Selective Bacteriocins: A Promising Treatment for Staphylococcus aureus Skin Infections Reveals Insights into Resistant Mutants, Vancomycin Resistance, and Cell Wall Alterations
Abstract
1. Introduction
2. Results
2.1. Discovery of Selective Bacteriocins
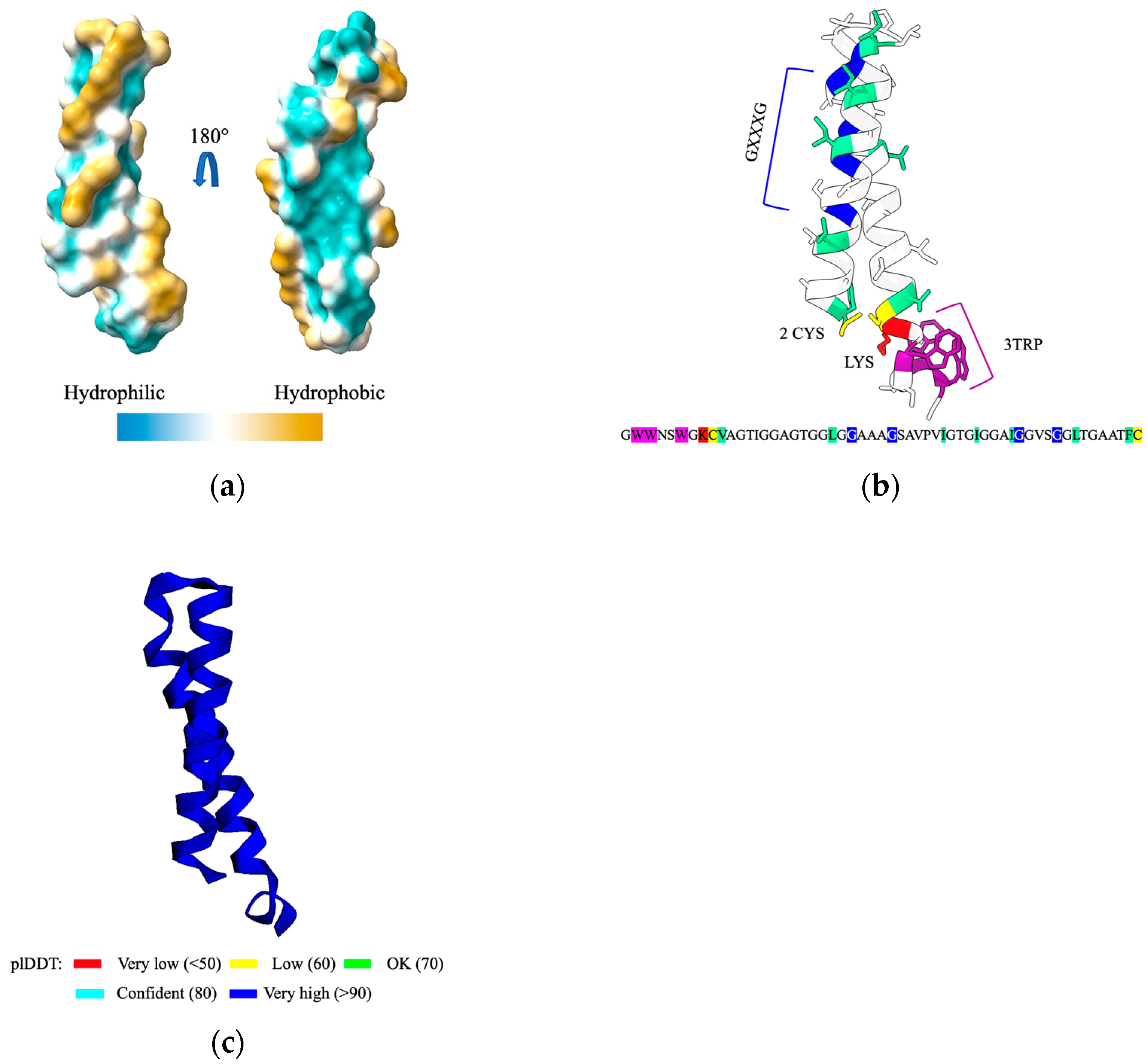
2.2. Cerein 7B, a Membrane Anchoring Bacteriocin
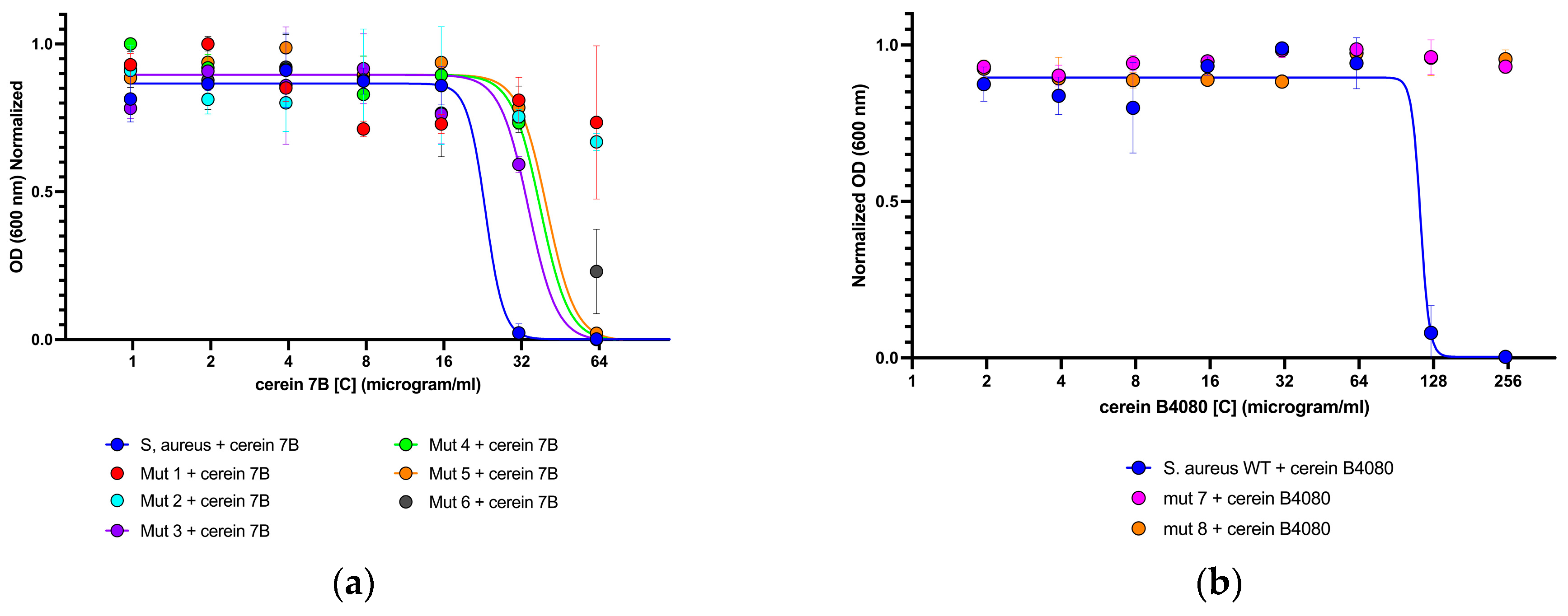
2.3. Isolation of Mutants Able to Grow in Presence of Bacteriocins
2.4. Resistance-Associated Mutations in Two Component Systems
2.5. Mutations Leading to Resistance against Cerein7b Are Associated to Decreased Sensitivity to Vancomycin
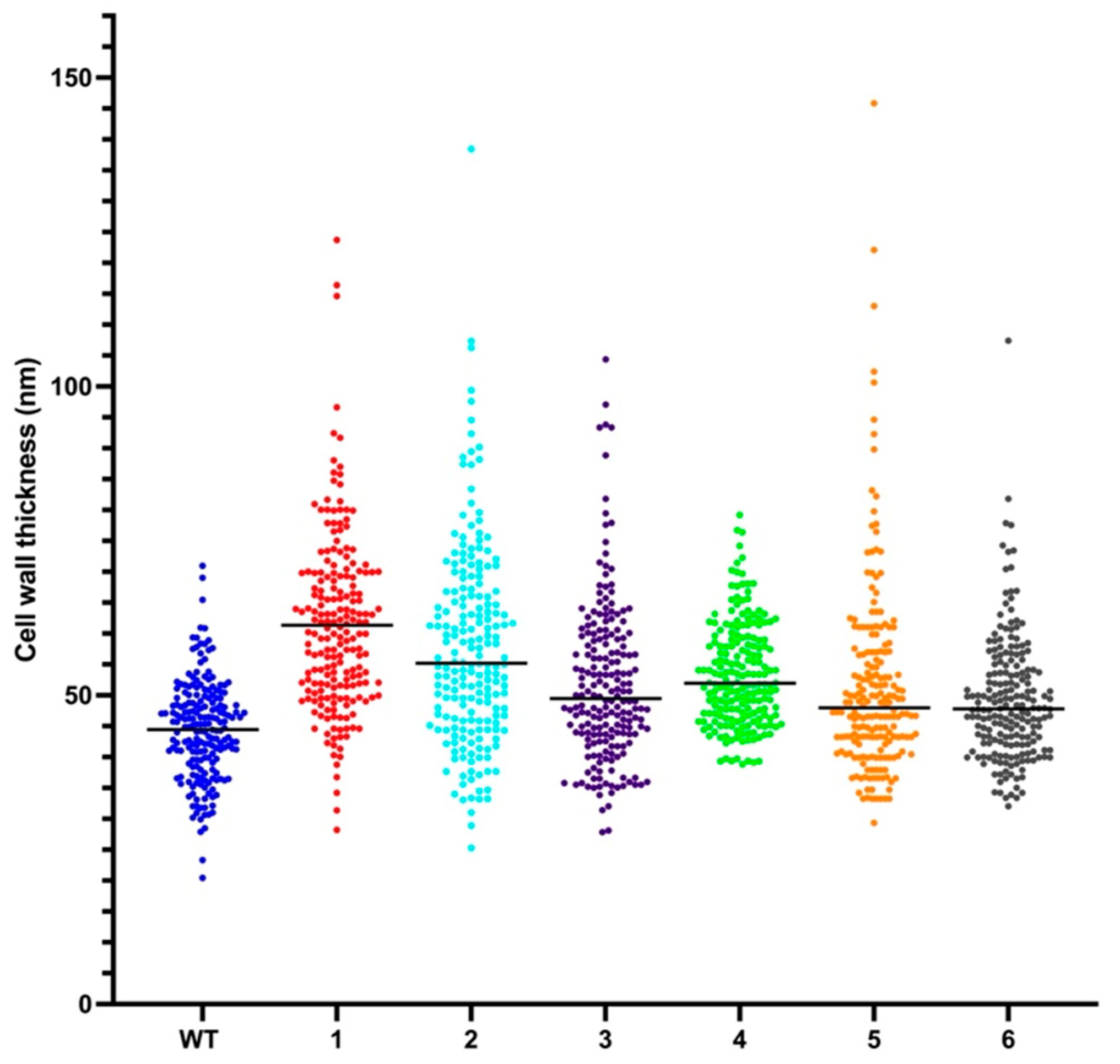
2.6. Resistant Mutant Display Cell Wall Thickening
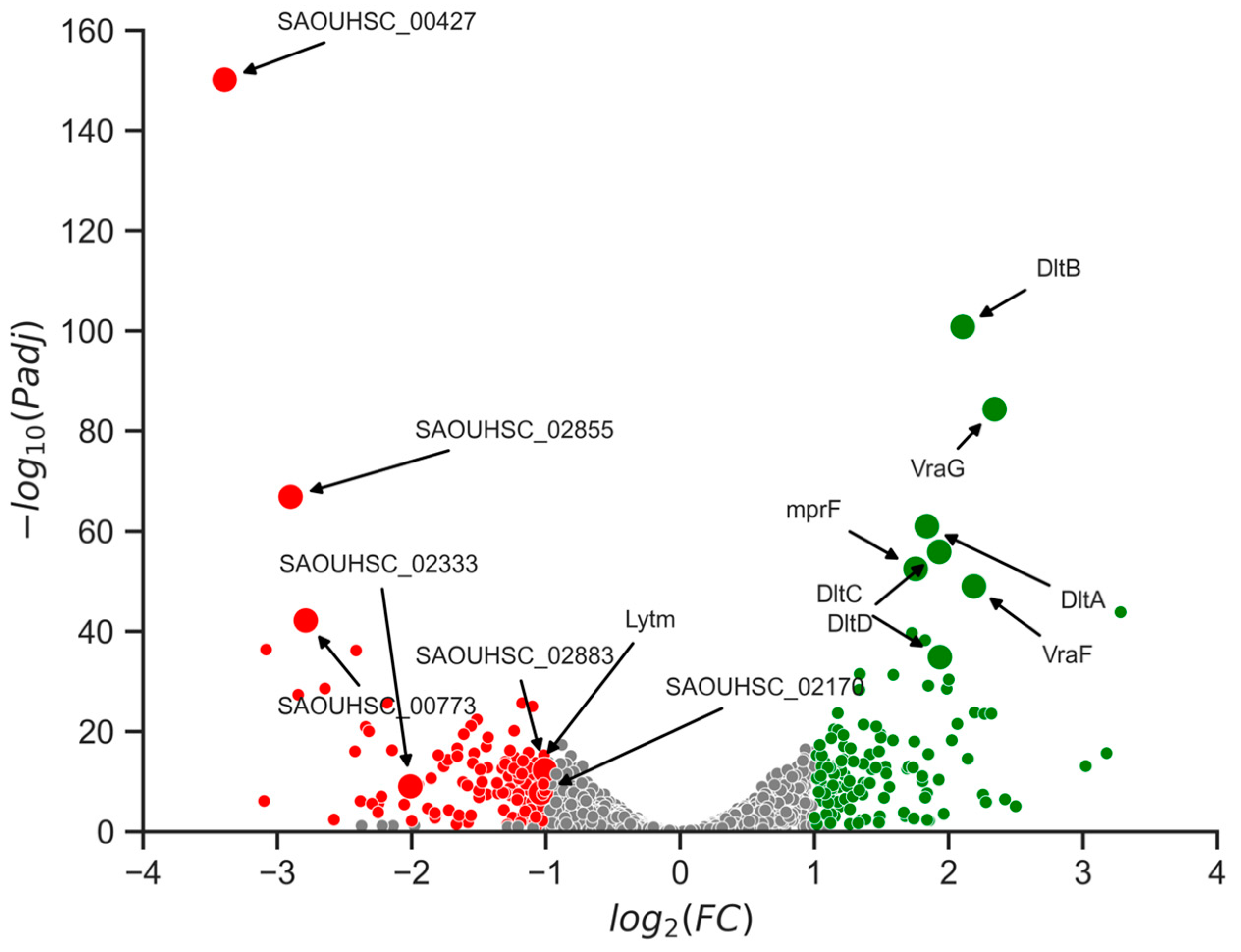
2.7. Changes in Gene Expression Levels Linked to Cell Wall Alterations
3. Discussion
3.1. The Study Findings
3.2. Significance of the Findings—Implication for the Field
3.3. Limitations of the Study
4. Materials and Methods
4.1. Spot Assay
4.2. Minimum Inhibition Concentraion (MIC)
4.3. Structural Prediction Alpha Fold
4.4. In Vitro Production of Cerein 7B Bacteriocin and Mutated Versions
4.5. Isolation of Mutants Able to Grow in Presence of Bacteriocins
4.6. S. aureus DNA Sequencing
4.7. Vancomycin Antibiogram
4.8. Transmission Electronic Microscopy (TEM)
4.9. RNA Sequencing and Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Lowy, F.D. Staphylococcus Aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Tong, Y.; Cheng, J.; Abbas, Z.; Li, Z.; Wang, J.; Zhou, Y.; Si, D.; Zhang, R. Biofilm and Small Colony Variants—An Update on Staphylococcus Aureus Strategies toward Drug Resistance. Int. J. Mol. Sci. 2022, 23, 1241. [Google Scholar] [CrossRef] [PubMed]
- Jang, S. Multidrug Efflux Pumps in Staphylococcus Aureus and Their Clinical Implications. J. Microbiol. 2016, 54, 1–8. [Google Scholar] [CrossRef]
- Watkins, K.E.; Unnikrishnan, M. Evasion of Host Defenses by Intracellular Staphylococcus Aureus. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 112, pp. 105–141. ISBN 978-0-12-820707-9. [Google Scholar]
- Appelbaum, P.C. Reduced Glycopeptide Susceptibility in Methicillin-Resistant Staphylococcus Aureus (MRSA). Int. J. Antimicrob. Agents 2007, 30, 398–408. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Naik, S.; Bouladoux, N.; Linehan, J.L.; Han, S.-J.; Harrison, O.J.; Wilhelm, C.; Conlan, S.; Himmelfarb, S.; Byrd, A.L.; Deming, C.; et al. Commensal–Dendritic-Cell Interaction Specifies a Unique Protective Skin Immune Signature. Nature 2015, 520, 104–108. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus Epidermidis Esp Inhibits Staphylococcus Aureus Biofilm Formation and Nasal Colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Bonelli, R.R.; Schneider, T.; Sahl, H.-G.; Wiedemann, I. Insights into In Vivo Activities of Lantibiotics from Gallidermin and Epidermin Mode-of-Action Studies. Antimicrob. Agents Chemother. 2006, 50, 1449–1457. [Google Scholar] [CrossRef]
- Lubelski, J.; Rink, R.; Khusainov, R.; Moll, G.N.; Kuipers, O.P. Biosynthesis, Immunity, Regulation, Mode of Action and Engineering of the Model Lantibiotic Nisin. Cell. Mol. Life Sci. 2008, 65, 455–476. [Google Scholar] [CrossRef]
- Watanabe, A.; Kawada-Matsuo, M.; Le, M.N.-T.; Hisatsune, J.; Oogai, Y.; Nakano, Y.; Nakata, M.; Miyawaki, S.; Sugai, M.; Komatsuzawa, H. Comprehensive Analysis of Bacteriocins in Streptococcus Mutans. Sci. Rep. 2021, 11, 12963. [Google Scholar] [CrossRef]
- Geng, M.; Ravichandran, A.; Escano, J.; Smith, L. Efficacious Analogs of the Lantibiotic Mutacin 1140 against a Systemic Methicillin-Resistant Staphylococcus Aureus Infection. Antimicrob. Agents Chemother. 2018, 62, e01626-18. [Google Scholar] [CrossRef]
- Grein, F.; Schneider, T.; Sahl, H.-G. Docking on Lipid II—A Widespread Mechanism for Potent Bactericidal Activities of Antibiotic Peptides. J. Mol. Biol. 2019, 431, 3520–3530. [Google Scholar] [CrossRef]
- Gabrielsen, C.; Brede, D.A.; Hernández, P.E.; Nes, I.F.; Diep, D.B. The Maltose ABC Transporter in Lactococcus Lactis Facilitates High-Level Sensitivity to the Circular Bacteriocin Garvicin ML. Antimicrob. Agents Chemother. 2012, 56, 2908–2915. [Google Scholar] [CrossRef]
- Tymoszewska, A.; Diep, D.B.; Wirtek, P.; Aleksandrzak-Piekarczyk, T. The Non-Lantibiotic Bacteriocin Garvicin Q Targets Man-PTS in a Broad Spectrum of Sensitive Bacterial Genera. Sci. Rep. 2017, 7, 8359. [Google Scholar] [CrossRef]
- Ramnath, M.; Arous, S.; Gravesen, A.; Hastings, J.W.; Héchard, Y. Expression of MptC of Listeria Monocytogenes Induces Sensitivity to Class IIa Bacteriocins in Lactococcus Lactis. Microbiology 2004, 150, 2663–2668. [Google Scholar] [CrossRef]
- Flores-Romero, H.; Ros, U.; Garcia-Saez, A.J. Pore Formation in Regulated Cell Death. EMBO J. 2020, 39, e105753. [Google Scholar] [CrossRef]
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, Potent Antimicrobial Peptides and the Fight against Multi Drug Resistant Species: Resistance Is Futile? Antibiotics 2020, 9, 32. [Google Scholar] [CrossRef]
- Gabant, P.; Borrero, J. PARAGEN 1.0: A Standardized Synthetic Gene Library for Fast Cell-Free Bacteriocin Synthesis. Front. Bioeng. Biotechnol. 2019, 7, 213. [Google Scholar] [CrossRef]
- Oscáriz, J.C.; Cintas, L.; Holo, H.; Lasa, Ã.; Nes, I.F.; Pisabarro, A.G. Purification and Sequencing of Cerein 7B, a Novel Bacteriocin Produced by Bacillus Cereus Bc7. FEMS Microbiol. Lett. 2006, 254, 108–115. [Google Scholar] [CrossRef]
- Lee, Y.B.; Byun, E.J.; Kim, H.S. Potential Role of the Microbiome in Acne: A Comprehensive Review. J. Clin. Med. 2019, 8, 987. [Google Scholar] [CrossRef]
- Situ, A.J.; Kang, S.-M.; Frey, B.B.; An, W.; Kim, C.; Ulmer, T.S. Membrane Anchoring of α-Helical Proteins: Role of Tryptophan. J. Phys. Chem. B 2018, 122, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Teese, M.G.; Langosch, D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry 2015, 54, 5125–5135. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, J.; Peng, Q.; Liu, Y.; Lei, L.; Zhang, H. The Role of Staphylococcus Aureus YycFG in Gene Regulation, Biofilm Organization and Drug Resistance. Antibiotics 2021, 10, 1555. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, M.; García, B.; Valle, J.; Rapún, B.; Ruiz De Los Mozos, I.; Solano, C.; Martí, M.; Penadés, J.R.; Toledo-Arana, A.; Lasa, I. Sensory Deprivation in Staphylococcus Aureus. Nat. Commun. 2018, 9, 523. [Google Scholar] [CrossRef]
- Peng, H.; Rao, Y.; Yuan, W.; Zheng, Y.; Shang, W.; Hu, Z.; Yang, Y.; Tan, L.; Xiong, K.; Li, S.; et al. Reconstruction of the Vancomycin-Susceptible Staphylococcus Aureus Phenotype From a Vancomycin-Intermediate S. Aureus XN108. Front. Microbiol. 2018, 9, 2955. [Google Scholar] [CrossRef]
- Cameron, D.R.; Jiang, J.-H.; Kostoulias, X.; Foxwell, D.J.; Peleg, A.Y. Vancomycin Susceptibility in Methicillin-Resistant Staphylococcus Aureus Is Mediated by YycHI Activation of the WalRK Essential Two-Component Regulatory System. Sci. Rep. 2016, 6, 30823. [Google Scholar] [CrossRef]
- Tan, X.; Ramond, E.; Jamet, A.; Barnier, J.-P.; Decaux-Tramoni, B.; Dupuis, M.; Euphrasie, D.; Tros, F.; Nemazanyy, I.; Ziveri, J.; et al. Transketolase of Staphylococcus Aureus in the Control of Master Regulators of Stress Response During Infection. J. Infect. Dis. 2019, 220, 1967–1976. [Google Scholar] [CrossRef]
- Lebrette, H.; Borezée-Durant, E.; Martin, L.; Richaud, P.; Boeri Erba, E.; Cavazza, C. Novel Insights into Nickel Import in Staphylococcus Aureus: The Positive Role of Free Histidine and Structural Characterization of a New Thiazolidine-Type Nickel Chelator. Metallomics 2015, 7, 613–621. [Google Scholar] [CrossRef]
- Bestebroer, J.; van Kessel, K.P.M.; Azouagh, H.; Walenkamp, A.M.; Boer, I.G.J.; Romijn, R.A.; van Strijp, J.A.G.; de Haas, C.J.C. Staphylococcal SSL5 Inhibits Leukocyte Activation by Chemokines and Anaphylatoxins. Blood 2009, 113, 328–337. [Google Scholar] [CrossRef]
- Howden, B.P.; Peleg, A.Y.; Stinear, T.P. The Evolution of Vancomycin Intermediate Staphylococcus Aureus (VISA) and Heterogenous-VISA. Infect. Genet. Evol. 2014, 21, 575–582. [Google Scholar] [CrossRef]
- Cui, L.; Ma, X.; Sato, K.; Okuma, K.; Tenover, F.C.; Mamizuka, E.M.; Gemmell, C.G.; Kim, M.-N.; Ploy, M.-C.; El Solh, N.; et al. Cell Wall Thickening Is a Common Feature of Vancomycin Resistance in Staphylococcus Aureus. J. Clin. Microbiol. 2003, 41, 5–14. [Google Scholar] [CrossRef]
- Singh, V.K.; Carlos, M.R.; Singh, K. Physiological Significance of the Peptidoglycan Hydrolase, LytM, in Staphylococcus Aureus: Staphylococcal LytM. FEMS Microbiol. Lett. 2010, 311, 167–175. [Google Scholar] [CrossRef]
- Stapleton, M.R.; Horsburgh, M.J.; Hayhurst, E.J.; Wright, L.; Jonsson, I.-M.; Tarkowski, A.; Kokai-Kun, J.F.; Mond, J.J.; Foster, S.J. Characterization of IsaA and SceD, Two Putative Lytic Transglycosylases of Staphylococcus Aureus. J. Bacteriol. 2007, 189, 7316–7325. [Google Scholar] [CrossRef]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a Widely Distributed Protein Motif for Binding to (Peptido)Glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef]
- Kawada-Matsuo, M.; Yoshida, Y.; Zendo, T.; Nagao, J.; Oogai, Y.; Nakamura, Y.; Sonomoto, K.; Nakamura, N.; Komatsuzawa, H. Three Distinct Two-Component Systems Are Involved in Resistance to the Class I Bacteriocins, Nukacin ISK-1 and Nisin A, in Staphylococcus Aureus. PLoS ONE 2013, 8, e69455. [Google Scholar] [CrossRef]
- Li, M.; Cha, D.J.; Lai, Y.; Villaruz, A.E.; Sturdevant, D.E.; Otto, M. The Antimicrobial Peptide-Sensing System Aps of Staphylococcus Aureus. Mol. Microbiol. 2007, 66, 1136–1147. [Google Scholar] [CrossRef]
- Falord, M.; Karimova, G.; Hiron, A.; Msadek, T. GraXSR Proteins Interact with the VraFG ABC Transporter To Form a Five-Component System Required for Cationic Antimicrobial Peptide Sensing and Resistance in Staphylococcus Aureus. Antimicrob. Agents Chemother. 2012, 56, 1047–1058. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Coculescu, B.-I. Antimicrobial Resistance Induced by Genetic Changes. J. Med. Life 2009, 2, 114. [Google Scholar]
- Mitrophanov, A.Y.; Groisman, E.A. Signal Integration in Bacterial Two-Component Regulatory Systems. Genes Dev. 2008, 22, 2601–2611. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of Neural Network Basecalling Tools for Oxford Nanopore Sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
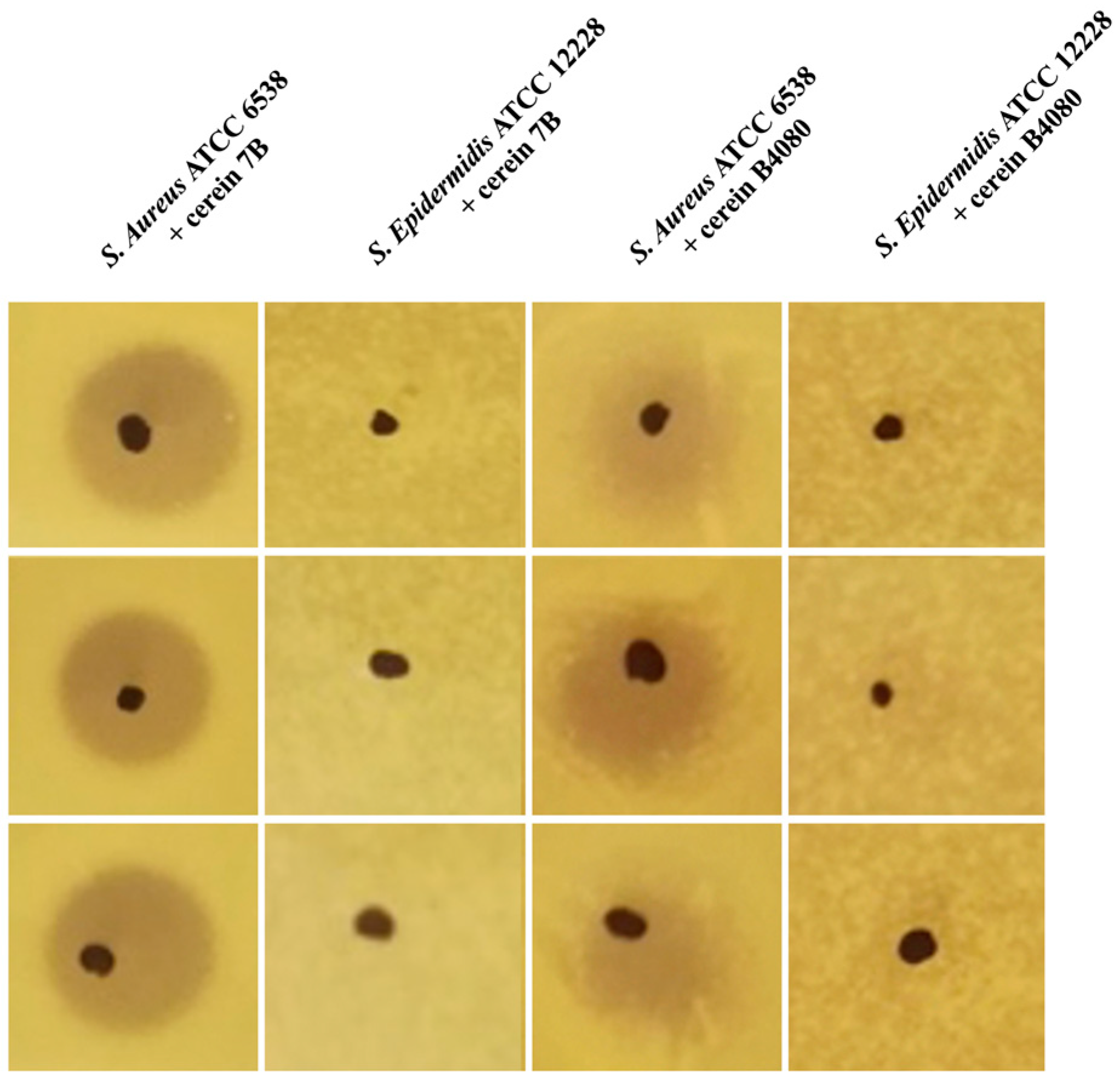


| Bacterial Species | Cerein 7B | Cerein B4080 |
|---|---|---|
| S. aureus ATCC 6538 | Active | Active |
| S. epidermidis ATCC 12228 | Non active | Non active |
| S. hominis ATCC 27844 | Non active | Active |
| Propionibacterium acnes ATCC 6919 | Non active | Non active |
| Escherichia coli MG 1655 | Non active | Non active |
| Lactococcus lactis IL1403 | Active | Active |
| Isolated Mutant | Mutation Position | Mutation | Amino Acid Mutation | Protein Affected |
|---|---|---|---|---|
| 1 | 26033 | t-g | I303S | WalK |
| 26252 | g-c | R376P | WalK | |
| 1621507 | g-t | H228N | AroE | |
| 2 | 26033 | t-g | I303S | WalK |
| 25075 | c-t | R222V | WalR | |
| 1621507 | g-t | H228N | AroE | |
| 3 | 24743 | c-t | A111V | WalR |
| 116151 | c-a | Y446Q | Hypothetical DNA-binding protein | |
| 1329116 | a-t | K385I | Tkt | |
| 4 | 26033 | t-g | I303S | WalK |
| 233744 | c-a | Y339T | NikA | |
| 995921 | g-a | V304M | MenD | |
| 1621507 | g-t | H228N | AroE | |
| 5 | 26033 | t-g | I303S | WalK |
| 1621507 | g-t | H228N | AroE | |
| 6 | 25793 | g-c | G223A | WalK |
| 26033 | t-g | I303S | WalK | |
| 1621507 | g-t | H228N | AroE | |
| 7 | 26033 | t-g | I303S | WalK |
| 26272 | g-a | V383I | WalK | |
| 1621507 | g-t | H228N | AroE | |
| 407356 | g-t | V196L | SSL5-like | |
| 241630 | c-t | H12Y | Hypothetical alcohol dehydrogenase | |
| 8 | 24724 | a-c | S105R | WalR |
| 27103 | g-c | A55P | Yych | |
| 1329116 | a-t | K385I | Tkt |
| Bacterial Species | Vancomycin MIC (µg/mL) |
|---|---|
| S. aureus ATCC 6538 | ≤0.5 |
| Isolated mutant 1 | 1 |
| Isolated mutant 2 | 2 |
| Isolated mutant 3 | ≤0.5 |
| Isolated mutant 4 | ≤0.5 |
| Isolated mutant 5 | ≤0.5 |
| Isolated mutant 6 | 1 |
| Isolated mutant 7 | 1 |
| Isolated mutant 8 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaumaux, F.; Petit, K.; Martin, A.; Rodriguez-Villalobos, H.; Vermeersch, M.; Perez-Morga, D.; Gabant, P. Selective Bacteriocins: A Promising Treatment for Staphylococcus aureus Skin Infections Reveals Insights into Resistant Mutants, Vancomycin Resistance, and Cell Wall Alterations. Antibiotics 2023, 12, 947. https://doi.org/10.3390/antibiotics12060947
Jaumaux F, Petit K, Martin A, Rodriguez-Villalobos H, Vermeersch M, Perez-Morga D, Gabant P. Selective Bacteriocins: A Promising Treatment for Staphylococcus aureus Skin Infections Reveals Insights into Resistant Mutants, Vancomycin Resistance, and Cell Wall Alterations. Antibiotics. 2023; 12(6):947. https://doi.org/10.3390/antibiotics12060947
Chicago/Turabian StyleJaumaux, Félix, Kenny Petit, Anandi Martin, Hector Rodriguez-Villalobos, Marjorie Vermeersch, David Perez-Morga, and Philippe Gabant. 2023. "Selective Bacteriocins: A Promising Treatment for Staphylococcus aureus Skin Infections Reveals Insights into Resistant Mutants, Vancomycin Resistance, and Cell Wall Alterations" Antibiotics 12, no. 6: 947. https://doi.org/10.3390/antibiotics12060947
APA StyleJaumaux, F., Petit, K., Martin, A., Rodriguez-Villalobos, H., Vermeersch, M., Perez-Morga, D., & Gabant, P. (2023). Selective Bacteriocins: A Promising Treatment for Staphylococcus aureus Skin Infections Reveals Insights into Resistant Mutants, Vancomycin Resistance, and Cell Wall Alterations. Antibiotics, 12(6), 947. https://doi.org/10.3390/antibiotics12060947





