Abstract
Linezolid is an antibiotic of last resort for the treatment of infections caused by Gram-positive bacteria, including vancomycin-resistant enterococci. Enterococcus faecalis, a member of enterococci, is a significant pathogen in nosocomial infections. E. faecalis resistance to linezolid is frequently related to the presence of optrA, which is often co-carried with fex, phenicol exporter genes, and erm genes encoding macrolide resistance. Therefore, the common use of antibiotics in veterinary might promote the occurrence of optrA in livestock settings. This is a cross-sectional study aiming to investigate the prevalence of optrA positive E. faecalis (OPEfs) in 6 reservoirs in farms in Ha Nam province, Vietnam, and its associated factors and to explore genetic relationships of OPEfs isolates. Among 639 collected samples, the prevalence of OPEfs was highest in flies, 46.8% (51/109), followed by chickens 37.3% (72/193), dogs 33.3% (17/51), humans 18.7% (26/139), wastewater 16.4% (11/67) and pigs 11.3%, (14/80). The total feeding area and total livestock unit of the farm were associated with the presence of OPEfs in chickens, flies, and wastewater. Among 186 OPEfs strains, 86% were resistant to linezolid. The presence of optrA was also related to the resistant phenotype against linezolid and levofloxacin of E. faecalis isolates. Close genotypic relationships identified by Pulsed Field Gel Electrophoresis between OPEfs isolates recovered from flies and other reservoirs including chickens, pigs, dogs, and wastewater suggested the role of flies in the transmission of antibiotic-resistant pathogens. These results provided warnings of linezolid resistance although it is not used in livestock.
1. Introduction
Linezolid is the first synthetic antimicrobial agent of the oxazolidinone class. It inhibits primary ribosome synthesis and protein synthesis of many species of Gram-positive bacteria by targeting the 50S ribosomal subunits and acting on its binding affinity to N-formylmethionyl-tRNA [1]. Because of its broad antibacterial spectrum, linezolid has been widely used as one of the main options for the treatment of infectious diseases caused by multidrug-resistant Gram-positive pathogens, including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), penicillin-resistant Streptococcus, and Mycobacteria [2]. However, there is an alarming increase in resistance to linezolid, especially in livestock farming. In Korea, during the period of 2010–2019, the rate of linezolid-resistant E. faecalis escalated from 0% to 5.7% in pigs and from 0% to 2.2% in chicken [3].
Although members of the Enterococcus genus are considered commensal bacteria, they can also be opportunistic pathogens in favorable environmental conditions. Hospital-acquired intestinal infections have become a cause of global concern due to their increasing morbidity and resistance to several important classes of antibiotics, such as vancomycin, linezolid, and fluoroquinolone. Notably, these resistances in many cases came from farm animals [4,5]. E. faecalis is ubiquitous bacteria in the intestinal tract of mammals and it is also a significant pathogen in foodborne infections and antibiotic resistance. With many virulent factors that help to adhere, penetrate, and evade the immune response, the pathogenic potential of E. faecalis is higher than other Enterococcus species. It can be responsible for endocarditis, sepsis, wound infections, and urinary tract infections [6,7,8,9,10]. Due to its intrinsic resistance to antibiotics such as cephalosporins, sulfonamides, and clindamycin and acquisition of other resistance genes through mobile genetic elements (plasmids, transposons) such as vanA, vanB (vancomycin resistance), ermA, ermB (macrolide resistance), fexA (phenicol resistance) or by mutations under the selective pressure of antibiotics (daptomycin, fluoroquinolone resistance), its resistance is even more worrying [11,12]. Multiple-drug resistant (MDR) E. faecalis is reported worldwide and presents an increasing trend in both hospitals and communities, in animal food and farming settings [3,8,13,14]. In Vietnam, Indonesia, and Thailand (2007), 73–100% of E. faecalis isolates obtained from chickens exhibited MDR [15].
Linezolid resistance can be caused by (i) mutations in the V domain of 23S rRNA and rplC/rplD genes encoding the L3/L4 ribosomal proteins, (ii) acquisition of oxazolidinone resistance genes such as cfr, which encodes a methyltransferase-modified 23S rRNA, or (iii) acquisition of optrA and poxtA, two genes encoding the ABC-F protein that presumably protects the ribosomal target from binding to antibiotics [16]. Among genes encoding resistance to linezolid reported in E. faecalis, optrA is the most common and was reported worldwide [17,18,19]. This gene was detected in both plasmids, chromosomes, and different genetic environments [14,18,19]. Multiple variants of the gene have been described, such as optrA2, optrA5, optrA7, etc., demonstrating the plasticity of this resistance region [20,21]. OptrA confers transferable resistance to linezolid and is often co-carried with fexA, fexB (phenicol exporter genes), and ermA, ermB (conferring macrolide-licosamide-streptogramin B resistance) [17,21,22,23]. Therefore, although linezolid is not permitted in livestock, common use of other antibiotics in veterinary might promote the occurrence of optrA in animal reservoirs [24,25,26]. Furthermore, OptrA regularly locates on plasmids with a capacity to transmit horizontally between bacteria, creating a greater risk of getting resistance genes [22,27].
The use of antibiotics in livestock increases selection pressure on bacteria and generates the development of antibiotic resistance in animals. In addition, poor living conditions (improper lavatories, untreated wastewater, poor hygiene, etc.) and close interactions between farmers with food animals and the farm environment can facilitate the transmission of resistant bacteria, including Enterococcus, from animals to humans and vice versa [28,29,30,31]. The emergence of antimicrobial resistance in the livestock industry, especially resistance to critically important antimicrobials such as linezolid [32], poses a serious threat to public health [33]. Linezolid-resistant E. faecalis and optrA-positive E. faecalis (OPEfs) have been detected in many countries in livestock and animal food [18,27,34] but data on associations between OPEfs infection in the community and its associated factors are scarce. Only some reports on the correlation between linezolid resistance and clinical epidemiological factors were published [8,9,35].
In Vietnam, linezolid is not approved for veterinary use in livestock [36]. The data regarding linezolid resistance in communities and farms are, thus, relatively scarce and mostly from clinical settings. Furthermore, studies on antibiotic resistance in healthy humans or animals typically concentrate on Gram-negative pathogens such as E. coli or Salmonella [37,38] while only a few studies focus on enterococci. Data on linezolid-resistant E. faecalis and optrA gene are even more limited. Therefore, it is essential to understand the presence and transmission of optrA-positive E. faecalis among humans and reservoirs connected to farm animals. This study aims to (1) investigate the prevalence of optrA-positive E. faecalis (OPEfs) in humans, chickens, pigs, dogs, wastewater, and flies on farms and (2) identify factors associated with the presence of this pathogen and genetic relationships of OPEfs isolates in livestock settings.
2. Results
2.1. Demographic Characteristics
Out of 139 enrolled farmers 78 were male (56.1%). Most of them were over 40 years old (82%), in secondary school in literacy (64.7%). More than two-thirds of farmers (70.5%) worked at household farms, 17.3% on small farms, and 12.2% on medium farms. There was no large farm in our study. Among 70 investigated farms, 72.9% were households, 17.1% were small and 10% were medium farms. The total livestock unit of farms ranges from 0.2 to 37.0 with a mean of 9.6. Poultry farms took 65.7% (46 farms), only 4 pig farms and 20 mixed farms (Table 1).

Table 1.
Characteristics of investigated population and farms.
2.2. Prevalence of E. faecalis and OPEfs in Collected Samples
A total of 639 samples from investigated farms were collected, including feces samples from humans (n = 139, from 70 farms), chickens (n = 193, from 66 farms), pigs (n = 80 from 24 farms), and dogs (n = 51 from 51 farms); wastewater and flies (n = 67 and 109, from 67 farms). Out of 639 tested samples, 336 (52.6%) samples contained E. faecalis and 186 (29.1%) samples contained optrA-positive E. faecalis (OPEfs). Figure 1 shows the distribution of E. faecalis and OPEfs in 6 sample types. The highest occurrence of E. faecalis and OPEfs was observed in flies at 74.3% (81/109) and 46.8% (51/109), respectively), compared to chickens (56.5% (109/193) and 37.3% (72/193)), dogs (56.9% (29/51) and 33.3% (17/51)), and in wastewater (40.3% (27/67) and 16.4% (11/67)). These proportions were lowest in pigs (17.5% (14/80) and 11.3% (9/80)). The prevalences in humans were 54.7% (76/139) and 18.7% (26/139), respectively. By age, 21.5% of farmers in the 41–60 age group (17/62) harbored OPEfs, higher than other groups. By education level, farmers with high school and above had the lowest OPEfs infection rate (11.4%, 3/41) (Table 2).
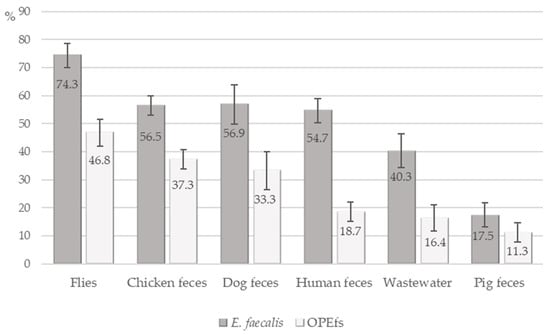
Figure 1.
Detection of E. faecalis and OptrA-positive E. faecalis in samples.

Table 2.
OptrA-positive E. faecalis infection in samples and associated factors.
At the farm level, the prevalence of OPEfs was also highest in fly samples with 41 of 67 investigated farms having OPEfs positive samples (61.2%). The lowest prevalence was in wastewater (16.4%, 11/67 farms). Twenty-six of 70 farms (37.1%) had farmers harboring OPEfs (Figure 2).
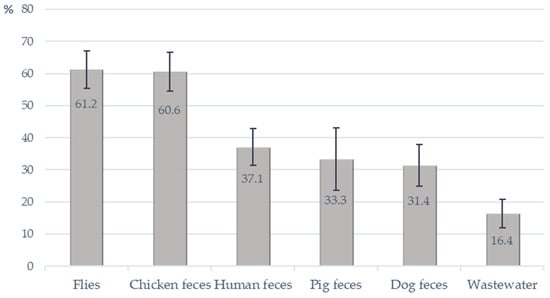
Figure 2.
Detection of OptrA-positive E. faecalis from investigated farms.
2.3. Associated Epidemiology Factors of OPEfs in Different Sample Types
Epidemiological factors were analyzed for 6 investigated objects (Table 2). In humans, there was no statistically significant association between the investigated epidemiological factors and OPEfs carriage except the farm scale factor. Farmers working on small farms were 2.77 times more likely to be infected with OPEfs than those working on household farms (OR = 2.77, p = 0.049).
The total livestock unit of the farm did not affect OPEfs status in humans but was associated with carriage status in chickens, flies, and wastewater. An increase of the total livestock unit of a farm by 1 unit was likely to increase the possibility of OPEfs carriage in these objects by 3% (p = 0.02), 7% (p = 0.002), and 8% (p = 0.005), respectively. Univariate analysis showed that feeding area was a factor frequently associated with OPEfs infection in 3 reservoirs, excluding human, dog, and pig fecal samples. Particularly, chickens on farms with over 1000 square meters had a nearly 2 times greater risk of getting OPEfs than those on smaller farms (OR = 1.94, p = 0.04). The risks were even higher for flies (OR = 2.85, p = 0.02) and wastewater samples (OR = 4.4, p = 0.03). However, the multivariate logistic regression model rejected the association of these factors with infection status in chickens and wastewater (p > 0.5). In flies, total livestock unit of farm and antibiotic use in animals were association factors for OPEfs. When the total livestock unit of farm increased by one unit, flies were 6% more likely to be infected with OPEfs (p = 0.04) and flies in farms where farmers used antibiotics in livestock brought OPEfs more than 3.35 times than farms that did not use (p = 0.02). There was no statistically significant relationship between investigated factors and OPEfs infection in pigs and dogs. Therefore, data from these samples are not shown.
2.4. OptrA-Positive E. faecalis Status and Antibiotic Resistance
The antibiotic susceptibility of 336 E. faecalis strains obtained from all samples was tested by using the agar dilution method. There was no strain resistant to penicillin, ampicillin, and vancomycin. However, 53 strains (15.8%) showed an intermediate level of sensitivity to vancomycin. Antibiotic resistance rates to the remaining 10 antibiotics were presented in Table 3. The highest resistance rate was observed for tetracyclin (85.7%), followed by doxcycyclin (76.8%) and erythromycin (61.9%). The highest resistance rate specified by sample types also belonged to tetracycline. Resistance rates were higher in the optrA-positive group for all tested antibiotics and all types of samples.

Table 3.
Prevalence of antimicrobial resistance phenotype in E. faecalis.
Overall resistance and intermediate rate to linezolid in all E. faecalis strains were 48.8% (164/336) and 3.6% (12/336), respectively. The resistance rate to linezolid was highest in pigs (64.3%), followed by chickens (60.6%) and flies (54.3%). In humans, linezolid-resistant E. faecalis was 31.6% (24/76) of obtained strains. Among 186 OPEfs strains, 86% (160) isolates were resistant to linezolid while only 2.7% (4/150) optrA-negative-E. faecalis strains had resistance phenotype.
Multidrug resistance (MDR) E. faecalis was detected as shown in Table S1. The majority of OPEfs isolates (86.0%, 160/186 strains) had an MDR pattern. 82.3% of these isolates were resistant to 5 and above tested antibiotics, and 19 isolates were resistant to 9 over 13 tested antibiotics from different classes including tetracycline, macrolide, phenicol, fluoroquinolone, and linezolid. Two OPEfs isolates, recovered from flies, were found to be resistant to 10 antibiotics. The rate of MDR in OPEfs was significantly higher than in the optrA-negative E. faecalis group. Only 37 out of 150 (24.7%) optrA-negative strains were MDR, with 1 strain being resistant to 8 antibiotics. 19.3% of this group were not resistant to any antibiotics, while only 1.1% of OPEfs strains had this phenotype. Generally, the MDR rate in all E. faecalis isolates was 58.6% (197/336).
Presence of optrA on E. feacalis was significantly associated with resistant phenotype to linezolid (OR = 448) and also to other antibiotics (OR ranged from 2.15 for ciprofloxacin to 9.07 for high-level streptomycin resistance), except for vancomycin. Multivariate analysis confirmed positive associations between the presence of optrA on E. feacalis and their resistant phenotype to linezolid (adjusted OR = 540) and levofloxacin (adjusted OR = 5.37) (Table 4).

Table 4.
Relationship between harboring optrA in E. faecalis and resistance phenotype.
2.5. Molecular Typing of optrA-Positive E. faecalis by PFGE
114 OPEfs isolates obtained from 59 farms were chosen for PFGE. Genetic relationships among them were illustrated in Figure 3. PFGE analysis with a similar cut-off value at 90% revealed 72 types of pulse patterns with a low degree of homology between lines. 19 clusters were shared pulse types (designated as I through XIX) and 53 were treated as unique. A total of 34 pulsotypes for 44 isolates from flies, 7 for 11 isolates from humans, and 8 for 9 isolates from wastewater were obtained. In animal fecal samples, 31 isolates from chickens, 10 from dogs, and 9 from pigs were divided into 24, 7, and 8 pulsotypes respectively. There were differences in PFGE patterns between isolates from flies and humans but had overlapping PFGE types between flies and pigs (clusters III, V, VII), flies and dogs (clusters III, VIII, XIII), flies and chickens (clusters II, III, XVII), and flies and wastewater (clusters X, XIX). In cluster II, 3 isolates of flies had a similar type of pulse to 2 isolates from chickens. There were up to 12 OPEfs strains from dogs (4 strains), chickens (2 strains), pigs (2 strains), and flies (4 strains) obtained from 12 farms that had similar pulse patterns after electrophoresis in cluster III. Only types IV, VI, XII, XIV, XV and XVI had shared pulse types between the same sample types (2 samples per cluster). Isolates from the same farm did not cluster together with the exception of 4 isolates from chickens in cluster I (farm 18) and XVII (farm 2). In humans, there were no share PFGE patterns of OPEfs strains discovered from dogs, pigs, and flies but had a close genetic relationship between 4 isolates collected from farmers on farms 50, 53 and chicken on farm 18 (cluster I), 6 isolates from farms 50, 54, 60 (humans), farm 7,8 (wastewater) and farm 8 (chickens) (cluster XI). Farmers in farms 59 and 61 harbored OPEfs strains with identical pulsotype.
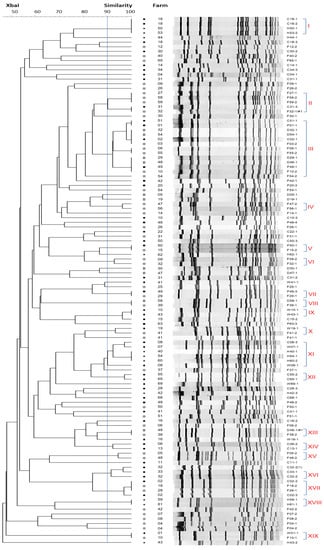
Figure 3.
Genetic relatedness of the 114 optrA-positive E. faecalis examined, based on the PFGE banding patterns of the isolates. Strain codes and PFGE subtypes are depicted. A total of 19 pulsotypes were identified at 90% similarity, designated as I through XIX. (C: chickens, D: dogs, F: flies, H: humans, P: pigs, W: wastewater).
3. Discussion
This is one among a few studies in Vietnam utilizing the One Health approach in investigating the situation and transmission of antibiotic-resistant bacteria in livestock settings. One Health concept was first mentioned in 2003–2004 in accordance with the emergence of SARS and avian influenza H5N1 [39]. One Health approach recognizes the interconnection between humans with shared environments with animals and plants. This approach is getting more critical in recent years and its health issues include emerging and re-emerging zoonotic diseases, vector-borne diseases as well as other health threats to humans, animals, and the environment [40]. Antibiotic resistance is also an issue of One Health. The global action plan on antimicrobial resistance in 2015 confirms that antibiotic resistance affects all areas of health and has impacts on various sectors and on the whole of society. The plan also underscores the roles of various sectors and resources to combat antibiotic resistance [41].
First identified in China in E. faecalis and E. faecium isolates in 2015 [17], resistance to oxazolidinones mediated by optrA is now detected worldwide, from both clinical resources, healthy humans, and animals [14,18,42]. However, in Vietnam, intensive research on the optrA gene and linezolid-resistant E. faecalis in clinical and community as well as associated factors are limited. According to our best knowledge, this is the first report that demonstrates the prevalence of OPEfs in livestock in our country. E. faecalis and OPEfs were present in all types of samples, from farmers, and animals (dogs, chickens, pigs) to flies and wastewater. It showed a higher prevalence of optrA in isolates from animals rather than humans. According to other studies [21,43,44], whole genome sequencing results indicated that fexA was co-located with optrA in E. faecalis strains, suggesting that optrA, a gene associated with resistance to linezolid, may be selected due to non-oxazolidinone antibiotics usage, such as phenicol (thiamphenicol, florfenicol, etc.). The popular use of phenicol in prevention and treatment of diseases in animals in Vietnam may explain the high percentage of OPEfs in this study [24,25,26].
In humans, the study indicated 18.7% of healthy farmers were infected with OPEfs, which is higher than 2.31% (36/1558) for adults and 3.47% (66/1900) for children in Hangzhou, China (2015) [44]. Study on fecal samples of humans and animals collected from 1998–2014 in five provinces/cities (Shandong, Henan, Tibet, Guangdong, and Shanghai), Wang reported a low proportion of optrA. Only 1.68% (10/595) of humans and 14.1% (41/290) animals (pigs n = 33; chickens n = 8) had OPEfs [17]. Compared to E. faecalis isolated from clinical infections in Spain in 2016–2017, and Korea in 2020, our results were higher [45,46]. In animal fecal samples, OPEfs prevalence in this study seemed more emergency than what was observed in cattle and pigs in Portugal in 2017 (0/201 and 6/249 samples) [34]. Notably, the results identified flies as the biggest host of this pathogen with 46.8% of samples infected by OPEfs. This prevalence was higher than the prevalence reported in a study on the carriage of mcr-1-positive Escherichia coli in Vietnam (36.7%) [29]. Carriage of resistant pathogens in flies was also reported in China [47]. The presence of OPEfs as well as other pathogens in flies might be due to exposure to various contaminated reservoirs and this implied contribution of flies to the spread of OPEfs by their borderless mobility.
No association between demographic factors such as age, sex, and education level and OPEfs infection in humans was identified in this study. Xiaoyu Ma’s study also reported no association of age with the carriage of linezolid-resistant E. faecalis isolated from patients with urinary tract infections in China from 2010 to 2015 [9]. However other studies reported an association of age with antibiotic-resistant bacteria infection status in humans. Jiaqi Zou studied 1902 samples that were collected in the Hospital of Chongqing Medical University for 5 years (2014–2018) and reported that patients with linezolid-resistant Enterococcus infections tended to be older than the control group [8]. Similarly, the older age group was also identified as at higher risk of other pathogens, such as mcr-1-positive Escherichia coli in a study in Vietnam [29]. The unique factor associated with OPEfs in humans was the farm scale (OR = 2.77, p = 0.049). The association of farm scale to infection status in farmers was also previously reported on antibiotic-resistant E. coli in Vietnam and Thailand [29,48].
The total livestock unit of the farm was also associated with the OPEfs infection in various reservoirs, including chicken, flies, and wastewater. Following that, an increase of the total livestock unit of the farm by 1 unit added a 3% chance of acquiring OPEfs in chicken, 7% in files, and 8% in wastewater. Similarly, the feeding area was also correlated with OPEfs status in these reservoirs. Chicken raised in over 1000 square meters of farms was 2.46 times more likely to get an infection than those in smaller farms. This ratio in flies was 2.85 and in wastewater was 4.40. It can be explained by the fact that the larger the animal feed, the more antibiotic may be used. The study in Thailand reported that medium-scale farms (having 100–500 sows) used a greater diversity of antimicrobials than small-scale farms (with a maximum of 20 sows) and also administered antimicrobials to a higher extent [48]. Similarly, two studies in Ghana identified antibiotic use in 100% and 97% of commercial farms while the percentage in backyard farms and domestic farms was only one-quarter and nearly one-half, respectively. Higher demand for productivity and a large number of animal feed in commercial farms can explain a higher use of antibiotics to maintain the wellness of animals [49,50].
Interestingly, antibiotic use in livestock did not create a statistically significant difference with OPEfs carrier status in humans and livestock but was associated with OPEfs carriage in flies. Multivariate analysis confirmed that flies captured in farms using antibiotics in livestock had a 3.35 times higher prevalence of OPEfs than those in farms that did not (p = 0.02). Exposure to a wide variety of samples in the environment, such as antibiotic-resistant bacteria in animal feces, wastewater, and food containing antibiotics might create this difference. Flies carrying bacteria that resist important antibiotics pose a potential threat to human health. A previous study in Vietnam also reported the potential roles of flies in the transmission of resistant pathogens in farming settings where associations of infection status with mcr-1-positive Escherichia coli in flies and in food animals in farms were identified [29]. They not only contact with animals, and the environment but also get into human food and water. Flies have been shown to be important vectors of the spread of antibiotic-resistant bacteria [51] and they are considered the most important non-biting insect pests in the medical and veterinary field because of the huge number of pathogens carriage [52]. Our PFGE data has confirmed this point. PFGE results demonstrated a genotypic diversity and wide transmission among OPEfs strains collected from a variety of samples in different farms (Figure 3). Among 114 OPEfs, 19 of 72 PFGE types were identified in at least 2 isolates or more. More than half of them (12/19 pulsotypes) contained OPEfs isolates from flies, which had the same PFGE subtype as others on distant farms. Flies in farms 27, 58, 59 (cluster II), farms 6, 55 (cluster III), farms 47, 56 (cluster IV), farms 09, 32 (cluster VI), and farms 05, 48 (cluster XV) provided OPEfs isolates with similar pulse types. There was no genetic relationship between OPEfs from flies and humans, but overlapping genotype between flies and dogs (2 pairs, farms (38, 48), farms (39, 58)), flies and pigs (2 pairs, farms (15, 50), farms (29, 49)), flies and chicken (2 pairs, farm (2, 18); (31, 32)), flies and wastewater (1 pair, farms 1, 10) were observed. According to Tenover et al., each pair of OPEfs strains had indistinguishable band patterns that could be “considered to represent the same strain” [53]. Especially, in cluster III, 12 isolates were collected from flies, dogs, pigs, and chickens in farms 1, 2, 3, 6, 10, 32, 48, 49, 51, 54, and 55 displayed genetic correlations at 96% to 100%. These farms were approximately 0.3 to 5.4 km distance from each other, suggesting the wide spread of resistant strains in the environment most likely through the mobility of files. Numerous studies also have demonstrated that flies can acquire, harbor, and transmit other resistant pathogens [29,54,55]. The high proportion of E. faecalis with optrA in flies in our study has strengthened the potential threat of flies to public health. Due to their unrestricted movement, and their attraction to residential areas, flies can play an important role in the ecology and transmission of bacteria, including enterococci with antibiotic-resistance genes [51,56].
In the remaining 53 unique pulse types, E. faecalis strains carrying the optrA gene were genetically heterogeneous indicating that a vast majority of them were not derived from a single clone, such as G. Dicuonzo’s study in Italy (reported in 2001) and Tamang MD’s study in Korea (published 2017) [13,27]. Exposure to physical and chemical stresses may have led to the evolution of a wide range of traits, which are necessary for the adaptation of E. faecalis to different environments. The evolutionary process, such as mutation, selection, and recombination might have played a role in the evolution of environmental stress tolerance, resulting in the observed high diversity [57].
Determination of sensitivity to linezolid of OPEfs strains showed resistant or intermediate ratio up to 92.5% (172 over 186 strains), proving the critical role of optrA to phenotypic resistance to linezolid. Univariate binary logistic regression analysis identified that E. feacalis strains carrying optrA were 448 times more likely to resist or reduce sensitivity to linezolid, multivariate analysis even strengthened this association (adjusted OR = 540, 95%CI 134-2175). Only 4 optrA negative E. faecalis resisted linezolid, which can be explained by other resistant mechanisms in these strains. Our results showed a contrast to the reported clinical data of a previous study in patients and pigs from 2008–2010 in Ha Noi (Vietnam) which reported no linezolid-resistant E. faecalis strain [5]. This raised an alarm about the presence and spread of resistant strains in the community. Compared to clinical data, the rate of linezolid resistance in this study is truly concerning [8,9,10].
Susceptibility testing results identified that OPEfs was not only resistant to linezolid but also to many other antibiotics commonly used in clinical settings. For vancomycin, a critical antibiotic in the treatment of positive gram bacteria infections, although there was no resistant strain identified, 15.8% of strains showed intermediate resistance to this antibiotic. Antibiotic sensitivity testing also raised an alarm of MDR E. faecalis, especially OPEfs strains, that showed resistance to important antibiotics such as fluoroquinolones. High-level resistance of aminoglycosides and macrolides of these strains was also reported (Table S1). Compared to a study by M. Usui et al. in 2015 on poultry farms in Southeast Asian countries, MDR rates in E. faecalis in our study were lower. The study pointed out that 100% of E. faecalis isolates in Vietnam were MDRO and was found to be resistant to 2–8 different antimicrobials, while 49 out of 58 isolates (84.5%) recovered from Indonesia exhibited MDR and resisted 2–6 antibacterial agents; 27 out of 37 isolates (73.0%) obtained from Thailand resisted to 2–7 different antibiotics [15]. However, in that study, resistance to linezolid and fluoroquinolone was not detected, and a higher MRD E. faecalis rate could be explained by a difference in criteria used to define multidrug-resistant bacteria. Moreover, it was noteworthy that resistance prevalence was high in flies, pigs, and chickens. As discussed above, flies are an important vector in the spread of antibiotic-resistant bacteria while chickens and pigs are 2 common sources of food in the Vietnamese diet. Therefore, MDR E. faecalis and OPEfs can be transmitted from animals to humans through flies or throughout the food chain. A similar MDR rate of E. faecalis (52.5%) was also detected in South Korea from cattle, chickens, and pigs during the period of 2010–2019 [3]. Although all OPEfs strains were susceptible to first-line treatment (ampicillin, penicillin), they were still threats to public health as reservoirs for many antibiotic-resistant genes, including optrA, which may horizontally spread through plasmids or transposons to other Enterococcus. Vice versa, OPEfs can receive other genes, for example, bla-, which makes resistance more worrisome.
Analyzing the relationship between optrA carriage status in E. faecalis isolates and their resistant phenotype to other antibiotics, we also found significant associations. The presence of the optrA gene was associated with phenotypic resistance of E. faecalis isolates against almost all tested antibiotics, excluding vancomycin. Multivariate analysis confirmed positive associations between the presence of optrA on E. feacalis and their resistant phenotype to levofloxacin (adjusted OR = 5.37) but rejected associations with phenotypic resistance to remaining antibiotics (Table 4). Levofloxacin is not on the list of livestock drugs allowed for trade in Vietnam [36] but there were 25% (84/336) of E. faecalis strains obtained in the study resisted or reduced sensitivity to this antibiotic, and there was an association between the presence of optrA to resistant phenotype to this antibiotic. These 2 points suggested the need to research resistance to levofloxacin in livestock settings.
The small sample size was a limitation of the study. Due to limited resources, the study was conducted in a small population (a commune). This influenced the representativeness of results on OPEfs prevalence. This might also be the reason why we were unable to identify any statistically significant relationship between OPEfs carriage in humans and in other objects such as chickens or pigs, neither could we identify any associations between any of our tested independent variables and OPEfs infection status in dog feces and pig feces samples. We believe that such associations should be identified with a larger sample size.
4. Materials and Methods
4.1. Setting and Population
A cross-sectional study was conducted in 2019 on livestock farms in a commune in Ha Nam, a province of northern Vietnam. The commune was selected as one of the communes with the largest number of domesticated animals in the province and that did not participate in any previous antibiotic resistance studies. The farming areas of this commune are geographically separated from residential areas. All farms and all their residents over the age of 18 engaged in farming were invited to participate in the study. The commune Veterinary Medicine Office made a list of farms and farmers from which 70 farms with 139 farmers who provided consent were enrolled in the study. The study created no potential harm to participants’ health as no invasive practice was conducted. They were provided with complete information on the study and provided consent to participate in the study. The study also strictly followed the ethical criteria of the Declaration of Helsinki, as well as receiving full benefits.
4.2. Sample Collection and Laboratory Analysis
Feces samples were collected from farmers, chickens, pigs, and dogs raised in these farms. Each farmer received a sterilized container with a spoon attached to the lid, gloves, and a biohazard bag and study staff demonstrated for home collection of their stool samples. Pooled feces of animals were collected by investigators. Dog feces sample was collected from several dunghills in each farm and then pooled in 1. Two to five pooled chicken feces samples were collected from each farm depending on the number of chickens in the farm (<1000: 2 samples, from 2000 to under 3000: 3 samples, from 3000 to under 10,000: 4 samples, from 10,000 and above 5 samples.), each sample was also pooled from feces collected from several dunghills in different areas in barns. Similarly, 3 to 5 pooled pig feces samples were collected from each farm (<10 pigs: 2 samples, from 10 to under 30: 4 samples, from 30 and above 5 samples). Additional samples from flies and wastewater were also collected on the same farms. Flies were retained using a glue board, then aseptically and individually transferred into 1.5 my Eppendorf tubes which contained 1 mL LB broth medium then pulverized using disposable plastic sticks (SPL Kore). 100 mL of wastewater was collected into a sterilized plastic bottle from each farm. All samples were stored at 4 °C after collection and transferred within 14 h to the Antimicrobial Resistance Laboratory of the National Institute of Hygiene and Epidemiology (NIHE) Vietnam for testing. Laboratory technicians took a part of about 1 g of each feces samples to cryotubes. 0.22 μm filters were used to filter wastewater samples, and filters were stored in cryotubes. All feces samples flies, and filters were stored at −80 °C until further use to ensure all samples remain stable for testing.
Each sample was spread onto Enterococcus Differential Agar Base (TITG Agar Base, Himedia, Mumbai, Maharashtra, India) with TTC Solution 1% (FD057) as a supplement. For wastewater samples, after filtering, a piece of the filter was taken and cultured in 10 mL LB broth medium at 37 °C for 6–8 h. Then, 1 full loop (10 µL) of the culture was speared onto the surface of a TITG agar. After 24 h at 37 °C, selected colonies with a deep red center and a narrow white periphery were sampled to identify E. faecalis by the MALDI Biotyper system (Bruker Daltonik GmbH, Bremen, Germany).
E. faecalis DNA was extracted by the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Detection of the optrA gene was performed with 2 primers (F: AGGTGGTCAGCGAACTAA, R: ATCAACTGTTCCCATTCA) and with the following PCR condition: 5 min at 94 °C, (1 min at 94 °C, 1 min at 48 °C, 1 min at 72 °C) × 34 cycles, 7 min at 72 °C [17]. Linezolid-resistant strain, E. faecalis R29-1-1 was used as a positive control, which was whole genome sequenced and confirmed optrA-positive by the Antimicrobial Resistance Laboratory of NIHE.
Samples that did not find out E. faecalis or optrA negative E. faecalis were considered OPEfs negative. A farm will be considered positive for OPEfs in chickens if at least 1 among 2–5 collected chicken feces samples collected in this farm harbors OPEfs. A similar manner is applied to the 5 remaining sample types.
4.3. Antimicrobial Susceptibility Testing
Antibiotic susceptibility of E. faecalis strains was assessed by the minimum inhibitory concentration method on Muller-Hinton agar and Brain heart infusion agar (BHI, for high-level aminoglycoside resistance) [58]. The bacteria were tested against 13 antimicrobial agents which are commonly prescribed for enterococcal infections and frequently used in farms, including ampicillin, penicillin (β-lactam), vancomycin (glycopeptide), chloramphenicol (phenicol), tetracycline, minocycline, doxycycline (tetracycline), ciprofloxacin, levofloxacin (fluoroquinolone), erythromycin (macrolide), high-level resistant aminoglycoside (gentamicin, streptomycin) as well as linezolid (oxazolidinone). Isolates showing intermediate/resistant levels of susceptibility were classified as non-susceptible [59,60]. An isolate was classified as MDR when it exhibited resistance to at least 3 antibiotics classes [59]. E. faecalis ATCC 29212 and E. faecalis ATCC 51299 were used as control strains. Susceptibility tests were interpreted according to CLSI 2022 (32 edition) [61].
These data were analyzed to identify the relationship to otrpA carrying on E. faecalis isolates.
4.4. Epidemiological Data Collection and Analysis
Registered participants were interviewed using a structured questionnaire. Investigated variables included demographic information (age, sex, education) and characteristics of farms (e.g., type of farm, feeding area, farms scale, farm livestock unit, use of antibiotic, …). Total livestock units of farm and farm scale were specified following the guidance of Decree 13/2020/ND-CP Detail the Law on Livestock issued by the Vietnam Government. It specifies the livestock unit coefficient for each type of food animal, ranging from 0.0003 to 1. This coefficient multiplied by the number of each respective type of animal equals “livestock unit”. The total livestock units of a farm is the sum of livestock units of all food animals types raised and it is used to categorize farm scale (Household: ≤10 units, small: from 10 to less than 30 units, medium: from 30 to less than 300, large: ≥300 units) [62]. Completed interviews and laboratory results were entered using Epidata Entry v3.1 (Denmark). SPSS 20.0 software was used for data analysis. Qualitative variables were presented as frequency, percentage (%), and 95% confidence interval (95%CI), while quantitative variables were presented as minimum, maximum, mean, and standard deviation (SD). χ2 test, binary logistic regression (for univariate and multivariate analysis) was performed to identify the association between OPEfs carriage of various reservoirs (humans, chickens, pigs, dogs, flies, and wastewater) and independent variables. The odds ratio (OR) and 95% confidence interval (CI) were calculated to assess the strength of the association. The Hosmer-Lemeshow test was used to evaluate the goodness of fit of the logistic regression model. The associations were considered significant with a p-value < 0.05. Variables that have p-values ≥ 0.05 in the univariate analysis were not included in the multivariable analysis.
4.5. Pulsed-Field Gel Electrophoresis (PFGE)
Pulsed-field gel electrophoresis was used to analyze clonal relatedness among the OPEfs isolates. We selected clinically important strains which had resistant phenotype to linezolid and at least two important antimicrobial agents for Gram-positive bacteria in the clinic, including quinolone resistance, macrolide resistance, high-level aminoglycoside resistance (HLAR) or non-sensitive to vancomycin [32]. PFGE for SmaI-digested genomic DNA was performed as described by US CDC [63] with some modifications in the DNA preparation. In brief, the bacteria cells were lysed with a combination of lysozyme (20 mg/mL) and 5 μL of recombinant lysostaphin (1 mg/mL). Plugs were incubated in cell lysis buffer (6 mM Tris HCl, 1 M NaCl, 100 mM EDTA, 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% sodium lauroylsarcosine) and proteinase K (20 mg/mL) for 3 h at 54 °C with vigorous (160 RPM) agitation. Slices of DNA plugs were digested for 4 h with 25UI SmaI at 25 °C and XbaI at 37 °C. PFGE was run using CHEF-DR II system (Bio-Rad, Hercules, California, US), pulse times 3.5 s to 23.5 s for 20 h (block 1, 6 V, 120°), running temperature 14 °C. XbaI-digested Salmonella enterica serovar Braenderup H9812 was used as the size marker. The PFGE profiles were analyzed by BioNumerics version 6.6 software (Applied Maths, Kortrijk, Belgium).
Simpson’s index of diversity (D) was calculated [64] to assess the differentiation of E. faecalis pulsotypes by PFGE. PFGE analysis was based on the Dice similarity coefficient and unweighted pair group method using arithmetic averages (UPGMA) clustering with position tolerance and optimization coefficient of 1.0%.
5. Conclusions
Our study is one of a few providing data on antibiotic resistance infection in various objects in livestock in Vietnam. This data is important and in accordance with Objective 2 of the Global action plan 2015 which emphasizes gaps in knowledge on antibiotic resistance that need to be filled to guide local, national, and regional actions [41]. The results provided strong evidence of the presence of resistant genes to last-resort antibiotics for humans such as linezolid in livestock settings although these antibiotics are not used for food animals and possibility to transmit to humans of resistant genes. This suggested a need to establish surveillance programs to monitor resistance to critically important antibiotics of commensal bacteria from farm animals to prevent their transmission to humans. Results on the role of flies in the transmission of resistance isolations among long-distance farms recommend that cleaning the barn and handling and managing animal manure properly to restrict the growth of this insect is also a way to limit the spread of antibiotic-resistant bacteria, including OPEfs. The next step of our study is performing molecular research to further investigate transmission, variants, and genetic context of optrA as well as find out other mechanisms of linezolid resistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12060954/s1. Supplemental Table S1. Resistance patterns of E. faecalis isolates.
Author Contributions
T.H.H., H.R.v.D., A.-L.B., P.T.N., D.D.A. and T.N.D. designed the study and directed study implementation. T.H.H., T.T.M.H., P.T.L.N., P.D.T. and N.T.M. managed the fieldwork. T.H.H., H.T.A.H., N.H.T. (Nguyen Ha Thanh) and T.H.A. performed laboratory work. H.T.A.H., P.T.L.N., T.T.M.H., P.D.T., V.T.N.B., N.T.H.H. and N.T.M. managed data. T.H.H., H.T.A.H., P.T.L.N., L.A.T., B.T.T., T.H.M.L., M.C., D.P.T., S.-N.T.M., H.C.T., N.H.T. (Nguyen Hoai Thu) and M.S. analyzed data. T.H.H., H.T.A.H., P.T.L.N. and T.H.A. developed the first draft of the paper. N.V.T. writing-review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 108.02-2017.320, by GCRF One Health Poultry Hub (Royal Veterinary College, UK) under grant number BB/S001269/1, by the Japan Agency for Medical Research and Development (AMED) under grants numbers JP23gm1610003, JP23fk0108642, JP23fk0108665, JP23fk0108683, JP23wm0325037, and JP23wm0225022, by the Wellcome Africa Asia Programme grant Vietnam, 2015–2020: 106680, and by IRD and LMI DRISA.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Hanoi Medical University (protocol code NCS05/HMU-IRB and date of approval 29 March 2019).
Informed Consent Statement
Informed consent was obtained from all farmers involved in the study.
Data Availability Statement
Data is contained within the article or Supplementary Materials.
Acknowledgments
We are grateful to colleagues in Ha Nam province for their assistance with collecting samples in the field. We express our appreciation to Fiona Tomley, Experimental Parasitology, Royla Veterinary College, The UK, Director of UKRI GCRF One Health Poultry Hub for her scientific advisory during this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wilson, D.N.; Schluenzen, F.; Harms, J.M.; Starosta, A.L.; Connell, S.R.; Fucini, P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. USA 2008, 105, 13339–13344. [Google Scholar] [CrossRef]
- Moellering, R.C. Linezolid: The first oxazolidinone antimicrobial. Ann. Intern. Med. 2003, 138, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Moon, D.C.; Kim, S.J.; Mechesso, A.F.; Song, H.J.; Kang, H.Y.; Choi, J.H.; Yoon, S.S.; Lim, S.K. Nationwide Surveillance on Antimicrobial Resistance Profiles of Enterococcus faecium and Enterococcus faecalis Isolated from Healthy Food Animals in South Korea, 2010 to 2019. Microorganisms 2021, 9, 925. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M. Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.L.; Bisgaard, M.; Son, N.T.; Trung, N.V.; An, H.M.; Dalsgaard, A. Enterococcus faecalis clones in poultry and in humans with urinary tract infections, Vietnam. Emerg. Infect. Dis. 2012, 18, 1096–1100. [Google Scholar] [CrossRef]
- Gaglio, R.; Couto, N.; Marques, C.; de Fatima Silva Lopes, M.; Moschetti, G.; Pomba, C.; Settanni, L. Evaluation of antimicrobial resistance and virulence of enterococci from equipment surfaces, raw materials, and traditional cheeses. Int. J. Food Microbiol. 2016, 236, 107–114. [Google Scholar] [CrossRef]
- Fozouni, L.; Askari, H.; Pordeli, H.R. Frequency Distribution of Fluoroquinolones-Resistant Enterococcus faecalis Isolates from Patients with Prostatitis in Golestan Province, Iran. Med. Lab. J. 2019, 13, 29–33. [Google Scholar] [CrossRef]
- Zou, J.; Xia, Y. Molecular characteristics and risk factors associated with linezolid-resistant Enterococcus faecalis infection in Southwest China. J. Glob. Antimicrob. Resist. 2020, 22, 504–510. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, F.; Bai, B.; Lin, Z.; Xu, G.; Chen, Z.; Sun, X.; Zheng, J.; Deng, Q.; Yu, Z. Linezolid Resistance in Enterococcus faecalis Associated with Urinary Tract Infections of Patients in a Tertiary Hospitals in China: Resistance Mechanisms, Virulence, and Risk Factors. Front. Public Health 2021, 9, 570650. [Google Scholar] [CrossRef]
- Anh Tram, Q. Study on antibiotic resistance characteristics of gram-positive strains of bacteria causing urinary tract infections in Nghe An General Friendship Hospital. Tạp chí Y học Việt Nam 2022, 517, 257–261. [Google Scholar] [CrossRef]
- Miller, W.R.; Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti-Infect. Ther. 2014, 12, 1221–1236. [Google Scholar] [CrossRef] [PubMed]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Dicuonzo, G.; Gherardi, G.; Lorino, G.; Angeletti, S.; Battistoni, F.; Bertuccini, L.; Creti, R.; Di Rosa, R.; Venditti, M.; Baldassarri, L. Antibiotic resistance and genotypic characterization by PFGE of clinical and environmental isolates of enterococci. FEMS Microbiol. Lett. 2001, 201, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Daniel, D.S.; Lee, S.M.; Gan, H.M.; Dykes, G.A.; Rahman, S. Genetic diversity of Enterococcus faecalis isolated from environmental, animal and clinical sources in Malaysia. J. Infect. Public Health 2017, 10, 617–623. [Google Scholar] [CrossRef]
- Usui, M.; Ozawa, S.; Onozato, H.; Kuge, R.; Obata, Y.; Uemae, T.; Ngoc, P.T.; Heriyanto, A.; Chalemchaikit, T.; Makita, K.; et al. Antimicrobial susceptibility of indicator bacteria isolated from chickens in Southeast Asian countries (Vietnam, Indonesia and Thailand). J. Vet. Med. Sci. 2014, 76, 685–692. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Feßler, A.T.; Hanke, D.; Eichhorn, I.; Azcona-Gutiérrez, J.M.; Pérez-Moreno, M.O.; Seral, C.; Aspiroz, C.; Alonso, C.A.; Torres, L.; et al. Mechanisms of Linezolid Resistance Among Enterococci of Clinical Origin in Spain-Detection of optrA- and cfr(D)-Carrying E. faecalis. Microorganisms 2020, 8, 1155. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef]
- He, T.; Shen, Y.; Schwarz, S.; Cai, J.; Lv, Y.; Li, J.; Feßler, A.T.; Zhang, R.; Wu, C.; Shen, J.; et al. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J. Antimicrob. Chemother. 2016, 71, 1466–1473. [Google Scholar] [CrossRef]
- Yi, M.; Zou, J.; Zhao, J.; Tang, Y.; Yuan, Y.; Yang, B.; Huang, J.; Xia, P.; Xia, Y. Emergence of optrA-Mediated Linezolid Resistance in Enterococcus faecium: A Molecular Investigation in a Tertiary Hospital of Southwest China from 2014–2018. Infect. Drug Resist. 2022, 15, 13–20. [Google Scholar] [CrossRef]
- Sadowy, E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid 2018, 99, 89–98. [Google Scholar] [CrossRef]
- Freitas, A.R.; Tedim, A.P.; Novais, C.; Lanza, V.F.; Peixe, L. Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb. Genom. 2020, 6, e000350. [Google Scholar] [CrossRef]
- Yoon, S.; Kim, Y.B.; Seo, K.W.; Ha, J.S.; Noh, E.B.; Lee, Y.J. Characteristics of linezolid-resistant Enterococcus faecalis isolates from broiler breeder farms. Poult. Sci. 2020, 99, 6055–6061. [Google Scholar] [CrossRef]
- Almeida, L.M.; Lebreton, F.; Gaca, A.; Bispo, P.M.; Saavedra, J.T.; Calumby, R.N.; Grillo, L.M.; Nascimento, T.G.; Filsner, P.H.; Moreno, A.M.; et al. Transferable Resistance Gene optrA in Enterococcus faecalis from Swine in Brazil. Antimicrob. Agents Chemother. 2020, 64, e00142-20. [Google Scholar] [CrossRef]
- Kim, D.P.; Saegerman, C.; Douny, C.; Dinh, T.V.; Xuan, B.; Vu, B.D.; Hong, N.P.; Scippo, M.-L. First Survey on the Use of Antibiotics in Pig and Poultry Production in the Red River Delta Region of Vietnam. Food Public Health 2013, 3, 247–256. [Google Scholar]
- Di, K.N.; Pham, D.T.; Tee, T.S.; Binh, Q.A.; Nguyen, T.C. Antibiotic usage and resistance in animal production in Vietnam: A review of existing literature. Trop. Anim. Health Prod. 2021, 53, 340. [Google Scholar] [CrossRef]
- Luu, Q.H.; Nguyen, T.L.A.; Pham, T.N.; Vo, N.G.; Padungtod, P. Antimicrobial use in household, semi-industrialized, and industrialized pig and poultry farms in Viet Nam. Prev. Vet. Med. 2021, 189, 105292. [Google Scholar] [CrossRef]
- Tamang, M.D.; Moon, D.C.; Kim, S.R.; Kang, H.Y.; Lee, K.; Nam, H.M.; Jang, G.C.; Lee, H.S.; Jung, S.C.; Lim, S.K. Detection of novel oxazolidinone and phenicol resistance gene optrA in enterococcal isolates from food animals and animal carcasses. Vet. Microbiol. 2017, 201, 252–256. [Google Scholar] [CrossRef]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. Biol. Sci. 2018, 285, 20180332. [Google Scholar] [CrossRef]
- Nguyen, P.T.L.; Tran, H.T.M.; Tran, H.A.; Pham, T.D.; Luong, T.M.; Nguyen, T.H.; Nguyen, L.T.P.; Nguyen, T.T.T.; Hoang, H.T.A.; Nguyen, C.; et al. Carriage of Plasmid-Mediated Colistin Resistance-1-Positive Escherichia coli in Humans, Animals, and Environment on Farms in Vietnam. Am. J. Trop. Med. Hyg. 2022, 107, 65–71. [Google Scholar] [CrossRef]
- Rawat, N.; Anjali; Shreyata; Sabu, B.; Jamwal, R.; Devi, P.P.; Yadav, K.; Raina, H.S.; Rajagopal, R. Understanding the role of insects in the acquisition and transmission of antibiotic resistance. Sci. Total Environ. 2023, 858, 159805. [Google Scholar] [CrossRef]
- Fletcher, S. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ. Health Prev. Med. 2015, 20, 243–252. [Google Scholar] [CrossRef] [PubMed]
- WHO. Critically Important Antimicrobials for Human Medicine, 6th Revision. 2018, p. 14. Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf (accessed on 18 February 2023).
- Daniel, D.S.; Lee, S.M.; Dykes, G.A.; Rahman, S. Public Health Risks of Multiple-Drug-Resistant Enterococcus spp. in Southeast Asia. Appl. Environ. Microbiol. 2015, 81, 6090–6097. [Google Scholar] [CrossRef] [PubMed]
- Gião, J.; Leão, C.; Albuquerque, T.; Clemente, L.; Amaro, A. Antimicrobial Susceptibility of Enterococcus Isolates from Cattle and Pigs in Portugal: Linezolid Resistance Genes optrA and poxtA. Antibiotics 2022, 11, 615. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Pan, H.; Lou, Y.; Wu, Z.; Zhang, J.; Huang, Y.; Yu, W.; Qiu, Y. Epidemiological characteristics and genetic structure of linezolid-resistant Enterococcus faecalis. Infect. Drug Resist. 2018, 11, 2397–2409. [Google Scholar] [CrossRef]
- MRAD. Vietnam Ministry of Agriculture & Rural Development, Circulars 10/2016TT-BNNPTN, List of Antibiotics Allowed to Be Used in Animal Feed. Available online: https://vanban.chinhphu.vn/default.aspx?pageid=27160&docid=186403 (accessed on 22 March 2023).
- Nguyen, V.T.; Carrique-Mas, J.J.; Ngo, T.H.; Ho, H.M.; Ha, T.T.; Campbell, J.I.; Nguyen, T.N.; Hoang, N.N.; Pham, V.M.; Wagenaar, J.A.; et al. Prevalence and risk factors for carriage of antimicrobial-resistant Escherichia coli on household and small-scale chicken farms in the Mekong Delta of Vietnam. J. Antimicrob. Chemother. 2015, 70, 2144–2152. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Nguyen, L.T.; Chau, T.T.H.; Nguyen, T.T.; Tran, B.N.; Taniguchi, T.; Hayashidani, H.; Ly, K.T.L. Prevalence and antibiotic resistance of Salmonella isolated from poultry and its environment in the Mekong Delta, Vietnam. Vet. World 2021, 14, 3216–3223. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Jeggo, M. The One Health Approach-Why Is It So Important? Trop. Med. Infect. Dis. 2019, 4, 88. [Google Scholar] [CrossRef]
- CDC. One Health Basics. Available online: https://www.cdc.gov/onehealth/basics/index.html#print (accessed on 26 April 2023).
- World Health Organization. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015. [Google Scholar]
- Tyson, G.H.; Sabo, J.L.; Hoffmann, M.; Hsu, C.H.; Mukherjee, S.; Hernandez, J.; Tillman, G.; Wasilenko, J.L.; Haro, J.; Simmons, M.; et al. Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J. Antimicrob. Chemother. 2018, 73, 3254–3258. [Google Scholar] [CrossRef]
- McHugh, M.P.; Parcell, B.J.; Pettigrew, K.A.; Toner, G.; Khatamzas, E.; El Sakka, N.; Karcher, A.M.; Walker, J.; Weir, R.; Meunier, D.; et al. Presence of optrA-mediated linezolid resistance in multiple lineages and plasmids of Enterococcus faecalis revealed by long read sequencing. Microbiology 2022, 168, 001137. [Google Scholar] [CrossRef]
- Cai, J.; Schwarz, S.; Chi, D.; Wang, Z.; Zhang, R.; Wang, Y. Faecal carriage of optrA-positive enterococci in asymptomatic healthy humans in Hangzhou, China. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2019, 25, 630.e1–630.e6. [Google Scholar] [CrossRef]
- Càmara, J.; Camoez, M.; Tubau, F.; Pujol, M.; Ayats, J.; Ardanuy, C.; Domínguez, M. Detection of the Novel optrA Gene Among Linezolid-Resistant Enterococci in Barcelona, Spain. Microb. Drug Resist. 2019, 25, 87–93. [Google Scholar] [CrossRef]
- Park, K.; Jeong, Y.S.; Chang, J.; Sung, H.; Kim, M.N. Emergence of optrA-Mediated Linezolid-Nonsusceptible Enterococcus faecalis in a Tertiary Care Hospital. Ann. Lab. Med. 2020, 40, 321–325. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017, 2, 16260. [Google Scholar] [CrossRef]
- Ström, G.; Halje, M.; Karlsson, D.; Jiwakanon, J.; Pringle, M.; Fernström, L.L.; Magnusson, U. Antimicrobial use and antimicrobial susceptibility in Escherichia coli on small- and medium-scale pig farms in north-eastern Thailand. Antimicrob. Resist. Infect. Control 2017, 6, 75. [Google Scholar] [CrossRef]
- Nkansa, M.; Agbekpornu, H.; Kikimoto, B.; Chandler, C. Antibiotic Use Among Poultry Farmers in the Dormaa Municipality, Ghana; Report for Fleming Fund Fellowship Programme; London School of Hygiene and Tropical Medicine: London, UK, 2020. [Google Scholar]
- Paintsil, E.K.; Ofori, L.A.; Akenten, C.W.; Fosu, D.; Ofori, S.; Lamshöft, M.; May, J.; Danso, K.O.; Krumkamp, R.; Dekker, D. Antimicrobial Usage in Commercial and Domestic Poultry Farming in Two Communities in the Ashanti Region of Ghana. Antibiotics 2021, 10, 800. [Google Scholar] [CrossRef]
- Doud, C.W.; Scott, H.M.; Zurek, L. Role of house flies in the ecology of Enterococcus faecalis from wastewater treatment facilities. Microb. Ecol. 2014, 67, 380–391. [Google Scholar] [CrossRef]
- Sruthi, B.; Ibrahim, S.S. Insect pests of medical importance and their management. In Insect Pests of Crops and Their Eco-Friendly Management; Bhumi Publishing: Maharashtra, India, 2023; pp. 211–214. [Google Scholar]
- Tenover, F.C.; Arbeit, R.D.; Goering, R.V.; Mickelsen, P.A.; Murray, B.E.; Persing, D.H.; Swaminathan, B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J. Clin. Microbiol. 1995, 33, 2233–2239. [Google Scholar] [CrossRef]
- Alves, T.D.S.; Lara, G.H.B.; Maluta, R.P.; Ribeiro, M.G.; Leite, D.D.S. Carrier flies of multidrug-resistant Escherichia coli as potential dissemination agent in dairy farm environment. Sci. Total Environ. 2018, 633, 1345–1351. [Google Scholar] [CrossRef]
- Onwugamba, F.C.; Fitzgerald, J.R.; Rochon, K.; Guardabassi, L.; Alabi, A.; Kühne, S.; Grobusch, M.P.; Schaumburg, F. The role of ‘filth flies’ in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 2018, 22, 8–17. [Google Scholar] [CrossRef]
- Macovei, L.; Zurek, L. Ecology of antibiotic resistance genes: Characterization of enterococci from houseflies collected in food settings. Appl. Environ. Microbiol. 2006, 72, 4028–4035. [Google Scholar] [CrossRef]
- Baureder, M.; Reimann, R.; Hederstedt, L. Contribution of catalase to hydrogen peroxide resistance in Enterococcus faecalis. FEMS Microbiol. Lett. 2012, 331, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Cockerill, F.R.; Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically: Approved Standard, 9th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- CDC. The National Antimicrobial Resistance Monitoring System for Enteric Bacteria. Glossary of Terms Related to Antibiotic Resistance. Available online: https://www.cdc.gov/narms/resources/glossary.html#print (accessed on 22 March 2023).
- Loftus, M.J.; Everts, R.J.; Cheng, A.C.; Eti, P.; Fakasiieiki, T.; Isaia, L.; Isopo, E.; Jenney, A.W.J.; Lameko, V.; Leaupepe, H.; et al. Antimicrobial susceptibility of bacterial isolates from clinical specimens in four Pacific Island countries, 2017–2021. Lancet Reg. Health—West. Pac. 2023, 32, 100677. [Google Scholar] [CrossRef] [PubMed]
- CLSI. M100-ED32:2022 Performance Standards for Antimicrobial Susceptibility Testing, 32nd Edition. Available online: http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSI%20M100%20ED32:2022&xormat=SPDF&src=BB (accessed on 28 June 2022).
- Vietnamese Government. Decree 13/2020/ND-CP Detail Guideline of Livestock Law; Vietnamese Government: Hanoi, Vietnam, 2020.
- Centers for Disease Control and Prevention (CDC). Unified Pulsed-Field Gel Electrophoresis (PFGE) Protocol for Gram Positive Bacteria. Available online: https://www.cdc.gov/hai/pdfs/labsettings/unified_pfge_protocol.pdf (accessed on 10 February 2023).
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).