Genomic Characterization of Carbapenem-Resistant Bacteria from Beef Cattle Feedlots
Abstract
1. Introduction
2. Results
2.1. Recovery of Carbapenem-Resistant Isolates and Species Identification
2.2. Phenotypic Characterization
2.3. Genomic Characterization
2.4. Comparative Genomic Analysis of Pseudomonas aeruginosa Isolates from Bovine and Human Clinical Origin
3. Discussion
4. Materials and Methods
4.1. Sampling, Isolation and Identification
4.2. Phenotypic Characterization
4.3. Whole-Genome Sequencing, Assembly and Annotation
4.4. Comparative Genomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bonfiglio, G.; Russo, G.; Nicoletti, G. Recent Developments in Carbapenems. Expert Opin. Investig. Drugs 2002, 11, 529–544. [Google Scholar] [PubMed]
- Nicolau, D.P. Carbapenems: A Potent Class of Antibiotics. Expert Opin. Pharmacother. 2008, 9, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Wiebe, R.; Dilay, L.; Thomson, K.; Rubinstein, E.; Hoban, D.J.; Noreddin, A.M.; Karlowsky, J.A. Comparative Review of the Carbapenems. Drugs 2007, 67, 1027–1052. [Google Scholar] [CrossRef] [PubMed]
- El-Gamal, M.I.; Brahim, I.; Hisham, N.; Aladdin, R.; Mohammed, H.; Bahaaeldin, A. Recent Updates of Carbapenem Antibiotics. Eur. J. Med. Chem. 2017, 131, 185–195. [Google Scholar] [CrossRef]
- Zhanel, G.G.; Lawrence, C.K.; Adam, H.; Schweizer, F.; Zelenitsky, S.; Zhanel, M.; Lagacé-Wiens, P.R.S.; Walkty, A.; Denisuik, A.; Golden, A.; et al. Imipenem-Relebactam and Meropenem-Vaborbactam: Two Novel Carbapenem-Β-Lactamase Inhibitor Combinations. Drugs 2018, 78, 65–98. [Google Scholar] [CrossRef]
- O’Donnell, J.N.; Miglis, C.M.; Lee, J.Y.; Tuvell, M.; Lertharakul, T.; Scheetz, M.H. Carbapenem Susceptibility Breakpoints, Clinical Implications with the Moving Target. Expert Rev. Anti-Infect. Ther. 2016, 14, 389–401. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Β-Lactams and Β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef]
- Schweizer, H.P. Efflux as a Mechanism of Resistance to Antimicrobials in Pseudomonas aeruginosa and Related Bacteria: Unanswered Questions. Genet. Mol. Res. 2003, 2, 48–62. [Google Scholar]
- Meletis, G.; Exindari, M.; Vavatsi, N.; Sofianou, D.; Diza, E. Mechanisms Responsible for the Emergence of Carbapenem Resistance in Pseudomonas aeruginosa. Hippokratia 2012, 16, 303–307. [Google Scholar]
- Ambler, R.P.; Coulson, A.F.; Frère, J.M.; Ghuysen, J.M.; Joris, B.; Forsman, M.; Levesque, R.C.; Tiraby, G.; Waley, S.G. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 1991, 276, 269–270. [Google Scholar] [CrossRef]
- Franco, M.R.G.; Caiaffa-Filho, H.H.; Burattini, M.N.; Rossi, F. Metallo-Beta-Lactamases among Imipenem-Resistant Pseudomonas aeruginosa in a Brazilian University Hospital. Clinics 2010, 65, 825–829. [Google Scholar] [CrossRef]
- Elshamy, A.A.; Khaled, M.A. A Review on Bacterial Resistance to Carbapenems: Epidemiology, Detection and Treatment Options. Future Sci. 2020, 6, FSO438. [Google Scholar] [CrossRef] [PubMed]
- Webb, H.E.; Bugarel, M.; den Bakker, H.C.; Nightingale, K.K.; Granier, S.A.; Scott, H.M.; Loneragan, G.H. Carbapenem-Resistant Bacteria Recovered from Faeces of Dairy Cattle in the High Plains Region of the USA. PLoS ONE 2016, 11, e0147363. [Google Scholar] [CrossRef] [PubMed]
- Tello, M.; Oporto, B.; Lavín, J.L.; Ocejo, M.; Hurtado, A. Characterization of a carbapenem-resistant Escherichia coli from dairy cattle harbouring blaNDM-1 in an IncC plasmid. J. Antimicrob. Chemother. 2022, 77, 843–845. [Google Scholar] [CrossRef] [PubMed]
- Klotz, P.; Göttig, S.; Leidner, U.; Semmler, T.; Scheufen, S.; Ewers, C. Carbapenem-resistance and pathogenicity of bovine Acinetobacter indicus-like isolates. PLoS ONE 2017, 12, e0171986. [Google Scholar] [CrossRef]
- Alam, M.; Rasool, M.H.; Khan, I.; Khurshid, M.; Aslam, B. Multilocus Sequence Typing of Carbapenem-Resistant Acinetobacter baumannii Isolates Harboring blaOXA-23 and blaIMP in Cattle from Punjab, Pakistan. Microb. Drug Resist. 2022, 28, 997–1002. [Google Scholar] [CrossRef]
- Tshitshi, L.; Manganyi, M.C.; Montso, P.K.; Mbewe, M.; Ateba, C.N. Extended Spectrum Beta-Lactamase-Resistant Determinants among Carbapenem-Resistant Enterobacteriaceae from Beef Cattle in the North West Province, South Africa: A Critical Assessment of Their Possible Public Health Implications. Antibiotics 2020, 9, 820. [Google Scholar] [CrossRef]
- Fischer, J.; Rodríguez, I.; Schmoger, S.; Friese, A.; Roesler, U.; Helmuth, R.; Guerra, B. Escherichia coli producing VIM-1 carbapenemase isolated on a pig farm. J. Antimicrob. Chemother. 2012, 67, 1793–1795. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Mutters, N.T.; Schmithausen, R.M.; Kreyenschmidt, J.; García-Meniño, I.; Schmoger, S.; Käsbohrer, A.; Hammerl, J.A. Genetic Characterization of Carbapenem-Resistant Klebsiella spp. from Municipal and Slaughterhouse Wastewater. Antibiotics 2022, 11, 435. [Google Scholar] [CrossRef]
- Ahlstrom, C.A.; Frick, A.; Pongratz, C.; Spink, K.; Xavier, C.; Bonnedahl, J.; Ramey, A.M. Genomic comparison of carbapenem-resistant Enterobacteriaceae from humans and gulls in Alaska. J. Glob. Antimicrob. Resist. 2021, 25, 23–25. [Google Scholar] [CrossRef]
- Darwich, L.; Vidal, A.; Seminati, C.; Albamonte, A.; Casado, A.; López, F.; Molina-López, R.A.; Migura-Garcia, L. High prevalence and diversity of extended-spectrum β-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 2019, 14, e0210686. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention; National Center for Emerging and Zoonotic Infectious Diseases. Antibiotic/Antimicrobial Resistance. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 15 April 2022).
- Girlich, D.; Naas, T.; Nordmann, P. Biochemical Characterization of the Naturally Occurring Oxacillinase Oxa-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2004, 48, 2043–2048. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. In vitro activity of ertapenem: Review of recent studies. J. Antimicrob. Chemother. 2004, 53 (Suppl. 2), ii11–ii21. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Zhang, G.; Ju, Y.; Tang, N.; Li, J.; Jia, R.; Feng, J. Five novel carbapenem-hydrolysing OXA-type β-lactamase groups are intrinsic in Acinetobacter spp. J. Antimicrob. Chemother. 2018, 73, 3279–3284. [Google Scholar] [CrossRef]
- Figueiredo, S.; Bonnin, R.A.; Poirel, L.; Duranteau, J.; Nordmann, P. Identification of the Naturally Occurring Genes Encoding Carbapenem-Hydrolysing Oxacillinases from Acinetobacter haemolyticus, Acinetobacter johnsonii, and Acinetobacter calcoaceticus. Clin. Microbiol. Infect. 2012, 18, 907–913. [Google Scholar] [CrossRef]
- Figueiredo, S.; Poirel, L.; Seifert, H.; Mugnier, P.; Benhamou, D.; Nordmann, P. Oxa-134, a Naturally Occurring Carbapenem-Hydrolyzing Class D Beta-Lactamase from Acinetobacter lwoffii. Antimicrob. Agents Chemother. 2010, 54, 5372–5375. [Google Scholar] [CrossRef]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, W.; Cui, Q.; Niu, W.; Li, H.; Zhao, X.; Wei, X.; Wang, X.; Huang, S.; Dong, D.; et al. Prevalence and Detection of Stenotrophomonas maltophilia Carrying Metallo-Β-Lactamase Blal1 in Beijing, China. Front. Microbiol. 2014, 5, 692. [Google Scholar] [CrossRef]
- Gideskog, M.; Welander, J.; Melhus, Å. Cluster of S. maltophilia among patients with respiratory tract infections at an intensive care unit. Infect. Prev. Pract. 2020, 2, 100097. [Google Scholar] [CrossRef]
- Geller, M.; Nunes, C.P.; Oliveira, L.; Nigri, R.S. maltophilia pneumonia: A case report. Respir. Med. Case Rep. 2018, 24, 44–45. [Google Scholar] [CrossRef] [PubMed]
- Brooke, J.S. New Strategies against Stenotrophomonas maltophilia: A Serious Worldwide Intrinsically Drug-Resistant Opportunistic Pathogen. Expert Rev. Anti-Infect. Ther. 2014, 12, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio-Ferraz, S.; Ene, A.; Gomes, V.J.; Queiroz, C.O.; Maskeri, L.; Oliveira, A.P.; Putonti, C.; Barbosa-Stancioli, E.F. Escherichia coli and Pseudomonas aeruginosa Isolated from Urine of Healthy Bovine Have Potential as Emerging Human and Bovine Pathogens. Front. Microbiol. 2022, 13, 764760. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Wolfgang, M.C.; Kulasekara, B.R.; Liang, X.; Boyd, D.; Wu, K.; Yang, Q.; Miyada, C.G.; Lory, S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 8484–8489. [Google Scholar] [CrossRef]
- Mathee, K.; Narasimhan, G.; Valdes, C.; Qiu, X.; Matewish, J.M.; Koehrsen, M.; Rokas, A.; Yandava, C.N.; Engels, R.; Zeng, E.; et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 3100–3105. [Google Scholar] [CrossRef]
- Strom, M.S.; Lory, S. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 1993, 47, 565–596. [Google Scholar] [CrossRef]
- Graupner, S.; Frey, V.; Hashemi, R.; Lorenz, M.G.; Brandes, G.; Wackernagel, W. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J. Bacteriol. 2000, 182, 2184–2190. [Google Scholar] [CrossRef]
- Zhou, H.Q.; Ning, L.W.; Zhang, H.X.; Guo, F.B. Analysis of the relationship between genomic GC Content and patterns of base usage, codon usage and amino acid usage in prokaryotes: Similar GC content adopts similar compositional frequencies regardless of the phylogenetic lineages. PLoS ONE 2014, 9, e107319. [Google Scholar] [CrossRef]
- Kung, V.L.; Ozer, E.A.; Hauser, A.R. The accessory genome of Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 621–641. [Google Scholar] [CrossRef]
- Puzari, M.; Chetia, P. Rnd Efflux Pump Mediated Antibiotic Resistance in Gram-Negative Bacteria Escherichia coli and Pseudomonas aeruginosa: A Major Issue Worldwide. World J. Microbiol. Biotechnol. 2017, 33, 24. [Google Scholar] [CrossRef] [PubMed]
- Savinova, T.A.; Samchenko, A.A.; Bocharova, Y.A.; Mayansky, N.A.; Chebotar, I.V. Computer Program for Detection and Analyzing the Porin-Mediated Antibiotic Resistance of Bacteria. Sovrem. Tekhnol. Med. 2021, 13, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Nikaido, H.; Poole, K. Role of Mexa-Mexb-Oprm in Antibiotic Efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1995, 39, 1948–1953. [Google Scholar] [CrossRef] [PubMed]
- Köhler, T.; Kok, M.; Michea-Hamzehpour, M.; Plesiat, P.; Gotoh, N.; Nishino, T.; Curty, L.K.; Pechere, J.C. Multidrug Efflux in Intrinsic Resistance to Trimethoprim and Sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1996, 40, 2288–2290. [Google Scholar] [CrossRef]
- Rojo-Bezares, B.; Estepa, V.; Cebollada, R.; de Toro, M.; Somalo, S.; Seral, C.; Castillo, F.J.; Torres, C.; Sáenz, Y. Carbapenem-Resistant Pseudomonas aeruginosa Strains from a Spanish Hospital: Characterization of Metallo-Beta-Lactamases, Porin Oprd and Integrons. Int. J. Med. Microbiol. 2014, 304, 405–414. [Google Scholar] [CrossRef] [PubMed]
- López-García, A.; del Carmen Rocha-Gracia, R.; Bello-López, E.; Juárez-Zelocualtecalt, C.; Sáenz, Y.; Castañeda-Lucio, M.; López-Pliego, L.; González-Vázquez, M.C.; Torres, C.; Ayala-Nuñez, T.; et al. Characterization of Antimicrobial Resistance Mechanisms in Carbapenem-Resistant Pseudomonas aeruginosa Carrying Imp Variants Recovered from a Mexican Hospital. Infect. Drug Resist. 2018, 11, 1523–1536. [Google Scholar] [CrossRef]
- Noble, R.C.; Overman, S.B. Pseudomonas stutzeri infection. A review of hospital isolates and a review of the literature. Diagn. Microbiol. Infect. Dis. 1994, 19, 51–56. [Google Scholar] [CrossRef]
- Berglund, F.; Marathe, N.P.; Österlund, T.; Bengtsson-Palme, J.; Kotsakis, S.; Flach, C.F.; Larsson, D.G.J.; Kristiansson, E. Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome 2017, 5, 134. [Google Scholar] [CrossRef]
- Fallah, F.; Borhan, R.S.; Hashemi, A. Detection of bla(IMP) and bla(VIM) metallo-β-lactamases genes among Pseudomonas aeruginosa strains. Int. J. Burn. Trauma 2013, 3, 122–124. [Google Scholar]
- Senda, K.; Arakawa, Y.; Nakashima, K.; Ito, H.; Ichiyama, S.; Shimokata, K.; Kato, N.; Ohta, M. Multifocal outbreaks of metallo-beta-lactamase-producing Pseudomonas aeruginosa resistant to broad-spectrum beta-lactams, including carbapenems. Antimicrob. Agents Chemother. 1996, 40, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, J.; Morita, K.; Kitao, T.; Watanabe, N.; Okazaki, M.; Miyoshi-Akiyama, T.; Kanamori, M.; Kirikae, T. KHM-1, a novel plasmid-mediated metallo-beta-lactamase from a Citrobacter freundii clinical isolate. Antimicrob. Agents Chemother. 2008, 52, 4194–4197. [Google Scholar] [CrossRef]
- Azhar, E.I.; Papadioti, A.; Bibi, F.; Ashshi, A.M.; Raoult, D.; Angelakis, E. Pseudomonas saudiphocaensis’ sp. nov., a new bacterial species isolated from currency notes collected during the Hajj pilgrimage in 2012 at Makkah, Saudi Arabia. New Microbes New Infect. 2016, 15, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.B.; Risson, A.N.; Jauregui, R.; Maclean, P.; Brightwell, G. Draft Genome Sequence of Pseudomonas saudiphocaensis Strain AGROB56, Isolated from a Dairy Farm in New Zealand. Microbiol. Resour. Announc. 2021, 10, e01258-20. [Google Scholar] [CrossRef]
- Zaheer, R.; Cook, S.R.; Klima, C.L.; Stanford, K.; Alexander, T.; Topp, E.; Read, R.R.; McAllister, T.A. Effect of Subtherapeutic vs. Therapeutic Administration of Macrolides on Antimicrobial Resistance in Mannheimia haemolytica and Enterococci Isolated from Beef Cattle. Front. Microbiol. 2013, 4, 133. [Google Scholar] [CrossRef]
- Freire, B.; Ladra, S.; Parama, J.R. Memory-Efficient Assembly Using Flye. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 3564–3577. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap and Miniasm: Fast Mapping and De Novo Assembly for Noisy Long Sequences. Bioinformatics 2016, 32, 2103–2110. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. Quast: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. Vfdb 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H. Mob-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (Itol) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
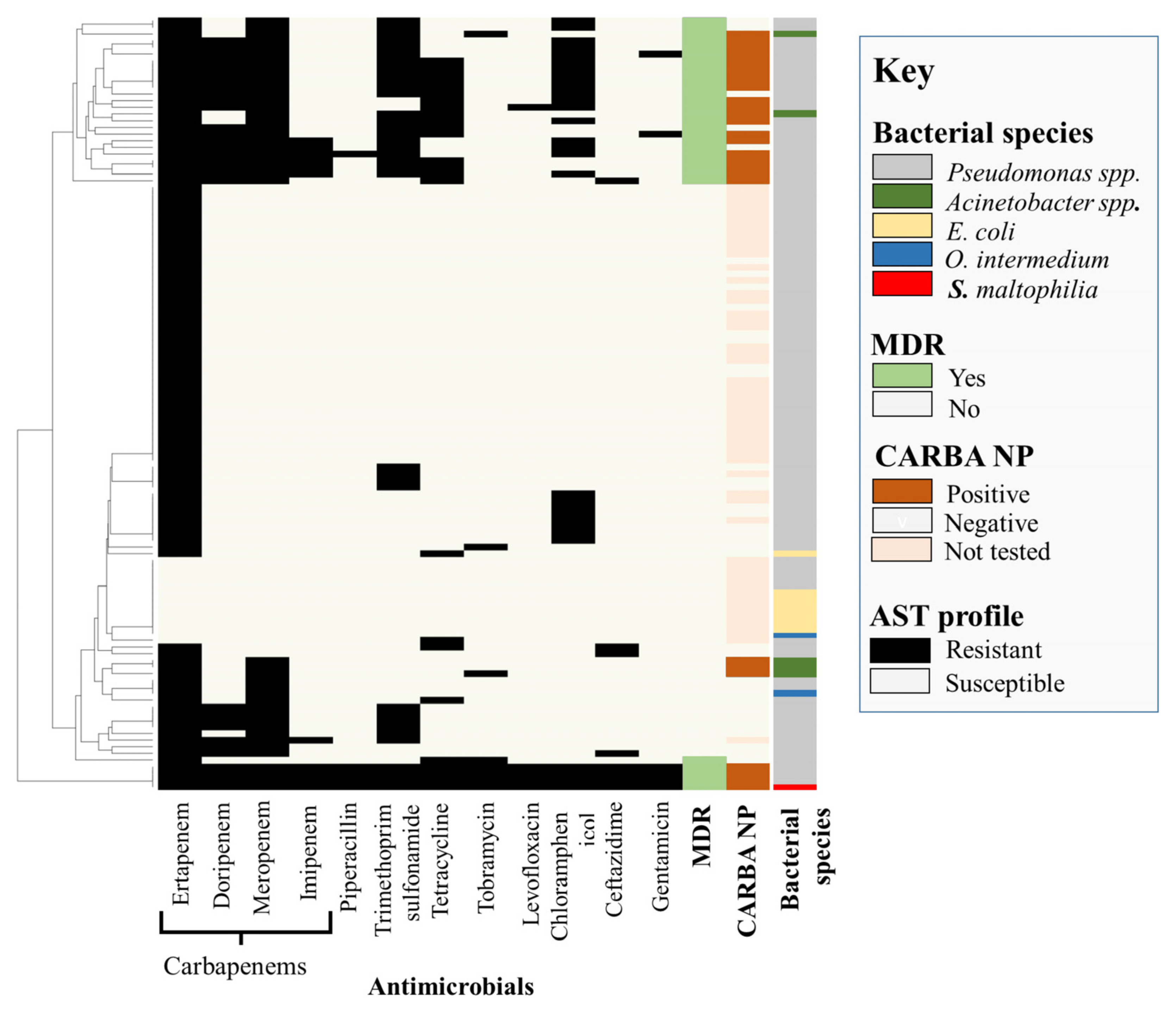
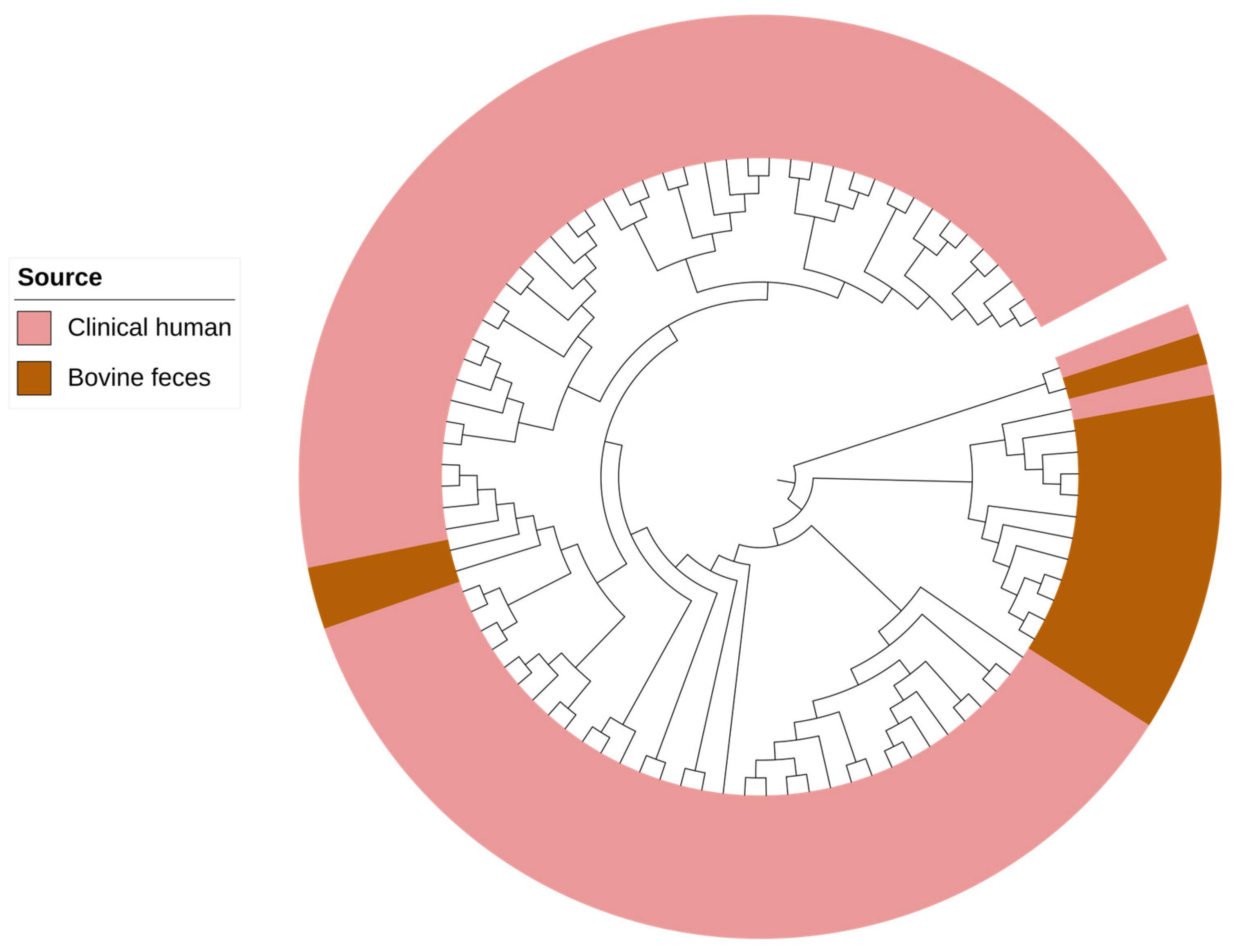
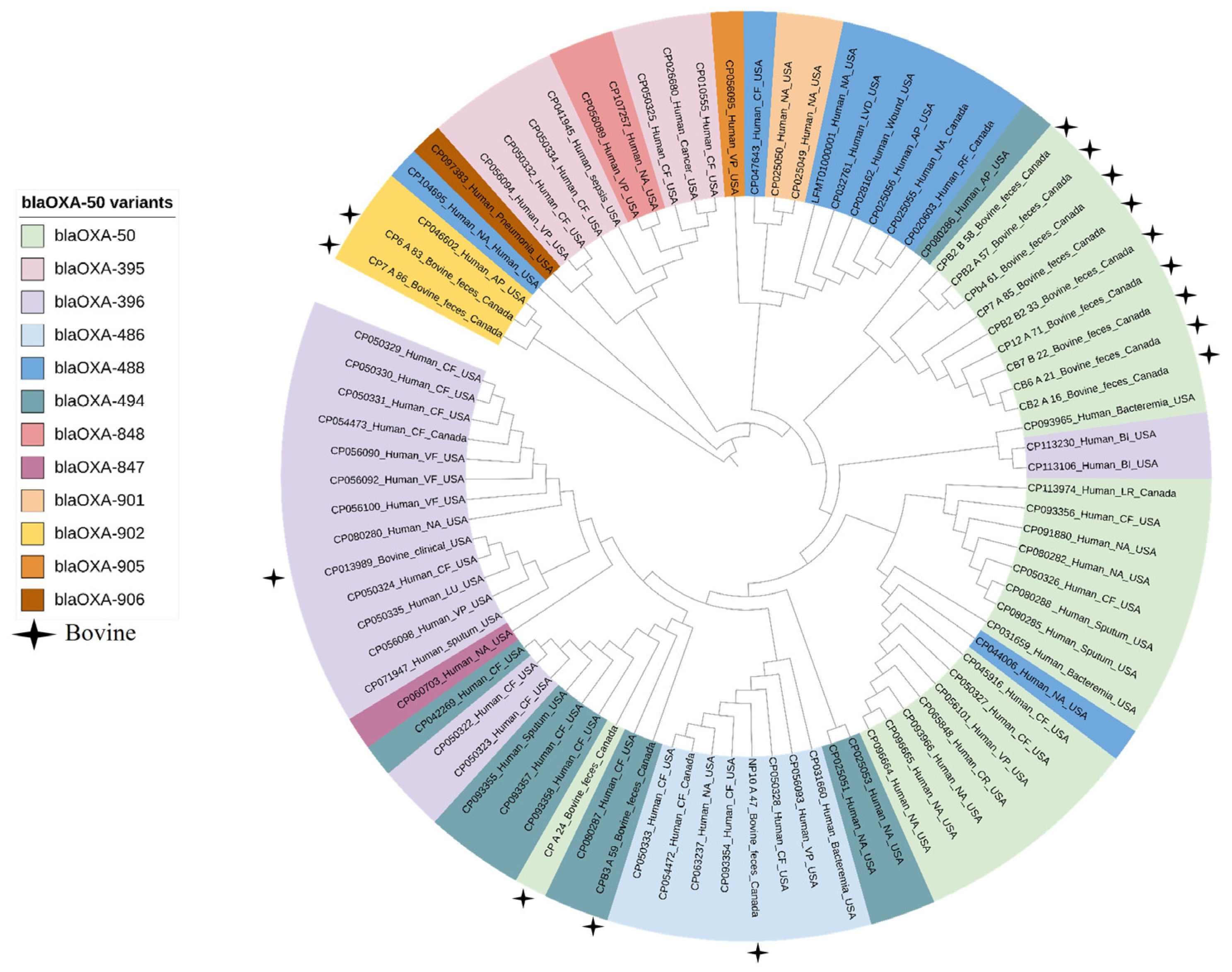
| Bacterial Species | Antimicrobial Resistance Genes | Genotype-Based Phenotype |
|---|---|---|
| E. coli (n = 1) | blaEC, blaCMY-2, aph(3″)-Ib, aph(6)-Id, sul2, tet(A), floR | Aminoglycosides, Chloramphenicol, Sulfisoxazole, Tetracycline |
| A. haemolyticus (n = 3) | blaOXA-265, aacA-ACI, blaPDC-197 | Carbapenem Aminoglycosides Cephalosporin |
| blaOXA-265, aacA-ACI1 | Carbapenem Aminoglycosides | |
| A. lwoffii (n = 1) | blaOXA-648 | Carbapenem |
| A. junii (n = 1) | blaOXA -278 | Carbapenem |
| S. maltophilia (n = 1) | blaL1,blaL2, aph(6)-Smalt, aph(3′)-IIc, oqxB9, oqxA10, floR2 | Carbapenem Aminoglycosides Phenicol Chloramphenicol |
| O. intermedium (n = 1) | floR, oqxB12, blaOCH-2 | Chloramphenicol Quinolone Cephalosporin |
| P. aeruginosa (n = 20) | blaOXA-50, blaPDC-197, aph(3′)-IIb, catB7, fosA | Carbapenem Cephalosporin Chloramphenicol Aminoglycoside Fosfomycin |
| blaOXA-50, blaPDC-197, aph(3′)-IIb, catB7, fosA, crpP | ||
| blaOXA-50, blaPDC-55, aph(3′)-IIb, catB7, fosA | ||
| blaOXA-50, blaPDC-66, aph(3′)-IIb, fosA, crpP, catB7 | ||
| blaOXA-486, blaPDC-374, aph(3′)-IIb, fosA, crpP, catB7 | ||
| blaOXA-486, blaPDC-374, aph(3′)-IIb, catB7, fosA | ||
| blaOXA-494, blaPDC-374, aph(3″)-la, aph(6)-ld, catB7 fosA, crpP | ||
| blaOXA-902, blaPDC-133, aph(3′)-IIb, catB7, fosA, crpP | ||
| blaOXA-902 | Carbapenem | |
| P. entomophila (n = 1) | blaPDC-33 | Cephalosporin |
| P. plecoglossicida (n = 9) | No gene | - |
| P. mosselii (n = 1) | No gene | - |
| P. putida (n = 1) | No gene | - |
| P. saudiphocaensis (n = 1) | blaPST-2, aadA1 | Carbapenem, Aminoglycoside |
| P. stutzeri (n = 1) | blaPST-2 | Carbapenems |
| Bacterial Species | Virulence Genes (%) 1 |
|---|---|
| P. aeruginosa (n = 20) | Biofilm and capsule synthesis: alg44 (21/24, 88%), alg8 (22/24, 92%), algA-G,I-L,P-R,U,W,X,Z (22/24, 92%), mucA-E,P (21/24, 88%) Pili and Fimbriae: chpA-E (92%), pilA (9%), pilB (88%), pilC (34%), pilE,F (84%), pilG-K,M-X,Y1,Y2 (92%), fimT-V (84%), fleI/flag (13%), fleN (100%), fleP (13%), fleQ (100%), fleR (88%), fleS (88%), flgA (84%), flgB (88%), flgC (100%), flgD-F (84%), flgG-I (24), flgJ,K (84%), flgL (13%), flgM,N (84%), flhA,B,F (100%), fliA,C (100%), fliD (13%), fliE-L (92%), fliM,N (100%), fliO-R (88%), fliS (13%), motA-D,Y (92%) Biosynthesis of small ferric-ion-chelating molecules: pchA-I,R (84%), fptA (84%), fpvA (21%), mbtH-like (100%), pvcA-D (84%), pvdA (84%), pvdD,E (21%), pvdF-H (88%), pvdI,J (21%), pvdL,M (100), pvdM (22), pvdN-Q (88%), pvdS (96%) Phenazine biosynthesis: phzA1-G1,M,S (80%), phzH (30%) Rhamnolipids: rhlA,B,I (84%), rhlC (75%) Type VI secretion system: clpV1 (96%), dotU1 (88%), flha1 (80%), hcp1 (96%), hsiA1 (84%), hsiB1/vipA (100%), hsiC1/vipB (96%), hsiE1,F1,H1 (84%), hsiG1 (96%), hsiJ1 (92%), icmF1/tssM1 (84%), tagQ (88%), tse1-3 (84%), vgrG1a (80%), vgrG1b (84%), ppkA(84%), tagR (100%) tagS (88%), tagT (88%), pppA (84%), tagF/pppB (84%) Type III secretion system: pscB-L, N-U (80%), popB,D,N (80%), pcr1-4,D,H,R,V (80%), exsA-E (80%), exoS,T,Y (80%), ptxR (84%) Type I secretion system: aprA (84%) Type II secretion system (Xcp) and exo-proteins: xcpP-Z (84%), xcpA/pilD (88%), lasA (88%), lasB (84%), plcH (84%), toxA (21%), lip1 (84%) Quorum sensing: lasI (88%) Lipopolysaccharide core biosynthesis: waaA,C (88%), waaF (96%), waaG,P (100%), wzy (17%), wzz (17%) |
| P. entomophila (n = 1) | Biofilm and capsule synthesis: algCBU, mucD Lipopolysaccharide core biosynthesis: waaF, wag Type VI secretion system: clpV1, hsiG1, hcp1, hsiC1/vipB, hsiB1/vipA, tagR Biosynthesis of small ferric-ion-chelating molecules: pvdH, pvdS, mbtH-like Pili and Fimbriae: motC, fleNQ, flhA, fliAIGMNPQ, flgCGHI, pilH |
| P. mosselii (n = 1) | Biofilm and capsule synthesis: algA-D,U,W,I (100%), alg8 (100%), mucD (100%) Biosynthesis of small ferric-ion-chelating molecules: mbtH-like (100%), pvdH,S,M (100%) Lipopolysaccharide core biosynthesis: waaF,G,P (100%) Pili and Fimbriae: flgC,G-I (100%), fliA,F,G,I,M,-Q (100%), fleN,Q (100%), motA-C (100%), pilH (100%) Type VI secretion system: tagR, dotU1 (100%), hsiB1/vipA (100%), hsiC1/vipB (100%), hcp1(100%), hsiG1 (100%), clpV1 (100%) |
| P. putida (n = 2) | Biofilm and capsule synthesis: algA-D,U,I (100%),alg8 (100%), mucD (100%) Biosynthesis of small ferric-ion-chelating molecules: mbtH-like (100%), pvdH,S (100%) Lipopolysaccharide core biosynthesis: waaF,G (100%) Pili and Fimbriae: flgC,G-I (100%), fliA,G,I,M,N,P,Q (100%), fleN,Q (100%), flhA (100%), motC,D (100%), pilH (100%) |
| P. saudiphocaensis (n = 1) | Biofilm and capsule synthesis: algC,R,U (100%) Pili and Fimbriae: flgC,G,I (100%), flhA (100%), fliE,G,I,M-P (100%), pilG,H,U,T (100%) Type II secretion system (Xcp): xcpT,R (100%) Lipopolysaccharide core biosynthesis: waaF |
| P. stutzeri (n = 1) | Biofilm and capsule synthesis: algA-C,R (100%) Pili and Fimbriae: flgG,I (100%), flhA (100%), fliA,E-G,I,M,N-R (100%), fleN,Q (100%), motA (100%), pilG,H,J,M,R,T,U (100%) Type II secretion system (Xcp): xcpT,R (100%) Lipopolysaccharide core biosynthesis: waaF,P (100%) |
| P. plecoglossicida (n = 9) | Biofilm and capsule synthesis: algB,C,U (100%), algW (67%), mucD (100%) Pili and Fimbriae: flgC,G,H,I (100%), flhA (100%), fliA,G,I,M,N,P,Q (100%), fleN,Q (100%), motC (100%), pilH (100%) Lipopolysaccharide core biosynthesis: waaF (67%), waaG (100%) Type VI secretion system: clpV1 (100%), hcp1 (100%), hsiB1/vipA (100%), hsiC1/vipB (100%), hsiG1 (100%), tagR (100%) Biosynthesis of small ferric-ion-chelating molecules: mbtH-like (100%), pvdSH (100%) |
| E. coli (n = 1) | Type II secretion system: gspC m (100%) Type III secretion system: espX2 (100%), ompA (100%), espR1(100%), espR4 (100%), espR3 (100%), espL1 (100%), espY3 (100%), espY2 (100%), espX1(100%), espY4 (100%), espL4 (100%), espX4 (100%), espX5 (100%), Curli biogenesis: csgB,D,F,G (100%) Iron import system: shuA,S,T,W,X (100%), chuUVW (100%) Pili and Fimbriae: fimA-H (100%), yagV/ecpE (100%), yagW/ecpD (100%), yagX/ecpC (100%), yagY/ecpB, yagZ/ecpA (100%), ykgK/ecpR (100%) Adhesion: fdeC (100%) Enterobactin: entA-E,F,S (100%), fepA-D,G (100%) |
| A. haemolyticus (n = 3) | No virulence gene |
| A. lwoffii (n = 1) | |
| A. junii (n = 1) | |
| S. maltophilia (n = 1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaidi, S.-e.-Z.; Zaheer, R.; Thomas, K.; Abeysekara, S.; Haight, T.; Saville, L.; Stuart-Edwards, M.; Zovoilis, A.; McAllister, T.A. Genomic Characterization of Carbapenem-Resistant Bacteria from Beef Cattle Feedlots. Antibiotics 2023, 12, 960. https://doi.org/10.3390/antibiotics12060960
Zaidi S-e-Z, Zaheer R, Thomas K, Abeysekara S, Haight T, Saville L, Stuart-Edwards M, Zovoilis A, McAllister TA. Genomic Characterization of Carbapenem-Resistant Bacteria from Beef Cattle Feedlots. Antibiotics. 2023; 12(6):960. https://doi.org/10.3390/antibiotics12060960
Chicago/Turabian StyleZaidi, Sani-e-Zehra, Rahat Zaheer, Krysty Thomas, Sujeema Abeysekara, Travis Haight, Luke Saville, Matthew Stuart-Edwards, Athanasios Zovoilis, and Tim A. McAllister. 2023. "Genomic Characterization of Carbapenem-Resistant Bacteria from Beef Cattle Feedlots" Antibiotics 12, no. 6: 960. https://doi.org/10.3390/antibiotics12060960
APA StyleZaidi, S.-e.-Z., Zaheer, R., Thomas, K., Abeysekara, S., Haight, T., Saville, L., Stuart-Edwards, M., Zovoilis, A., & McAllister, T. A. (2023). Genomic Characterization of Carbapenem-Resistant Bacteria from Beef Cattle Feedlots. Antibiotics, 12(6), 960. https://doi.org/10.3390/antibiotics12060960







