Staphylococcus aureus Small-Colony Variants from Airways of Adult Cystic Fibrosis Patients as Precursors of Adaptive Antibiotic-Resistant Mutations
Abstract
1. Introduction
2. Results
2.1. S. aureus Infections among CF Patients
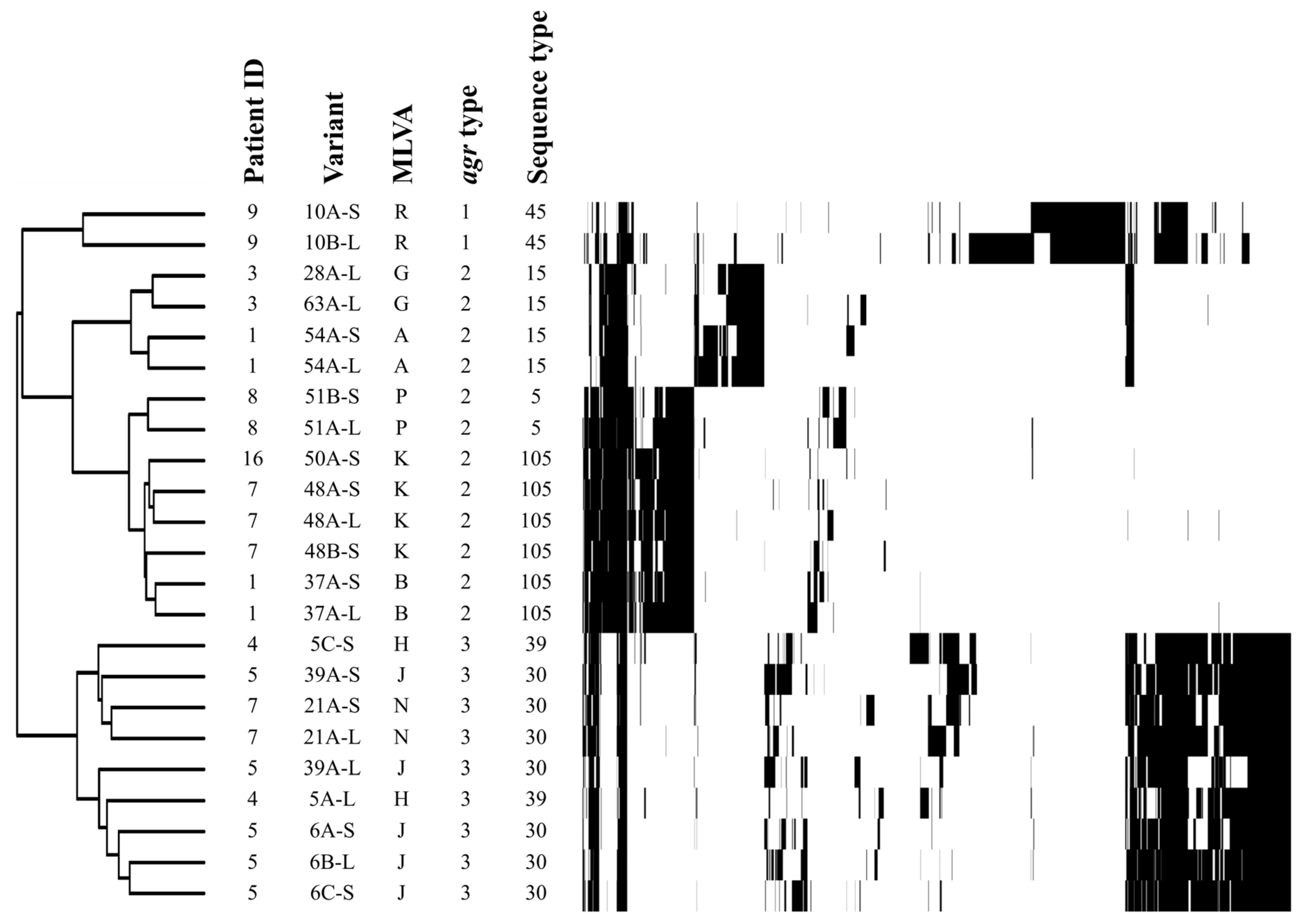
2.2. Genotype Profiles
2.3. Auxotrophy of SCVs
2.4. Antibiotic Susceptibility Profiles
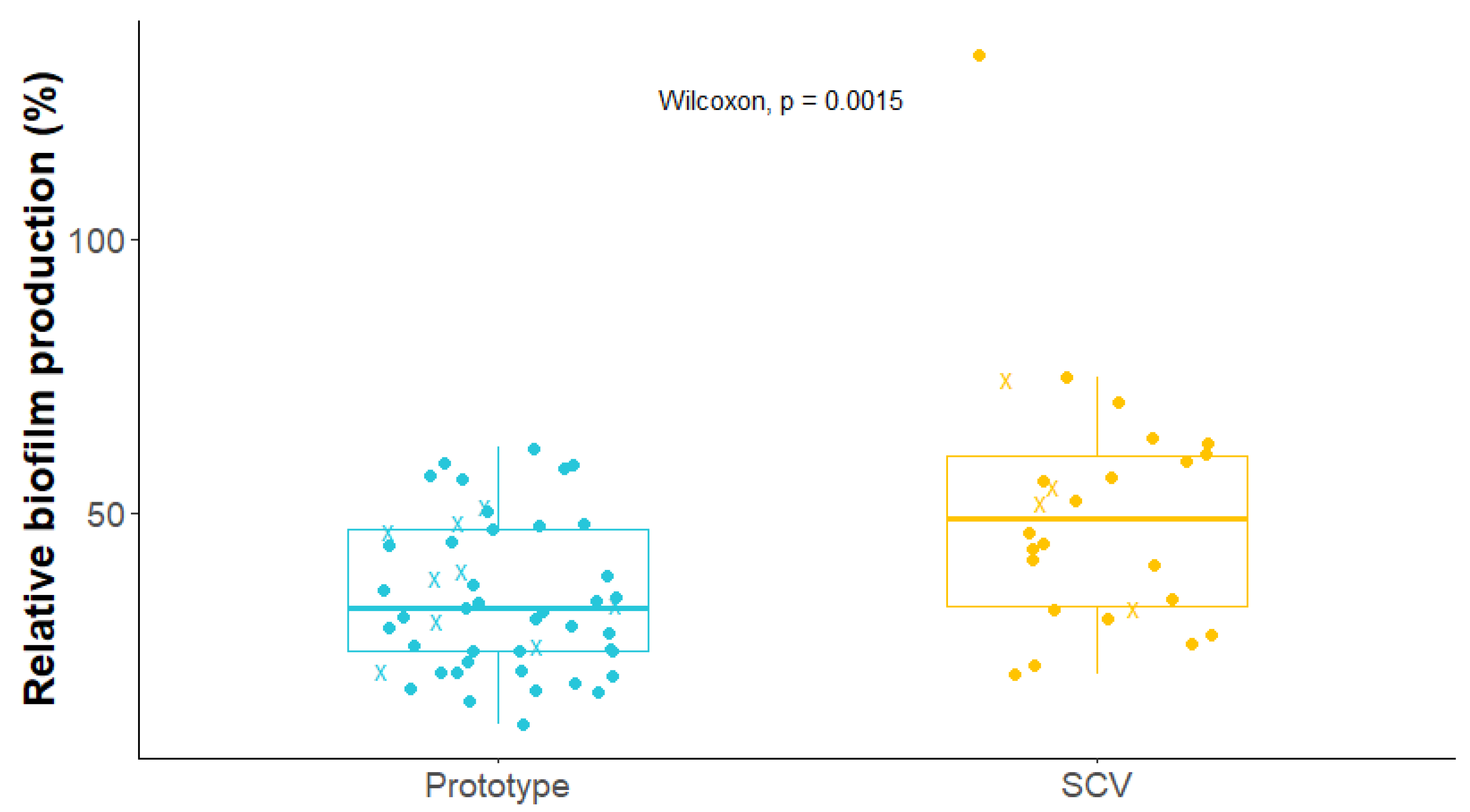
2.5. Biofilm Production by S. aureus Isolates
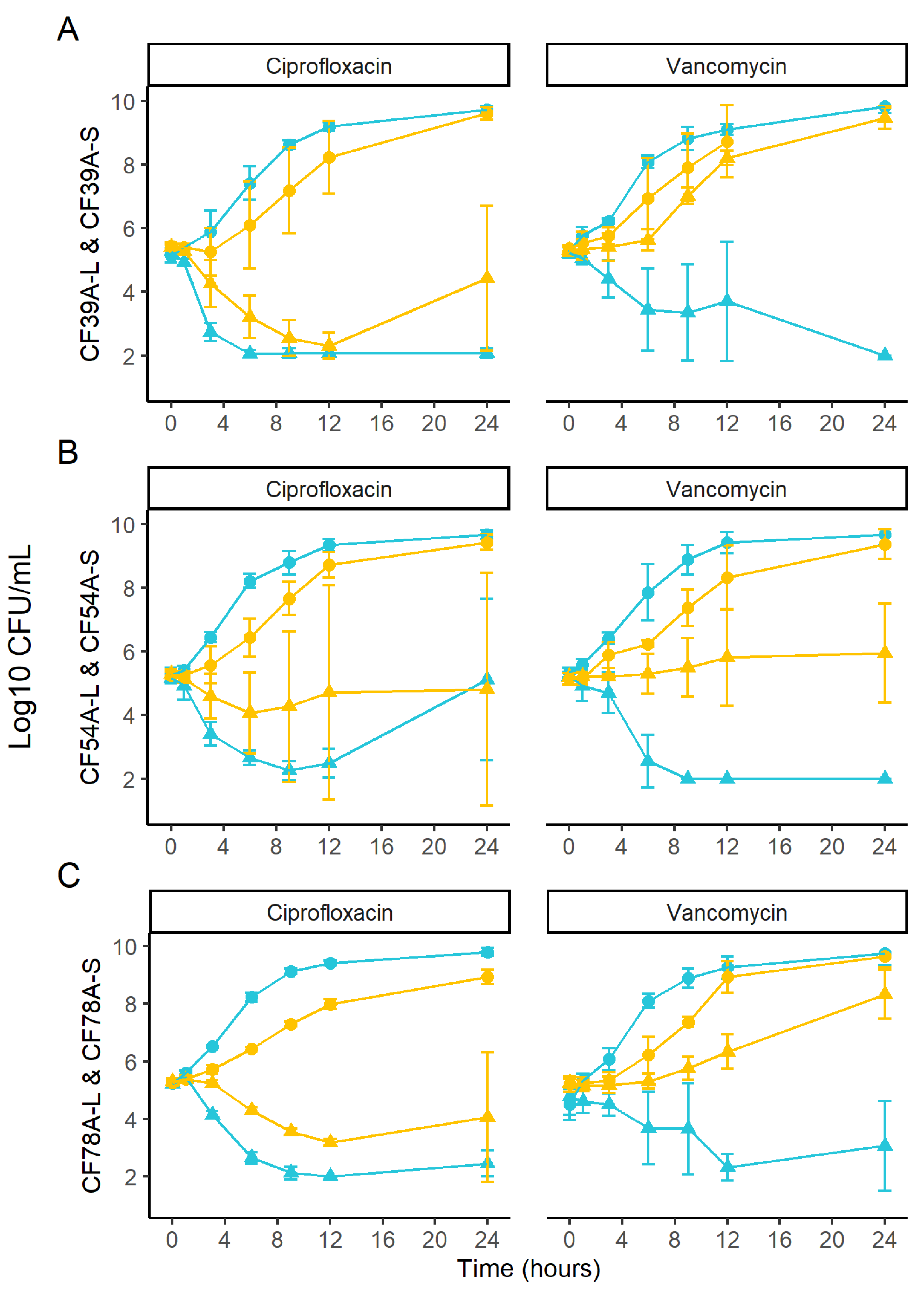
2.6. Kill Kinetics of Related SCV and Prototypic Isolates
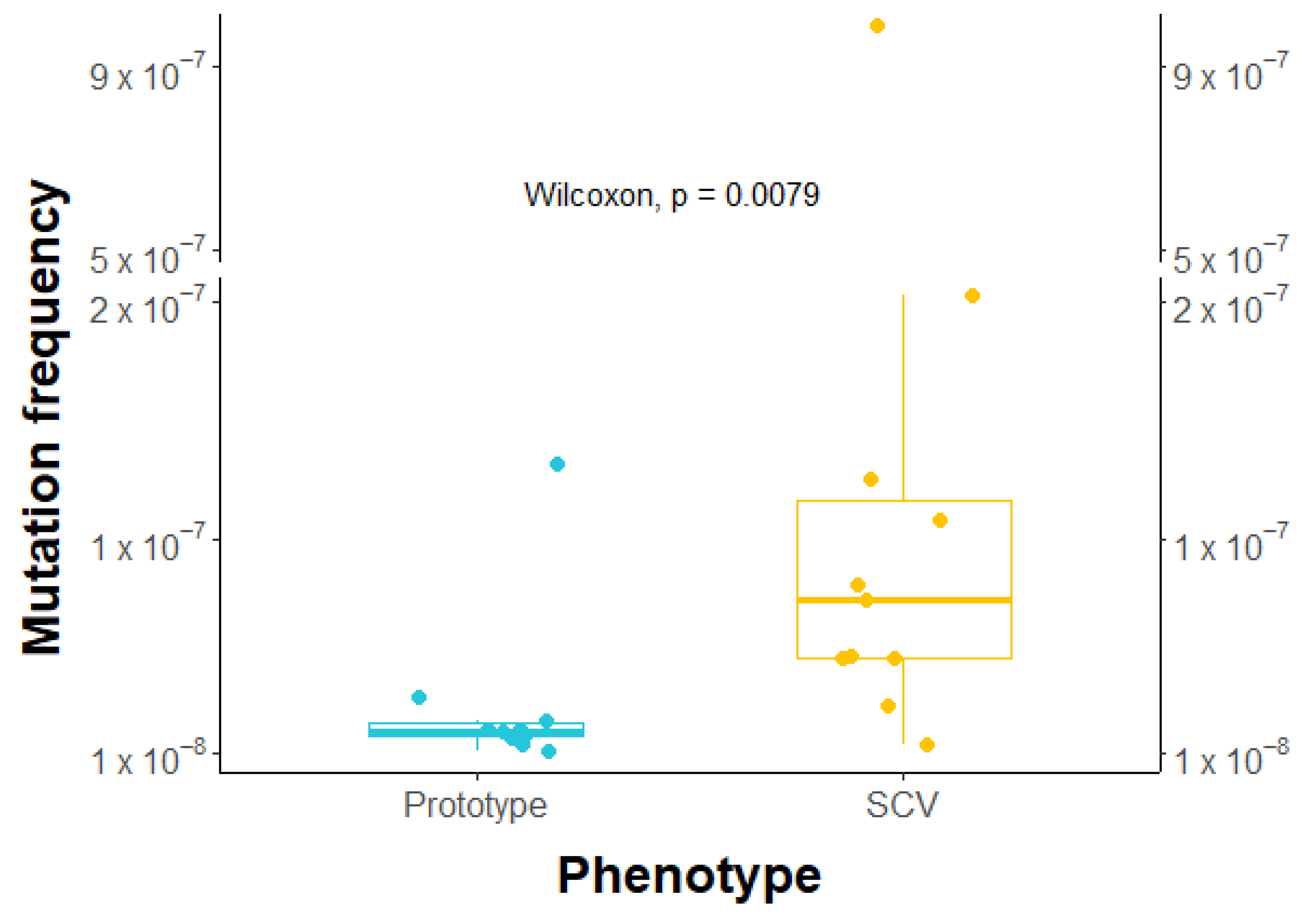
2.7. Higher Frequency of Rifampicin-Induced Resistant Mutants in SCVs
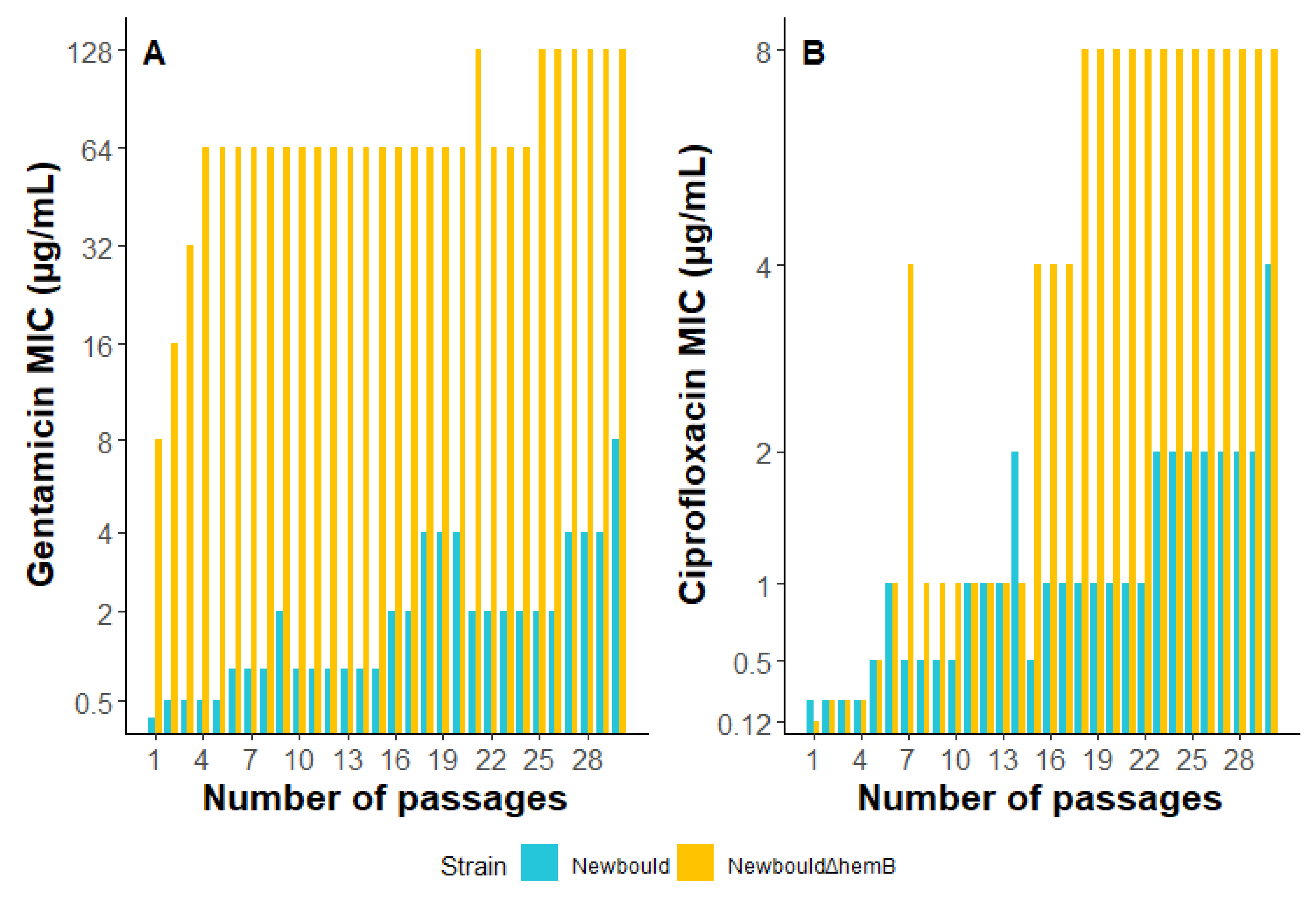
2.8. S. aureus Exposure to Sub-Inhibitory Concentrations of Gentamicin or Ciprofloxacin
3. Discussion
4. Materials and Methods
4.1. Reference Strains and Growth Conditions
4.2. Patients and Specimens
4.3. Microbiology of Clinical Samples
4.4. Genotyping and Selection of Representative Isolates
4.5. Genome Sequencing and Annotation
4.6. Minimal Inhibitory Concentrations (MICs)
4.7. Staphylococcal Cassette Chromosome mec (SCCmec) Typing
4.8. Aminoglycoside Resistance Determinants
4.9. Auxotrophy of SCVs
4.10. Biofilm Production
4.11. Kill Kinetics
4.12. Selection of Rifampicin-Resistant Mutants in Prototypic and SCV Backgrounds
4.13. Selection of Resistance Using Sub-Inhibitory Concentrations of Antibiotics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Canadian Cystic Fibrosis Registry Annual Report. In Cystic Fibrosis Canada [Internet]. 2023, pp. 1–54. Available online: www.cysticfibrosis.ca (accessed on 1 May 2023).
- 2021 Annual Data Report—Cystic Fibrosis Foundation Patient Registry. 2022. Available online: https://www.cff.org/medical-professionals/patient-registry (accessed on 1 May 2023).
- Armstrong, D.S.; Hook, S.M.; Jamsen, K.M.; Nixon, G.M.; Carzino, R.; Carlin, J.B.; Robertson, C.F.; Grimwood, K. Lower airway inflammation in infants with cystic fibrosis detected by newborn screening. Pediatr. Pulmonol. 2005, 40, 500–510. [Google Scholar] [CrossRef]
- Sagel, S.D.; Gibson, R.L.; Emerson, J.; McNamara, S.; Burns, J.L.; Wagener, J.S.; Ramsey, B.W. Inhaled Tobramycin in Young Children Study Group; Cystic Fibrosis Foundation Therapeutics Development Network. Impact of Pseudomonas and Staphylococcus infection on inflammation and clinical status in young children with cystic fibrosis. J. Pediatr. 2009, 154, 183–188. [Google Scholar] [CrossRef]
- Cogen, J.D.; Hall, M.; Faino, A.V.; Ambroggio, L.; Blaschke, A.J.; Brogan, T.V.; Cotter, J.M.; Gibson, R.L.; Grijalva, C.G.; Hersh, A.L.; et al. Antibiotics and outcomes of CF pulmonary exacerbations in children infected with MRSA and Pseudomonas aeruginosa. J. Cyst. Fibros. 2022, 22, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Ahlgren, H.G.; Benedetti, A.; Landry, J.S.; Bernier, J.; Matouk, E.; Radzioch, D.; Lands, L.C.; Rousseau, S.; Nguyen, D. Clinical outcomes associated with Staphylococcus aureus and Pseudomonas aeruginosa airway infections in adult cystic fibrosis patients. BMC Pulm. Med. 2015, 15, 67. [Google Scholar] [CrossRef] [PubMed]
- Schwerdt, M.; Neumann, C.; Schwartbeck, B.; Kampmeier, S.; Herzog, S.; Görlich, D.; Dübbers, A.; Große-Onnebrink, J.; Kessler, C.; Küster, P.; et al. Staphylococcus aureus in the airways of cystic fibrosis patients—A retrospective long-term study. Int. J. Med. Microbiol. 2018, 308, 631–639. [Google Scholar] [CrossRef]
- Wolter, D.J.; Emerson, J.C.; McNamara, S.; Buccat, A.M.; Qin, X.; Cochrane, E.; Houston, L.S.; Rogers, G.B.; Marsh, P.; Prehar, K.; et al. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin. Infect. Dis. 2013, 57, 384–391. [Google Scholar] [CrossRef]
- Junge, S.; Görlich, D.; den Reijer, M.; Wiedemann, B.; Tümmler, B.; Ellemunter, H.; Dübbers, A.; Küster, P.; Ballmann, M.; Koerner-Rettberg, C.; et al. Factors Associated with Worse Lung Function in Cystic Fibrosis Patients with Persistent Staphylococcus aureus. Omri A, editor. PLoS ONE 2016, 11, e0166220. [Google Scholar] [CrossRef]
- Wolter, D.J.; Onchiri, F.M.; Emerson, J.; Precit, M.R.; Lee, M.; McNamara, S.; Nay, L.; Blackledge, M.; Uluer, A.; Orenstein, D.M.; et al. Prevalence and clinical associations of Staphylococcus aureus small-colony variant respiratory infection in children with cystic fibrosis (SCVSA): A multicentre, observational study. Lancet Respir. Med. 2019, 7, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Harik, N.S.; Com, G.; Tang, X.; Melguizo Castro, M.; Stemper, M.E.; Carroll, J.L. Clinical characteristics and epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in children with cystic fibrosis from a center with a high MRSA prevalence. Am. J. Infect. Control. 2016, 44, 409–415. [Google Scholar] [CrossRef]
- Dasenbrook, E.C.; Checkley, W.; Merlo, C.A.; Konstan, M.W.; Lechtzin, N.; Boyle, M.P. Association between respiratory tract methicillin-resistant Staphylococcus aureus and survival in cystic fibrosis. JAMA 2010, 303, 2386–2392. [Google Scholar] [CrossRef]
- Kahl, B.C. Impact of Staphylococcus aureus on the pathogenesis of chronic cystic fibrosis lung disease. Int. J. Med. Microbiol. 2010, 300, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Kahl, B.C.; Becker, K.; Löffler, B. Clinical Significance and Pathogenesis of Staphylococcal Small Colony Variants in Persistent Infections. Clin. Microbiol. Rev. 2016, 29, 401–427. [Google Scholar] [CrossRef]
- Miller, M.H.; Edberg, S.C.; Mandel, L.J.; Behar, C.F.; Steigbigel, N.H. Gentamicin uptake in wild-type and aminoglycoside-resistant small-colony mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 1980, 18, 722–729. Available online: http://www.ncbi.nlm.nih.gov/pubmed/7447428 (accessed on 1 May 2023). [CrossRef]
- Proctor, R.A.; Von Eiff, C.; Kahl, B.C.; Becker, K.; McNamara, P.; Herrmann, M.; Peters, G. Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 2006, 4, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Brouillette, E.; Séguin, D.L.; Asselin, A.-E.; Jacob, C.L.; Malouin, F. A role for sigma factor B in the emergence of Staphylococcus aureus small-colony variants and elevated biofilm production resulting from an exposure to aminoglycosides. Microb. Pathog. 2010, 48, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Lamontagne, C.-A.; Brouillette, E.; Grondin, G.; Talbot, B.G.; Grandbois, M.; Malouin, F. Staphylococcus aureus SigB activity promotes a strong fibronectin-bacterium interaction which may sustain host tissue colonization by small-colony variants isolated from cystic fibrosis patients. Mol. Microbiol. 2008, 70, 1540–1555. [Google Scholar] [CrossRef]
- Moisan, H.; Brouillette, E.; Jacob, C.L.; Langlois-Bégin, P.; Michaud, S.; Malouin, F. Transcription of virulence factors in Staphylococcus aureus small-colony variants isolated from cystic fibrosis patients is influenced by SigB. J. Bacteriol. 2006, 188, 64–76. [Google Scholar] [CrossRef]
- Balwit, J.M.; van Langevelde, P.; Vann, J.M.; Proctor, R.A. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 1994, 170, 1033–1037. Available online: http://www.ncbi.nlm.nih.gov/pubmed/7930701 (accessed on 1 May 2023). [CrossRef]
- Tuchscherr, L.; Löffler, B.; Proctor, R.A. Persistence of Staphylococcus aureus: Multiple Metabolic Pathways Impact the Expression of Virulence Factors in Small-Colony Variants (SCVs). Front. Microbiol. 2020, 11, 1028. [Google Scholar] [CrossRef]
- Proctor, R. Respiration and Small Colony Variants of Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, M.; Rayner, O.; Smyth, A.R. Prophylactic anti-staphylococcal antibiotics for cystic fibrosis. Cochrane Database Syst. Rev. 2020, 2020, CD001912. [Google Scholar] [CrossRef]
- Lo, D.K.H.; Muhlebach, M.S.; Smyth, A.R. Interventions for the eradication of meticillin-resistant Staphylococcus aureus (MRSA) in people with cystic fibrosis. Cochrane Database Syst. Rev. 2018, 2018, CD009650. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.C.; Mall, M.A.; Gutierrez, H.; Macek, M.; Madge, S.; Davies, J.C.; Burgel, P.-R.; Tullis, E.; Castaños, C.; Castellani, C.; et al. The Lancet Respiratory Medicine Commission on the Future of Care of Cystic Fibrosis. Lancet Respir. Med. 2020, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Gläser, R.; Becker, K.; Von Eiff, C.; Meyer-Hoffert, U.; Harder, J. Decreased Susceptibility of Staphylococcus aureus Small-Colony Variants toward Human Antimicrobial Peptides. J. Invest. Dermatol. 2014, 134, 2347–2350. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019, 13, 1239–1251. [Google Scholar] [CrossRef]
- Kriegeskorte, A.; Lorè, N.I.; Bragonzi, A.; Riva, C.; Kelkenberg, M.; Becker, K.; Proctor, R.A.; Peters, G.; Kahl, B.C. Thymidine-Dependent Staphylococcus aureus Small-Colony Variants Are Induced by Trimethoprim-Sulfamethoxazole (SXT) and Have Increased Fitness during SXT Challenge. Antimicrob. Agents Chemother. 2015, 59, 7265–7272. [Google Scholar] [CrossRef]
- Vestergaard, M.; Paulander, W.; Ingmer, H. Activation of the SOS response increases the frequency of small colony variants. BMC Res. Notes. 2015, 8, 749. [Google Scholar] [CrossRef]
- Hoffman, L.R.; Deziel, E.; D’Argenio, D.A.; Lepine, F.; Emerson, J.; McNamara, S.; Gibson, R.L.; Ramsey, B.W.; Miller, S.I. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19890–19895. [Google Scholar] [CrossRef]
- Mitchell, G.; Séguin, D.L.; Asselin, A.-E.; Déziel, E.; Cantin, A.M.; Frost, E.H.; Michaud, S.; Malouin, F. Staphylococcus aureus sigma B-dependent emergence of small-colony variants and biofilm production following exposure to Pseudomonas aeruginosa 4-hydroxy-2-heptylquinoline-N-oxide. BMC Microbiol. 2010, 10, 33. [Google Scholar] [CrossRef]
- Lightbown, J.W.; Jackson, F.L. Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem. J. 1956, 63, 130–137. [Google Scholar] [CrossRef]
- Vesga, O.; Groeschel, M.C.; Otten, M.F.; Brar, D.W.; Vann, J.M.; Proctor, R.A. Staphylococcus aureus small colony variants are induced by the endothelial cell intracellular milieu. J. Infect. Dis. 1996, 173, 739–742. [Google Scholar] [CrossRef]
- Bui, L.M.G.; Turnidge, J.D.; Kidd, S.P. The induction of Staphylococcus aureus biofilm formation or Small Colony Variants is a strain-specific response to host-generated chemical stresses. Microbes Infect. 2015, 17, 77–82. [Google Scholar] [CrossRef]
- Gao, W.; Chua, K.; Davies, J.K.; Newton, H.J.; Seemann, T.; Harrison, P.; Holmes, N.E.; Rhee, H.-W.; Hong, J.-I.; Hartland, E.; et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010, 6, e1000944. [Google Scholar] [CrossRef] [PubMed]
- Painter, K.L.; Strange, E.; Parkhill, J.; Bamford, K.B.; Armstrong-James, D.; Edwards, A.M. Staphylococcus aureus adapts to oxidative stress by producing H2O2-resistant small-colony variants via the SOS response. Infect. Immun. 2015, 83, 1830–1844. [Google Scholar] [CrossRef] [PubMed]
- Taber, H.W.; Mueller, J.P.; Miller, P.F.; Arrow, A.S. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 1987, 51, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Wieneke, M.K.; Dach, F.; Neumann, C.; Görlich, D.; Kaese, L.; Thißen, T.; Dübbers, A.; Kessler, C.; Große-Onnebrink, J.; Küster, P.; et al. Association of Diverse Staphylococcus aureus Populations with Pseudomonas aeruginosa Coinfection and Inflammation in Cystic Fibrosis Airway Infection. mSphere. 2021, 6, e0035821. [Google Scholar] [CrossRef]
- Bernardy, E.E.; Petit, R.A.; Raghuram, V.; Alexander, A.M.; Read, T.D.; Goldberg, J.B. Genotypic and phenotypic diversity of Staphylococcus aureus isolates from cystic fibrosis patient lung infections and their interactions with Pseudomonas aeruginosa. MBio 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Kittinger, C.; Toplitsch, D.; Folli, B.; Landgraf, L.M.; Zarfel, G. Phenotypic Stability of Staphylococcus aureus Small Colony Variants (SCV) Isolates from Cystic Fibrosis (CF) Patients. Int. J. Environ. Res. Public. Health 2019, 16, 1940. [Google Scholar] [CrossRef]
- Grundstad, M.L.; Parlet, C.P.; Kwiecinski, J.M.; Kavanaugh, J.S.; Crosby, H.A.; Cho, Y.-S.; Heilmann, K.; Diekema, D.; Horswill, A.R. Quorum Sensing, Virulence, and Antibiotic Resistance of USA100 Methicillin-Resistant Staphylococcus aureus Isolates. mSphere 2019, 4, e00553-19. [Google Scholar] [CrossRef]
- Parkins, M.D.; Somayaji, R.; Waters, V.J. Epidemiology, Biology, and Impact of Clonal Pseudomonas aeruginosa Infections in Cystic Fibrosis. Clinical microbiology reviews. Clin. Microbiol. Rev. 2018, 31, e00019-18. [Google Scholar] [CrossRef] [PubMed]
- Thornton, C.S.; Parkins, M.D. Microbial Epidemiology of the Cystic Fibrosis Airways: Past, Present, and Future. In Seminars in Respiratory and Critical Care Medicine; Thieme Medical Publishers, Inc.: New York, NY, USA, 2023. [Google Scholar]
- Biggs, S.L.; Jennison, A.V.; Bergh, H.; Graham, R.; Nimmo, G.; Whiley, D. Limited evidence of patient-to-patient transmission of Staphylococcus aureus strains between children with cystic fibrosis, Queensland, Australia. PLoS ONE 2022, 17, e0275256. [Google Scholar] [CrossRef] [PubMed]
- Westphal, C.; Görlich, D.; Kampmeier, S.; Herzog, S.; Braun, N.; Hitschke, C.; Mellmann, A.; Peters, G.; Kahl, B.C.; Staphylococcal CF Study Group; et al. Antibiotic Treatment and Age Are Associated with Staphylococcus aureus Carriage Profiles during Persistence in the Airways of Cystic Fibrosis Patients. Front. Microbiol. 2020, 11, 230. [Google Scholar] [CrossRef] [PubMed]
- Rumpf, C.; Lange, J.; Schwartbeck, B.; Kahl, B.C. Staphylococcus aureus and cystic fibrosis—A close relationship. What can we learn from sequencing studies? Pathogens 2021, 10, 1177. [Google Scholar] [CrossRef]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.R.; Stinear, T.P.; Grayson, M.L. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: Resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, G.; Fugère, A.; Gaudreau, K.P.; Brouillette, E.; Frost, E.H.; Cantin, A.M.; Malouin, F. SigB Is a Dominant Regulator of Virulence in Staphylococcus aureus Small-Colony Variants. PLoS ONE 2013, 8, 1–14. [Google Scholar] [CrossRef]
- Morikawa, K.; Maruyama, A.; Inose, Y.; Higashide, M.; Hayashi, H.; Ohta, T. Overexpression of Sigma Factor B, Urges Staphylococcus aureus to Thicken the Cell Wall and to Resist β-Lactams. Biochem. Biophys. Res. Commun. 2001, 288, 385–389. [Google Scholar] [CrossRef]
- Chen, H.Y.; Chen, C.C.; Fang, C.S.; Hsieh, Y.T.; Lin, M.H.; Shu, J.C. Vancomycin activates σ B in vancomycin-resistant Staphylococcus aureus resulting in the enhancement of cytotoxicity. PLoS ONE 2011, 6, e24472. [Google Scholar] [CrossRef]
- von Eiff, C.; Peters, G.; Becker, K. The small colony variant (SCV) concept—The role of staphylococcal SCVs in persistent infections. Injury. 2006, 37 (Suppl. 2), S26–S33. [Google Scholar] [CrossRef]
- Schleimer, N.; Kaspar, U.; Drescher, M.; Seggewiß, J.; von Eiff, C.; Proctor, R.A.; Peters, G.; Kriegeskorte, A.; Becker, K. The energy-coupling factor transporter module EcfAA’T, a novel candidate for the genetic basis of fatty acid-auxotrophic small-colony variants of Staphylococcus aureus. Front. Microbiol. 2018, 9, 1863. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Jaiswal, N.K.; Kumar, M.; Ansari, M.A.A.; Sarfraz, A. Breast abscess in a case of duct atresia caused by CO2-auxotrophic small colony variants of Staphylococcus aureus: Case report and review of literature. J. Nat. Sci. Biol. Med. 2015, 6, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Gómez-González, C.; Acosta, J.; Villa, J.; Barrado, L.; Sanz, F.; Orellana, M.A.; Otero, J.R.; Chaves, F. Clinical and molecular characteristics of infections with CO2-dependent small-colony variants of Staphylococcus aureus. J. Clin. Microbiol. 2010, 48, 2878–2884. [Google Scholar] [CrossRef] [PubMed]
- Bazaid, A.S.; Forbes, S.; Humphreys, G.J.; Ledder, R.G.; O’Cualain, R.; McBain, A.J. Fatty Acid Supplementation Reverses the Small Colony Variant Phenotype in Triclosan-Adapted Staphylococcus aureus: Genetic, Proteomic and Phenotypic Analyses. Sci Rep. 2018, 8, 3876. [Google Scholar] [CrossRef]
- Zander, J.; Besier, S.; Saum, S.H.; Dehghani, F.; Loitsch, S.; Brade, V.; Wichelhaus, T.A. Influence of dTMP on the phenotypic appearance and intracellular persistence of Staphylococcus aureus. Infect. Immun. 2008, 76, 1333–1339. [Google Scholar] [CrossRef]
- Oliver, A. Mutators in cystic fibrosis chronic lung infection: Prevalence, mechanisms, and consequences for antimicrobial therapy. Int. J. Med. Microbiol. 2010, 300, 563–572. [Google Scholar] [CrossRef]
- Boehringer, D.; O’Farrell, H.C.; Rife, J.P.; Ban, N. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J. Biol. Chem. 2012, 287, 10453–10459. [Google Scholar] [CrossRef]
- Zhang-Akiyama, Q.-M.; Morinaga, H.; Kikuchi, M.; Yonekura, S.-I.; Sugiyama, H.; Yamamoto, K.; Yonei, S. KsgA, a 16S rRNA adenine methyltransferase, has a novel DNA glycosylase/AP lyase activity to prevent mutations in Escherichia coli. Nucleic Acids Res. 2009, 37, 2116–2125. [Google Scholar] [CrossRef]
- Ochi, K.; Kim, J.-Y.; Tanaka, Y.; Wang, G.; Masuda, K.; Nanamiya, H.; Okamoto, S.; Tokuyama, S.; Adachi, Y.; Kawamura, F. Inactivation of KsgA, a 16S rRNA methyltransferase, causes vigorous emergence of mutants with high-level kasugamycin resistance. Antimicrob. Agents Chemother. 2009, 53, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Lama, A.; Pané-Farré, J.; Chon, T.; Wiersma, A.M.; Sit, C.S.; Vederas, J.C.; Hecker, M.; Nakano, M.M. Response of methicillin-resistant Staphylococcus aureus to amicoumacin a. PLoS ONE 2012, 7, e34037. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhang, W.; Zhang, H.; Zhang, X.D.; Peng, B.; Zheng, J. Studies on aminoglycoside susceptibility identify a novel function of KsgA to secure translational fidelity during antibiotic stress. Antimicrob. Agents Chemother. 2018, 62, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Loughman, J.A.; Caparon, M.G. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 2006, 25, 5414–5422. [Google Scholar] [CrossRef]
- Loughman, J.A.; Caparon, M.G. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 2007, 64, 269–280. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, H.S.; Kim, D.J.; Yoon, H.-J.; Kim, K.H.; Yoon, J.Y.; Suh, S.W. Crystal structures of LacD from Staphylococcus aureus and LacD.1 from Streptococcus pyogenes: Insights into substrate specificity and virulence gene regulation. FEBS Lett. 2011, 585, 307–312. [Google Scholar] [CrossRef]
- Sheikh, S.; Britt, R.D.; Ryan-Wenger, N.A.; Khan, A.Q.; Lewis, B.W.; Gushue, C.; Ozuna, H.; Jaganathan, D.; McCoy, K.; Kopp, B.T. Impact of elexacaftor-tezacaftor-ivacaftor on bacterial colonization and inflammatory responses in cystic fibrosis. Pediatr. Pulmonol. 2023, 58, 825–833. [Google Scholar] [CrossRef]
- Brouillette, E.; Martinez, A.; Boyll, B.J.; Allen, N.E.; Malouin, F. Persistence of a Staphylococcus aureus small-colony variant under antibiotic pressure in vivo. FEMS Immunol. Med. Microbiol. 2004, 41, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Gilligan, P.H. Microbiology of airway disease in patients with cystic fibrosis. Clin. Microbiol. Rev. 1991, 4, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Hammerschlag, M.R.; Harding, L.; Macone, A.; Smith, A.L.; Goldmann, D.A. Bacteriology of sputum in cystic fibrosis: Evaluation of dithiothreitol as a mucolytic agent. J. Clin. Microbiol. 1980, 11, 552–557. Available online: http://www.ncbi.nlm.nih.gov/pubmed/6776135 (accessed on 1 May 2023). [CrossRef]
- Gibson, R.L.; Burns, J.L.; Ramsey, B.W. Pathophysiology and Management of Pulmonary Infections in Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2003, 168, 918–951. [Google Scholar] [CrossRef] [PubMed]
- Estevez, E.G. Bacteriologic plate media: Review of mechanisms of action. Lab. Med. 1984, 15, 258–262. [Google Scholar] [CrossRef]
- Murray, C. Manual of Clinical Microbiology, 7th ed.; American Society for Microbiology: Washington, DC, USA, 1999. [Google Scholar]
- Hwang, S.Y.; Kim, S.H.; Jang, E.J.; Kwon, N.H.; Park, Y.K.; Koo, H.C.; Jung, W.K.; Kim, J.M.; Park, Y.H. Novel multiplex PCR for the detection of the Staphylococcus aureus superantigen and its application to raw meat isolates in Korea. Int. J. Food Microbiol. 2007, 117, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Minamide, W.; Wada, K.; Nakamura, E.; Teraoka, H.; Watanabe, S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 1991, 29, 2240–2244. Available online: http://www.ncbi.nlm.nih.gov/pubmed/1939577 (accessed on 1 May 2023). [CrossRef] [PubMed]
- Sabat, A.; Krzyszton-Russjan, J.; Strzalka, W.; Filipek, R.; Kosowska, K.; Hryniewicz, W.; Travis, J.; Potempa, J. New Method for Typing Staphylococcus aureus Strains: Multiple-Locus Variable-Number Tandem Repeat Analysis of Polymorphism and Genetic Relationships of Clinical Isolates. J. Clin. Microbiol. 2003, 41, 1801–1804. [Google Scholar] [CrossRef]
- Gilot, P.; Lina, G.; Cochard, T.; Poutrel, B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 2002, 40, 4060–4067. [Google Scholar] [CrossRef]
- CLSI Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement; CLSI document M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Zhang, K.; McClure, J.-A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5026–5033. [Google Scholar] [CrossRef]
- Mitchell, G.; Lafrance, M.; Boulanger, S.; Séguin, D.L.; Guay, I.; Gattuso, M.; Marsault, É.; Bouarab, K.; Malouin, F. Tomatidine acts in synergy with aminoglycoside antibiotics against multiresistant Staphylococcus aureus and prevents virulence gene expression. J. Antimicrob. Chemother. 2012, 67, 559–568. [Google Scholar] [CrossRef]
- Boulanger, S.; Mitchell, G.; Bouarab, K.; Marsault, É.; Cantin, A.; Frost, E.H.; Déziel, E.; Malouin, F. Bactericidal effect of tomatidine-tobramycin combination against methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa is enhanced by interspecific small-molecule interactions. Antimicrob. Agents Chemother. 2015, 59, 7458–7464. [Google Scholar] [CrossRef]
| Patient ID | Visit | First Isolate | Variant | MLVA | agr | mecA | Antibiotic Susceptibility a | Aux b | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA | TOB | GEN | T-S | CIP | ERY | CLI | TET | VAN | RIF | ||||||||
| 1 | V1 | CF1A-L | A | 2 | S | S | S | S | S | S | S | S | S | S | |||
| V1 | CF1C-S | A | 2 | S | I | I | S | S | S | S | S | S | S | H | |||
| V1 | CF1D-S | A | 2 | nt | I | S | nt | S | S | R | S | S | S | U | |||
| V2 | CF37B-S | A | 2 | S | I | I | S | R | R | nt | S | S | S | U | |||
| V3 | CF54A-L * | A | 2 | S | S | S | S | S | S | nt | S | S | S | ||||
| V4 | CF54A-S * | A | 2 | S | R | R | S | S | S | nt | S | S | S | U | |||
| V2 | CF37A-L * | B | 2 | + | R | S | S | S | R | R | nt | S | S | S | |||
| V2 | CF37A-S * | B | 2 | + | R | S | S | S | R | R | nt | S | S | S | U | ||
| 2 | V1 | CF2A-L | C | 1 | S | S | S | S | S | S | S | S | S | S | |||
| V3 | CF62B-L | C | 1 | S | S | S | S | S | S | nt | S | S | S | ||||
| V1 | CF2C-L | D | 3 | S | S | S | S | S | S | S | S | S | S | ||||
| V1 | CF2B-S | D | 3 | S | I | S | S | S | S | S | S | S | S | H | |||
| V3 | CF62A-L | D | 3 | S | S | S | S | S | S | nt | S | S | S | ||||
| V2 | CF34A-L | E | 1 | S | S | S | S | S | S | nt | S | S | S | ||||
| V2 | CF34B-L | F | 3 | S | S | S | S | S | S | nt | S | S | S | ||||
| 3 | V1 | CF4B-L | A | 2 | S | S | S | S | S | S | S | S | S | S | |||
| V1 | CF4B-S | A | 2 | S | I | I | S | S | S | S | S | S | S | M | |||
| V2 | CF28B-L | A | 2 | S | S | S | S | S | S | nt | S | S | S | ||||
| V2 | CF28A-L * | G | 2 | S | S | S | S | S | R | nt | S | S | S | ||||
| V3 | CF63A-S | G | 2 | S | R | S | S | S | R | nt | S | S | S | U | |||
| V3 | CF63A-L * | G | 2 | S | R | I | S | R | R | nt | S | S | S | ||||
| 4 | V1 | CF5A-L * | H | 3 | S | S | S | S | S | S | S | S | S | S | |||
| V1 | CF5C-S * | H | 3 | nt | I | I | S | S | S | S | S | S | S | U | |||
| V1 | CF5E-S | I | 3 | S | I | I | S | S | S | S | S | S | S | H+M | |||
| 5 | V1 | CF6B-L * | J | 3 | S | R | R | S | S | S | S | S | S | S | |||
| V1 | CF6A-S * | J | 3 | S | R | R | R | S | S | S | S | S | S | T | |||
| V1 | CF6C-S * | J | 3 | S | R | R | S | S | S | S | S | S | S | H+M | |||
| V2 | CF39A-L * | J | 3 | S | R | R | R | S | S | nt | S | S | S | ||||
| V2 | CF39A-S * | J | 3 | S | R | R | R | S | I | nt | S | S | S | U | |||
| 6 | V1 | CF7A-L | K | 2 | + | R | R | S | S | R | R | R | S | S | S | ||
| V1 | CF7D-L | K | 2 | S | S | S | S | R | R | R | S | S | S | ||||
| V2 | CF27A-L | K | 2 | + | R | R | S | S | R | R | nt | S | S | S | |||
| V3 | CF49A-L | L | 3 | S | S | S | S | S | S | nt | S | S | S | ||||
| V3 | CF49A-S | M | 3 | S | I | S | S | S | S | nt | S | S | S | H | |||
| V3 | CF49B-S | M | 3 | S | I | S | S | S | S | nt | S | S | S | U | |||
| 7 | V1 | CF8A-L | N | 3 | S | S | S | S | S | S | S | S | S | S | |||
| V2 | CF21A-L * | N | 3 | S | S | S | S | S | S | nt | S | S | S | ||||
| V2 | CF21A-S * | N | 3 | S | S | S | S | S | S | nt | S | S | S | M | |||
| V1 | CF8C-L | O | 3 | S | S | S | S | S | S | S | S | S | S | ||||
| V1 | CF21D-L | O | 3 | S | S | S | S | S | S | nt | S | S | S | ||||
| V1 | CF8D-L | P | 3 | S | S | S | S | S | S | S | S | S | S | ||||
| V3 | CF48A-L * | K | 2 | + | R | R | S | S | R | R | nt | S | S | S | |||
| V3 | CF48A-S * | K | 2 | + | R | R | S | S | R | R | nt | S | S | S | U | ||
| V3 | CF48B-S * | K | 2 | S | R | I | S | R | R | nt | S | I | S | U | |||
| 8 | V1 | CF9A-L | K | 2 | + | R | R | S | S | R | R | R c | S | S | S | ||
| V1 | CF9F-L | K | 2 | S | R | R | S | R | R | I | S | S | S | ||||
| V2 | CF51A-L * | P | 2 | S | S | S | S | R | R | nt | S | S | S | ||||
| V2 | CF51B-S * | P | 2 | S | I | I | S | R | R | nt | S | I | S | U | |||
| V3 | CF75B-L | P | 2 | S | S | S | S | R | R | nt | S | S | S | ||||
| V3 | CF75A-L | Q | 1 | S | S | S | S | R | S | nt | S | S | S | ||||
| 9 | V1 | CF10B-L * | R | 1 | S | S | S | S | S | R | R | S | S | S | |||
| V1 | CF10A-S * | R | 1 | S | R | R | S | S | R | R | S | S | S | H + M | |||
| 10 | V1 | CF18A-L | S | 2 | S | S | S | S | S | R | R c | S | S | S | |||
| V2 | CF33A-L | S | 2 | S | S | S | S | I | R | nt | S | S | S | ||||
| V3 | CF78A-L | S | 2 | S | I | S | S | S | S | nt | S | S | S | ||||
| V3 | CF78A-S | S | 2 | S | I | S | S | S | S | nt | S | S | S | M | |||
| V3 | CF78B-S | S | 2 | S | I | S | S | S | S | nt | S | S | S | H | |||
| V1 | CF18E-L | T | 1 | S | S | S | S | S | S | S | S | S | S | ||||
| 11 | V1 | CF19A-L | T | 1 | S | S | S | S | R | S | S | S | S | S | |||
| V1 | CF19B-L | U | 1 | S | S | S | S | R | S | S | S | S | S | ||||
| V1 | CF19A-S | V | 1 | S | R | R | S | S | S | S | S | S | S | U | |||
| V2 | CF41A-L | W | 1 | S | S | S | S | R | R | nt | S | S | S | ||||
| V2 | CF41A-S | W | 1 | S | I | S | S | R | S | nt | S | S | S | U | |||
| V3 | CF58A-L | X | 1 | S | R | R | R | S | S | nt | S | S | S | ||||
| V3 | CF58B-L | Y | 1 | S | I | S | S | R | S | nt | S | S | S | ||||
| 12 | V1 | CF22A-L | Z | 1 | S | S | S | S | S | S | S | S | S | S | |||
| 13 | V1 | CF29A-L | AA | 1 | S | I | I | S | S | S | R | S | S | S | |||
| 14 | V1 | CF35A-L | K | 2 | + | R | S | S | S | R | R | R c | S | S | S | ||
| V2 | CF77A-L | K | 2 | + | R | S | S | S | R | R | nt | S | S | S | |||
| 15 | V1 | CF43A-L | BB | 3 | S | S | S | S | R | R | R | S | S | S | |||
| V1 | CF43B-L | CC | 3 | S | S | S | S | R | R | R | S | S | S | ||||
| 16 | V1 | CF50A-L | K | 2 | + | R | R | S | S | R | R | S | S | S | S | ||
| CF50A-S * | K | 2 | + | R | R | I | S | R | R | S | S | S | S | H | |||
| V2 | CF111A-L | DD | 2 | + | R | R | S | S | R | R | nt | S | S | S | |||
| 17 | V2 | CF86A-S | EE | 2 | S | S | S | S | S | S | nt | R | S | S | H | ||
| 18 | V1 | CF81A-L | FF | 3 | S | S | S | S | S | S | S | S | S | S | |||
| V2 | CF91A-L | GG | 1 | S | S | S | S | S | S | nt | S | I | S | ||||
| Strains | Patient | Visit | MLVA | mecA | MIC (µg/mL) a | AMEs b | |||
|---|---|---|---|---|---|---|---|---|---|
| GEN | TOB | AAC | ANT4 | ANT9 | |||||
| CF1A-L | 1 | V1 | A | 0.25–0.5 | 0.5–1 | - | - | - | |
| CF54A-L | 1 | V3 | A | 2–4 | 2–4 | - | - | - | |
| CF54A-S | 1 | V4 | A | >32 | >32 | - | - | - | |
| CF2C-L | 2 | V1 | D | 2 | 2 | - | - | - | |
| CF2B-S | 2 | V1 | D | 4 | 4–8 | - | - | - | |
| CF4B-L | 3 | V1 | A | 1 | 1 | - | - | - | |
| CF4B-S | 3 | V1 | A | 4–8 | 4–8 | - | - | - | |
| CF28A-L | 3 | V2 | G | 0.5 | 0.5 | - | - | - | |
| CF63A-S | 3 | V3 | G | 4 | >32 | - | - | - | |
| CF63A-L | 3 | V3 | G | 4–8 | >32 | - | - | - | |
| CF5A-L | 4 | V1 | H | 0.25 | 0.5 | - | - | - | |
| CF5C-S | 4 | V1 | H | 8 | 4–8 | - | - | - | |
| CF7A-L | 6 | V1 | K | + | 0.5 | >32 | + | + | - |
| CF27A-L | 6 | V2 | K | + | 2–4 | >32 | + | + | - |
| CF48A-L | 7 | V3 | K | + | 1 | >32 | - | - | + |
| CF48A-S | 7 | V3 | K | + | 4 | >32 | - | - | + |
| CF51A-L | 8 | V2 | P | 2–4 | 2–4 | - | - | - | |
| CF51B-S | 8 | V2 | P | 8–16 | 4–8 | - | - | - | |
| CF10B-L | 9 | V1 | R | 0.5–1 | 0.5–1 | - | - | - | |
| CF10A-S | 9 | V1 | R | >32 | >32 | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millette, G.; Séguin, D.L.; Isabelle, C.; Chamberland, S.; Lucier, J.-F.; Rodrigue, S.; Cantin, A.M.; Malouin, F. Staphylococcus aureus Small-Colony Variants from Airways of Adult Cystic Fibrosis Patients as Precursors of Adaptive Antibiotic-Resistant Mutations. Antibiotics 2023, 12, 1069. https://doi.org/10.3390/antibiotics12061069
Millette G, Séguin DL, Isabelle C, Chamberland S, Lucier J-F, Rodrigue S, Cantin AM, Malouin F. Staphylococcus aureus Small-Colony Variants from Airways of Adult Cystic Fibrosis Patients as Precursors of Adaptive Antibiotic-Resistant Mutations. Antibiotics. 2023; 12(6):1069. https://doi.org/10.3390/antibiotics12061069
Chicago/Turabian StyleMillette, Guillaume, David Lalonde Séguin, Charles Isabelle, Suzanne Chamberland, Jean-François Lucier, Sébastien Rodrigue, André M. Cantin, and François Malouin. 2023. "Staphylococcus aureus Small-Colony Variants from Airways of Adult Cystic Fibrosis Patients as Precursors of Adaptive Antibiotic-Resistant Mutations" Antibiotics 12, no. 6: 1069. https://doi.org/10.3390/antibiotics12061069
APA StyleMillette, G., Séguin, D. L., Isabelle, C., Chamberland, S., Lucier, J.-F., Rodrigue, S., Cantin, A. M., & Malouin, F. (2023). Staphylococcus aureus Small-Colony Variants from Airways of Adult Cystic Fibrosis Patients as Precursors of Adaptive Antibiotic-Resistant Mutations. Antibiotics, 12(6), 1069. https://doi.org/10.3390/antibiotics12061069







