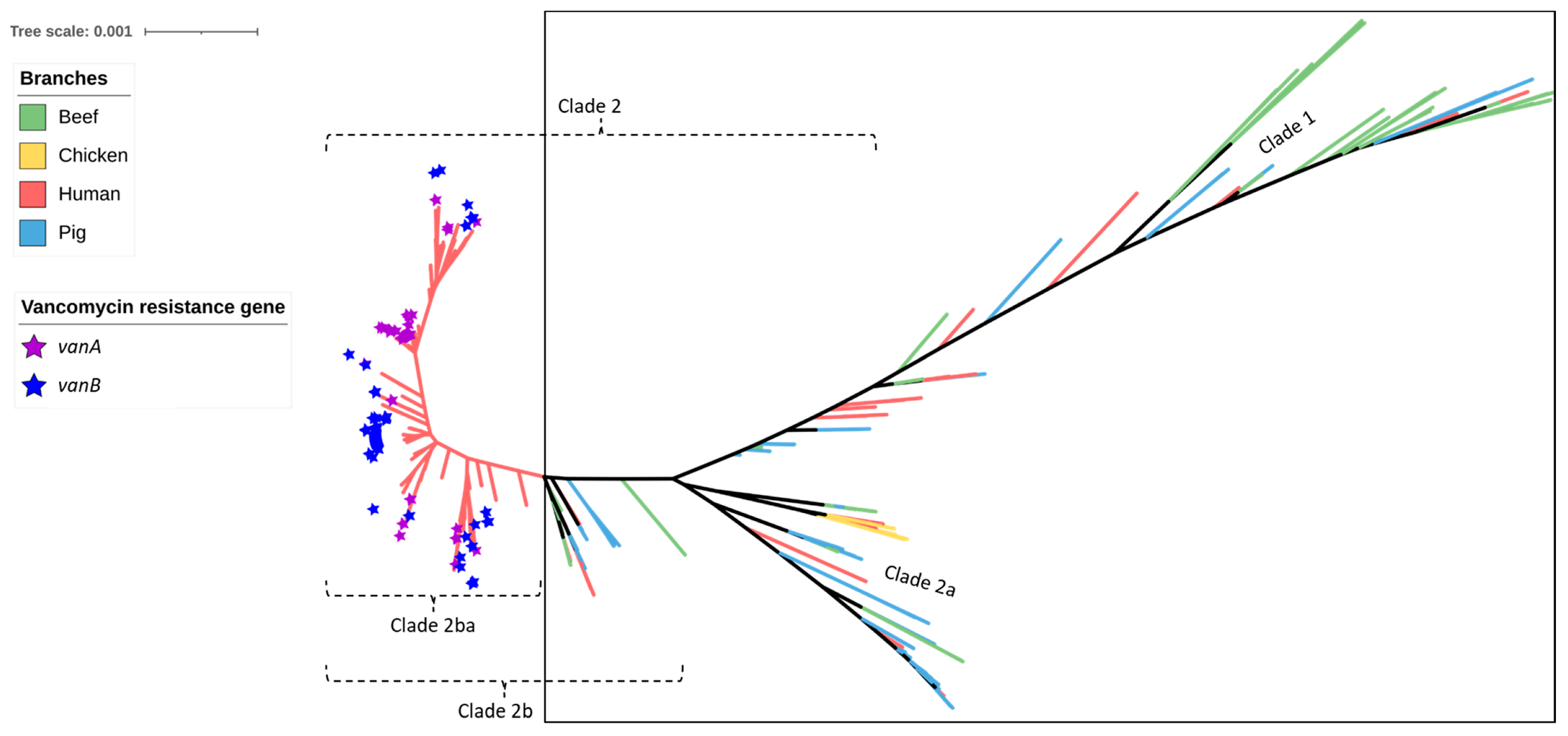
2.2. Antimicrobial Resistance Genes Including Those Encoding Resistance to Critically Important Antimicrobials
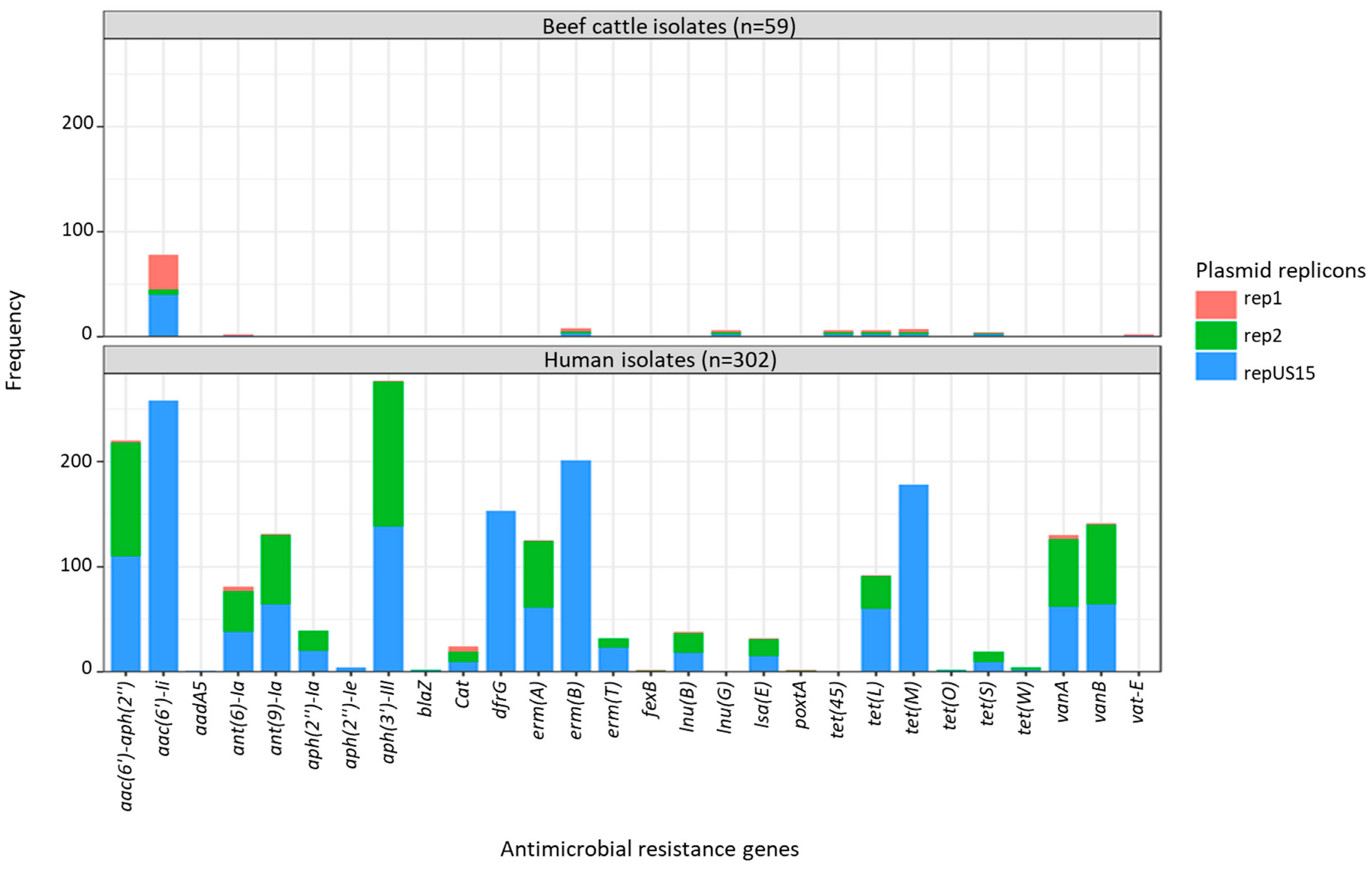
2.3. Plasmid Replicons
2.4. Agreement between Plasmid Replicons and ARG Content
3. Discussion
4. Materials and Methods
4.1. Genomic Analysis
4.2. Comparative Analysis of Virulence Genes, AMR Genes and Plasmids
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García-Solache, M.; Rice, L.B. The Enterococcus: A Model of Adaptability to Its Environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Fritzenwanker, M.; Kuenne, C.; Billion, A.; Hain, T.; Zimmermann, K.; Goesmann, A.; Chakraborty, T.; Domann, E. Complete Genome Sequence of the Probiotic Enterococcus faecalis Symbioflor 1 Clone DSM 16431. Genome Announc. 2013, 1, e00165-12. [Google Scholar] [CrossRef]
- O’Driscoll, T.; Crank, C.W. Vancomycin-resistant enterococcal infections: Epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 2015, 8, 217. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Pang, S.; Stegger, M.; Sahibzada, S.; Abraham, S.; Daley, D.; Coombs, G. A three-year whole genome sequencing perspective of Enterococcus faecium sepsis in Australia. PLoS ONE 2020, 15, e0228781. [Google Scholar] [CrossRef]
- Sava, I.G.; Heikens, E.; Huebner, J. Pathogenesis and immunity in enterococcal infections. Clin. Microbiol. Infect. 2010, 16, 533–540. [Google Scholar] [CrossRef]
- Jett, B.D.; Huycke, M.M.; Gilmore, M.S. Virulence of enterococci. Clin. Microbiol. Rev. 1994, 7, 462–478. [Google Scholar] [CrossRef]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection—Treatment and Antibiotic Resistance. Available online: https://pubmed.ncbi.nlm.nih.gov/24649502/2014 (accessed on 12 March 2023).
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–569. [Google Scholar] [CrossRef]
- Sanderson, H.; Gray, K.L.; Manuele, A.; Maguire, F.; Khan, A.; Liu, C.; Navanekere Rudrappa, C.; Nash, J.H.E.; Robertson, J.; Bessonov, K.; et al. Exploring the mobilome and resistome of Enterococcus faecium in a One Health context across two continents. Microb. Genom. 2022, 8, 000880. [Google Scholar] [CrossRef]
- Weaver, K.E.; Kwong, S.M.; Firth, N.; Francia, M.V. The RepA_N replicons of Gram-positive bacteria: A family of broadly distributed but narrow host range plasmids. Plasmid 2009, 61, 94–109. [Google Scholar] [CrossRef]
- Palmer, K.L.; Kos, V.N.; Gilmore, M.S. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 2010, 13, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.B.; Garcia-Migura, L.; Valenzuela, A.J.S.; Løhr, M.; Hasman, H.; Aarestrup, F. A classification system for plasmids from enterococci and other Gram-positive bacteria. J. Microbiol. Methods 2010, 80, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Rosvoll, T.C.; Pedersen, T.; Sletvold, H.; Johnsen, P.J.; Sollid, J.E.; Simonsen, G.S.; Jensen, L.B.; Nielsen, K.M.; Sundsfjord, A. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501-and pHTβ-related replicons associated with glycopeptide resistance and stabilizing toxin–antitoxin systems. FEMS Immunol. Med. Microbiol. 2010, 58, 254–268. [Google Scholar] [CrossRef]
- Qin, X.; Galloway-Peña, J.R.; Sillanpaa, J.; Roh, J.H.; Nallapareddy, S.R.; Chowdhury, S.; Bourgogne, A.; Choudhury, T.; Muzny, D.M.; Buhay, C.J.; et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012, 12, 135. [Google Scholar] [CrossRef]
- Markwart, R.; Willrich, N.; Eckmanns, T.; Werner, G.; Ayobami, O. Low Proportion of Linezolid and Daptomycin Resistance Among Bloodborne Vancomycin-Resistant Enterococcus faecium and Methicillin-Resistant Staphylococcus aureus Infections in Europe. Front. Microbiol. 2021, 12, 664199. [Google Scholar] [CrossRef]
- Yadav, G.; Thakuria, B.; Madan, M.; Agwan, V.; Pandey, A. Linezolid and Vancomycin Resistant Enterococci: A Therapeutic Problem. J. Clin. Diagn. Res. 2017, 11, GC07–GC11. [Google Scholar] [CrossRef]
- Fiedler, S.; Bender, J.K.; Klare, I.; Halbedel, S.; Grohmann, E.; Szewzyk, U.; Werner, G. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother. 2015, 71, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: Towards a common nomenclature. Drug Resist. Updates 2018, 40, 25–39. [Google Scholar] [CrossRef]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. J. Anal. Methods Chem. 2012, 18, 619–625. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Barbieri, R.; Goji, N.; Cameron, A.; Petkau, A.; Polo, R.O.; Tymensen, L.; Stamm, C.; Song, J.; et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020, 10, 3937. [Google Scholar] [CrossRef]
- Willems, R.J.L.; Top, J.; van Den Braak, N.; Van Belkum, A.; Endtz, H.; Mevius, D.; Stobberingh, E.; van den Bogaard, A.; Van Embden, J.D.A. Host Specificity of Vancomycin-Resistant Enterococcus faecium. J. Infect. Dis. 2000, 182, 816–823. [Google Scholar] [CrossRef]
- Trott, D.J.; Turnidge, J.; Kovac, J.H.; Simjee, S.; Wilson, D.; Watts, J. Comparative macrolide use in humans and animals: Should macrolides be moved off the World Health Organisation’s critically important antimicrobial list? J. Antimicrob. Chemother. 2021, 76, 1955–1961. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.D.; Pratt, R.; Hart, W.S. Antibiotic resistance in animals. Commun. Dis. Intell. 2003, 27, S121–S126. [Google Scholar]
- Barton, M.D. Antibiotic use in animal feed and its impact on human health. Nutr. Res. Rev. 2000, 13, 279–299. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.; O’Dea, M.; Page, S.W.; Trott, D.J. Current and future antimicrobial resistance issues for the Australian pig industry. Anim. Prod. Sci. 2017, 57, 2398–2407. [Google Scholar] [CrossRef]
- Barlow, R.S.; McMillan, K.E.; Duffy, L.L.; Fegan, N.; Jordan, D.; Mellor, G.E. Antimicrobial resistance status of Enterococcus from Australian cattle populations at slaughter. PLoS ONE 2017, 12, e0177728. [Google Scholar] [CrossRef]
- O’dea, M.; Sahibzada, S.; Jordan, D.; Laird, T.; Lee, T.; Hewson, K.; Pang, S.; Abraham, R.; Coombs, G.W.; Harris, T.; et al. Genomic, Antimicrobial Resistance, and Public Health Insights into Enterococcus spp. from Australian Chickens. J. Clin. Microbiol. 2019, 57, e00319-19. [Google Scholar] [CrossRef]
- Messele, Y.E.; Hasoon, M.F.; Trott, D.J.; Veltman, T.; McMeniman, J.P.; Kidd, S.P.; Low, W.Y.; Petrovski, K.R. Longitudinal Analysis of Antimicrobial Resistance among Enterococcus Species Isolated from Australian Beef Cattle Faeces at Feedlot Entry and Exit. Animals 2022, 12, 2690. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Abecia, L.; Angarita, E.; Aravena, P.; Nora Arenas, G.; Ariza, C.; et al. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Myer, P.R.; Freetly, H.C.; Wells, J.E.; Smith, T.P.L.; Kuehn, L.A. Analysis of the gut bacterial communities in beef cattle and their association with feed intake, growth, and efficiency. J. Anim. Sci. 2017, 95, 3215–3224. [Google Scholar] [CrossRef]
- Ramos, S.; Silva, V.; Dapkevicius, M.d.L.E.; Igrejas, G.; Poeta, P. Enterococci, from Harmless Bacteria to a Pathogen. Microorganisms 2020, 8, 1118. [Google Scholar] [CrossRef]
- Lee, T.; Jordan, D.; Sahibzada, S.; Abraham, R.; Pang, S.; Coombs, G.W.; O’dea, M.; Abraham, S. Antimicrobial Resistance in Porcine Enterococci in Australia and the Ramifications for Human Health. Appl. Environ. Microbiol. 2021, 87, e03037-20. [Google Scholar] [CrossRef]
- Trościańczyk, A.; Nowakiewicz, A.; Osińska, M.; Łagowski, D.; Gnat, S.; Chudzik-Rząd, B. Comparative characteristics of sequence types, genotypes and virulence of multidrug-resistant Enterococcus faecium isolated from various hosts in eastern Poland. Spread of clonal complex 17 in humans and animals. Res. Microbiol. 2022, 173, 103925. [Google Scholar] [CrossRef]
- Lee, R.S.; Gonçalves da Silva, A.; Baines, S.L.; Strachan, J.; Ballard, S.; Carter, G.P.; Kwong, J.C.; Schultz, M.B.; Bulach, D.M.; Seemann, T. The changing landscape of vancomycin-resistant Enterococcus faecium in Australia: A population-level genomic study. J. Antimicrob. Chemother. 2018, 73, 3268–3278. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Duarte, B.; Elghaieb, H.; Abbassi, M.S.; Hassen, A.; Read, A.; Alves, V.; Novais, C.; Peixe, L. Linezolid-resistant (Tn6246::fexB-poxtA) Enterococcus faecium strains colonizing humans and bovines on different continents: Similarity without epidemiological link. J. Antimicrob. Chemother. 2020, 75, 2416–2423. [Google Scholar] [CrossRef]
- Werner, G.; Fleige, C.; Feßler, A.T.; Timke, M.; Kostrzewa, M.; Zischka, M.; Peters, T.; Kaspar, H.; Schwarz, S. Improved identification including MALDI-TOF mass spectrometry analysis of group D streptococci from bovine mastitis and subsequent molecular characterization of corresponding Enterococcus faecalis and Enterococcus faecium isolates. Vet. Microbiol. 2012, 160, 162–169. [Google Scholar] [CrossRef]
- Rajendiran, S.; Veloo, Y.; Thahir, S.S.A.; Shaharudin, R. Resistance towards Critically Important Antimicrobials among Enterococcus faecalis and E. faecium in Poultry Farm Environments in Selangor, Malaysia. Antibiotics 2022, 11, 1118. [Google Scholar] [CrossRef]
- Jung, Y.-H.; Shin, E.S.; Kim, O.; Yoo, J.S.; Lee, K.M.; Yoo, J.I.; Chung, G.T.; Lee, Y.S. Characterization of Two Newly Identified Genes, vgaD and vatG, Conferring Resistance to Streptogramin A in Enterococcus faecium. Antimicrob. Agents Chemother. 2010, 54, 4744–4749. [Google Scholar] [CrossRef]
- Li, W.; Hu, J.; Li, L.; Zhang, M.; Cui, Q.; Ma, Y.; Su, H.; Zhang, X.; Xu, H.; Wang, M. New Mutations in cls Lead to Daptomycin Resistance in a Clinical Vancomycin- and Daptomycin-Resistant Enterococcus faecium Strain. Front. Microbiol. 2022, 13, 896916. [Google Scholar] [CrossRef]
- ASTAG. Importance Ratings and Summary of Antibacterial Uses in Human and Animal Health in Australia. Available online: https://www.amr.gov.au/resources/importance-ratings-and-summary-antibacterial-uses-human-and-animal-health-australia2018 (accessed on 12 April 2023).
- De Graef, E.; Decostere, A.; De Leener, E.; Goossens, H.; Baele, M.; Haesebrouck, F. Prevalence and Mechanism of Resistance against Macrolides, Lincosamides, and Streptogramins among Enterococcus faecium Isolates from Food-Producing Animals and Hospital Patients in Belgium. Microb. Drug Resist. 2007, 13, 135–141. [Google Scholar] [CrossRef]
- Isnard, C.M.; Malbruny, B.; Leclercq, R.; Cattoir, V. Genetic Basis for In Vitro and In Vivo Resistance to Lincosamides, Streptogramins A, and Pleuromutilins (LS A P Phenotype) in Enterococcus faecium. Antimicrob. Agents Chemother. 2013, 57, 4463–4469. [Google Scholar] [CrossRef]
- Wang, S.; Guo, Y.; Lv, J.; Qi, X.; Li, D.; Chen, Z.; Zhang, X.; Wang, L.; Yu, F. Characteristic of Enterococcus faecium clinical isolates with quinupristin/dalfopristin resistance in China. BMC Microbiol. 2016, 16, 246. [Google Scholar] [CrossRef]
- Badger, S.; Sullivan, K.; Jordan, D.; Caraguel, C.; Page, S.; Cusack, P.; Frith, D.; Trott, D. Antimicrobial use and stewardship practices on Australian beef feedlots. Aust. Vet. J. 2020, 98, 37–47. [Google Scholar] [CrossRef]
- Lean, I.; Page, S.; Rabiee, A.; Willams, S. A Survey of Antibacterial Product Use in the Australian Cattle Industry; Meat and Livestock Australia Report; Meat & Livestock Australia Limited: North Sydney, Australia, 2013. [Google Scholar]
- APVMA. Findings of the Reconsideration of the Registration of Products Containing Virginiamycin, and Their Labels; APVMA: Canberra, Australia, 2004. [Google Scholar]
- Sparo, M.; Delpech, G.; García Allende, N. Impact on Public Health of the Spread of High-Level Resistance to Gentamicin and Vancomycin in Enterococci. Front. Microbiol. 2018, 9, 3073. [Google Scholar] [CrossRef]
- Bates, J.; Jordens, J.Z.; Griffiths, D.T. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J. Antimicrob. Chemother. 1994, 34, 507–514. [Google Scholar] [CrossRef]
- Descheemaeker, P.R.; Chapelle, S.; Devriese, L.A.; Butaye, P.; Vandamme, P.; Goossens, H. Comparison of Glycopeptide-Resistant Enterococcus faecium Isolates and Glycopeptide Resistance Genes of Human and Animal Origins. Antimicrob. Agents Chemother. 1999, 43, 2032–2037. [Google Scholar] [CrossRef]
- Klare, I.; Heier, H.; Claus, H.; Reissbrodt, R.; Witte, W. vanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol. Lett. 1995, 125, 165–171. [Google Scholar] [CrossRef]
- Yoshimura, H.; Ishimaru, M.; Endoh, Y.S.; Suginaka, M.; Yamatani, S. Isolation of Glycopeptide-Resistant Enterococci from Chickens in Japan. Antimicrob. Agents Chemother. 1998, 42, 3333. [Google Scholar] [CrossRef]
- Aarestrup, F.M. Occurrence of Glycopeptide Resistance among Enterococcus faecium Isolates from Conventional and Ecological Poultry Farms. Microb. Drug Resist. 1995, 1, 255–257. [Google Scholar] [CrossRef]
- Bager, F.; Madsen, M.; Christensen, J.; Aarestrup, F.M. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant on Danish poultry and pig farms. Prev. Vet. Med. 1997, 31, 95–112. [Google Scholar] [CrossRef]
- Althaus, F.R. Ban on antimicrobial growth promoters: A safety advantage for consumers? Schweiz. Arch. Tierheilkd. 1999, 141, 103–107. [Google Scholar] [PubMed]
- Sabol, K.; Patterson, J.E.; Lewis, J.S.; Owens, A.; Cadena, J.; Jorgensen, J.H. Emergence of Daptomycin Resistance in Enterococcus faecium during Daptomycin Therapy. Antimicrob. Agents Chemother. 2005, 49, 1664–1665. [Google Scholar] [CrossRef] [PubMed]
- Werth, B.J.; Steed, M.E.; Ireland, C.E.; Tran, T.T.; Nonejuie, P.; Murray, B.E.; Rose, W.E.; Sakoulas, G.; Pogliano, J.; Arias, C.A.; et al. Defining Daptomycin Resistance Prevention Exposures in Vancomycin-Resistant Enterococcus faecium and E. faecalis. Antimicrob. Agents Chemother. 2014, 58, 5253–5261. [Google Scholar] [CrossRef] [PubMed]
- Lellek, H.; Franke, G.C.; Ruckert, C.; Wolters, M.; Wolschke, C.; Christner, M.; Büttner, H.; Alawi, M.; Kröger, N.; Rohde, H. Emergence of daptomycin non-susceptibility in colonizing vancomycin-resistant Enterococcus faecium isolates during daptomycin therapy. Int. J. Med. Microbiol. 2015, 305, 902–909. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Ning, J.; Sajid, A.; Cheng, G.; Yuan, Z.; Hao, H. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob. Resist. Infect. Control 2019, 8, 44. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Blanco, P.; Alcalde-Rico, M.; Corona, F.; Reales-Calderón, J.A.; Sánchez, M.B.; Martínez, J.L. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials. Drug Resist. Updates 2016, 28, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.E.; Jalal, S.; Wretlind, B. Alterations in GyrA and ParC Associated with Fluoroquinolone Resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 1999, 43, 947–949. [Google Scholar] [CrossRef] [PubMed]
- Bennett, P.M. Plasmid encoded antibiotic resistance: Acquisition and transfer of antibiotic resistance genes in bacteria. Br. J. Pharmacol. 2008, 153, S347–S357. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting Plasmids to Limit Acquisition and Transmission of Antimicrobial Resistance. Front. Microbiol. 2020, 11, 761. [Google Scholar] [CrossRef]
- Freitas, A.R.; Coque, T.M.; Novais, C.; Hammerum, A.M.; Lester, C.H.; Zervos, M.J.; Donabedian, S.; Jensen, L.B.; Francia, M.V.; Baquero, F.; et al. Human and Swine Hosts Share Vancomycin-Resistant Enterococcus faecium CC17 and CC5 and Enterococcus faecalis CC2 Clonal Clusters Harboring Tn on Indistinguishable Plasmids. J. Clin. Microbiol. 2011, 49, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Tedim, A.P.; Lanza, V.F.; Rodríguez, C.M.; Freitas, A.R.; Novais, C.; Peixe, L.; Baquero, F.; Coque, T.M. Fitness cost of vancomycin-resistant Enterococcus faecium plasmids associated with hospital infection outbreaks. J. Antimicrob. Chemother. 2021, 76, 2757–2764. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.C.; Jonas, D.; Huber, I.; Karygianni, L.; Wölber, J.; Hellwig, E.; Arweiler, N.; Vach, K.; Wittmer, A.; Al-Ahmad, A. Enterococcus faecalis from Food, Clinical Specimens, and Oral Sites: Prevalence of Virulence Factors in Association with Biofilm Formation. Front. Microbiol. 2016, 6, 1534. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Willems, R.J.; Fortún, J.; Top, J.; Diz, S.; Loza, E.; Cantón, R.; Baquero, F. Population Structure of Enterococcus faecium Causing Bacteremia in a Spanish University Hospital: Setting the Scene for a Future Increase in Vancomycin Resistance? Antimicrob. Agents Chemother. 2005, 49, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Galloway-Peña, J.R.; Nallapareddy, S.R.; Arias, C.A.; Eliopoulos, G.M.; Murray, B.E. Analysis of Clonality and Antibiotic Resistance among Early Clinical Isolates of Enterococcus faecium in the United States. J. Infect. Dis. 2009, 200, 1566–1573. [Google Scholar] [CrossRef]
- Werner, G.; Coque, T.M.; Hammerum, A.M.; Hope, R.; Hryniewicz, W.; Johnson, A.; Klare, I.; Kristinsson, K.G.; Leclercq, R.; Lester, C.H.; et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance 2008, 13, 19046. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Didelot, X.; Wilson, D.J. ClonalFrameML: Efficient Inference of Recombination in Whole Bacterial Genomes. PLoS Comput. Biol. 2015, 11, e1004041. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontéen, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Tetzschner, A.M.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Voldby Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- EFSA. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar] [CrossRef]


| Antimicrobial Classes | ARGs | Prevalence (%) | |||
|---|---|---|---|---|---|
| Beef (n = 59) | Chicken (n = 8) | Human (n = 302) | Pig (n = 60) | ||
| Fluoroquinolones | efmA | 21 (35.6) | 8 (100) | 279 (92.4) | 34 (56.7%) |
| gyrA | - | - | 273 (90.4) | - | |
| parC | - | - | 273 (90.4) | - | |
| Glycopeptides | vanA | - | - | 65 (21.5) | - |
| vanB | - | - | 86 (28.5) | - | |
| Lipopeptides | cls | - | - | 1 (0.3) | - |
| liaR | - | - | 23 (7.6) | - | |
| liaS | - | - | 23 (7.6) | - | |
| Streptogramin A | eatAv | 45 (76.3) | 7 (87.5) | 67(22.2) | 48 (80.0) |
| lsa(E) | - | - | 17 (5.6) | 19 (31.7) | |
| VatE | 1 (1.7) | 4 (50) | - | 1 (1.7) | |
| Streptogramins B | ermA | - | - | 63 (20.9) | 4 (6.7) |
| ermB | 3 (5.1) | - | 226 (74.8) | 50 (83.3) | |
| ermT | - | - | 25 (8.3) | 4 (6.7) | |
| msrC | 58 (98.3) | 6(75.0) | 298(98.7) | 59 (98.3) | |
| Clade | MLST | Isolate (n = 16) | Source of Isolate | No of SNPs | WGS Coverage | Antimicrobial Resistance Genes | Plasmid Replicons | Virulence Genes |
|---|---|---|---|---|---|---|---|---|
| 2b | 22 | 19MLAP075 | Beef cattle | 4579 | 62.1 | aac(6’)-Ii, efmA, msr(C), pbp5 | rep18b, repUS15 | acm, efaAfm |
| SRR10041104 | Human | 4332 | 33.5 | aac(6’)-Ii, efmA, msr(C), pbp5 | acm, efaAfm | |||
| SRR10041117 | 3627 | 28.5 | aac(6’)-Ii, efmA, msr(C), pbp5 | acm, efaAfm | ||||
| SRR12031114 | Pig | 2076 | 26.8 | aac(6’)-Ii, efmA, erm(B), lnu(G), msr(C), pbp5, tet(L), tet(M) | rep1, rep2, repUS1, repUS41 | |||
| 19MLAP293 | Beef cattle | 4048 | 55.5 | aac(6’)-Ii, efmA, msr(C), pbp5 | repUS15 | acm, efaAfm | ||
| 32 | 19MLAP294 | 3954 | 55.6 | aac(6’)-Ii, efmA, msr(C), pbp5 | repUS15 | acm, efaAfm | ||
| 19MLAP303 | 4495 | 71.8 | aac(6’)-Ii, efmA, msr(C), pbp5 | acm, efaAfm | ||||
| 19MLAP357 | 4221 | 50.4 | aac(6’)-Ii, efmA, msr(C), pbp5 | repUS15 | acm, efaAfm | |||
| 19MLAP364 | 4452 | 68.0 | aac(6’)-Ii, efmA, msr(C), pbp5 | acm, efaAfm | ||||
| 19MLAP366 | 4201 | 81.5 | aac(6’)-Ii, efmA, msr(C), pbp5 | repUS15 | acm, efaAfm | |||
| 19MLAP382 | 5123 | 77.7 | aac(6’)-Ii, efmA, msr(C), pbp5 | acm, efaAfm | ||||
| 19MLAP385 | 4366 | 50.7 | aac(6’)-Ii, efmA, msr(C), pbp5 | acm, efaAfm | ||||
| SRR10040901 | Human | 4544 | 28.5 | aac(6’)-Ii, efmA, msr(C), pbp5 | acm, efaAfm | |||
| 1 | 327 | 19MLAP257 | Beef cattle | 57,889 | 44.1 | aac(6’)-Ii, eatAv, msr(C) | acm, efaAfm | |
| 19MLAP391 | 59,188 | 64.1 | aac(6’)-Ii, eatAv, msr(C) | acm, efaAfm | ||||
| SRR10041173 | Human | 56,950 | 32.0 | aac(6’)-Ii, eatAv, msr(C) | acm, efaAfm |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Messele, Y.E.; Trott, D.J.; Hasoon, M.F.; Veltman, T.; McMeniman, J.P.; Kidd, S.P.; Petrovski, K.R.; Low, W.Y. Phylogeny, Virulence, and Antimicrobial Resistance Gene Profiles of Enterococcus faecium Isolated from Australian Feedlot Cattle and Their Significance to Public and Environmental Health. Antibiotics 2023, 12, 1122. https://doi.org/10.3390/antibiotics12071122
Messele YE, Trott DJ, Hasoon MF, Veltman T, McMeniman JP, Kidd SP, Petrovski KR, Low WY. Phylogeny, Virulence, and Antimicrobial Resistance Gene Profiles of Enterococcus faecium Isolated from Australian Feedlot Cattle and Their Significance to Public and Environmental Health. Antibiotics. 2023; 12(7):1122. https://doi.org/10.3390/antibiotics12071122
Chicago/Turabian StyleMessele, Yohannes E., Darren J. Trott, Mauida F. Hasoon, Tania Veltman, Joe P. McMeniman, Stephen P. Kidd, Kiro R. Petrovski, and Wai Y. Low. 2023. "Phylogeny, Virulence, and Antimicrobial Resistance Gene Profiles of Enterococcus faecium Isolated from Australian Feedlot Cattle and Their Significance to Public and Environmental Health" Antibiotics 12, no. 7: 1122. https://doi.org/10.3390/antibiotics12071122
APA StyleMessele, Y. E., Trott, D. J., Hasoon, M. F., Veltman, T., McMeniman, J. P., Kidd, S. P., Petrovski, K. R., & Low, W. Y. (2023). Phylogeny, Virulence, and Antimicrobial Resistance Gene Profiles of Enterococcus faecium Isolated from Australian Feedlot Cattle and Their Significance to Public and Environmental Health. Antibiotics, 12(7), 1122. https://doi.org/10.3390/antibiotics12071122







