Optimization of Empirical Antimicrobial Therapy in Enterobacterales Bloodstream Infection Using the Extended-Spectrum Beta-Lactamase Prediction Score
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics and Microbiology
2.2. Time to Appropriate Antimicrobial Therapy
3. Discussion
3.1. Clinical Interpretation of Study Findings
3.2. Potential Applications of the ESBL Prediction Score
3.3. Clinical Prediction Tools versus Advanced Rapid Diagnostics
3.4. Strengths and Limitations
4. Methods
4.1. Setting
4.2. Case Ascertainment
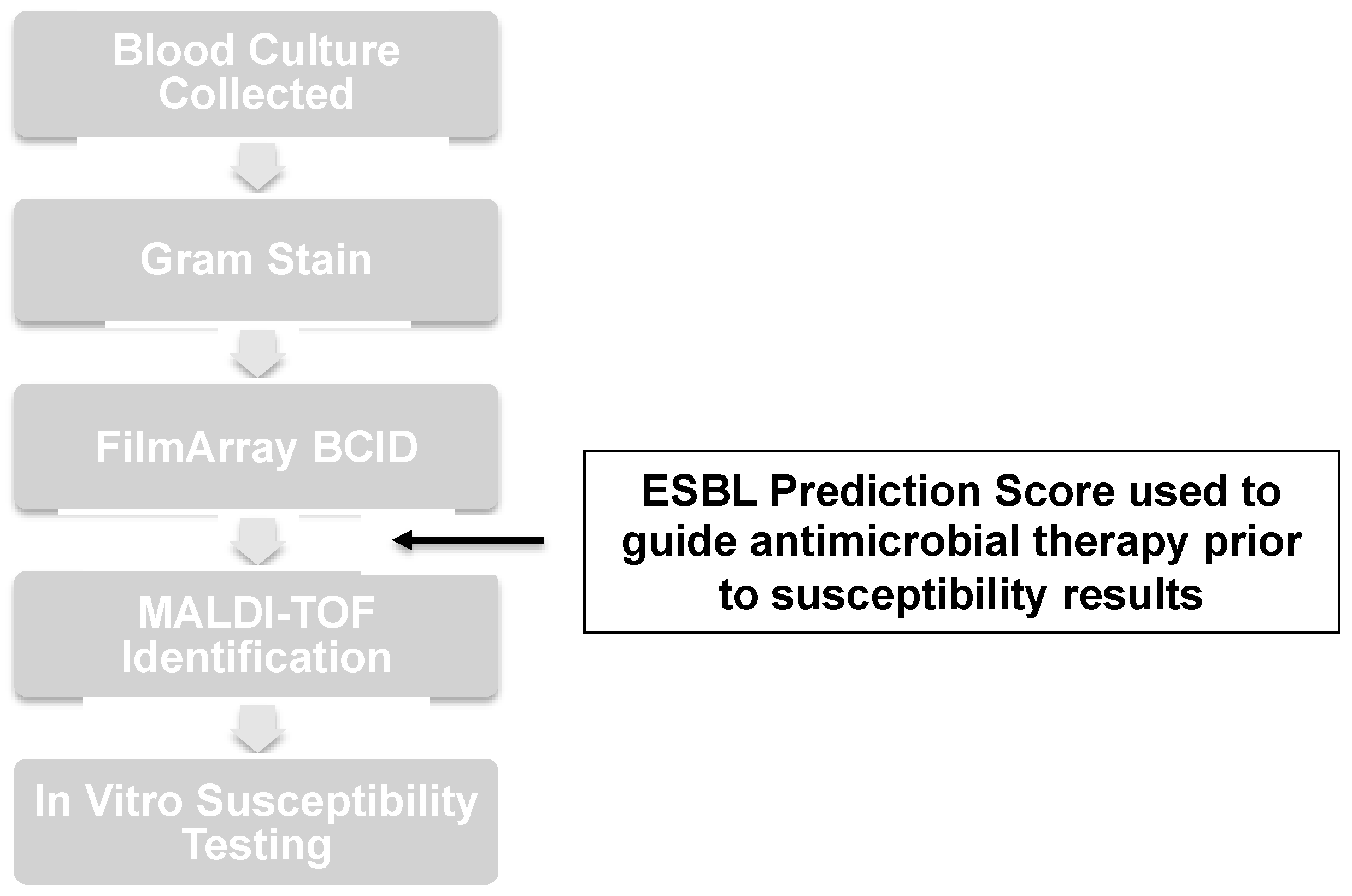
4.3. Microbiology Techniques
4.4. Antimicrobial Stewardship Intervention
4.5. Outcomes and Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Goto, M.; Al-Hasan, M.N. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin. Microbiol. Infect. 2013, 19, 501–509. [Google Scholar] [CrossRef]
- Jacoby, G.A. Extended-spectrum beta-lactamases and other enzymes providing resistance to oxyimino-beta-lactams. Infect. Dis. Clin. N. Am. 1997, 11, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 3 May 2023).
- Kern, W.V.; Rieg, S. Burden of bacterial bloodstream infection-a brief update on epidemiology and significance of multidrug-resistant pathogens. Clin. Microbiol. Infect. 2020, 26, 151–157. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Thaden, J.T.; Fowler, V.G.; Sexton, D.J.; Anderson, D.J. Increasing Incidence of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Community Hospitals throughout the Southeastern United States. Infect. Control. Hosp. Epidemiol. 2016, 37, 49–54. [Google Scholar] [CrossRef] [PubMed]
- McDanel, J.; Schweizer, M.; Crabb, V.; Nelson, R.; Samore, M.; Khader, K.; Blevins, A.E.; Diekema, D.; Chiang, H.-Y.; Nair, R.; et al. Incidence of Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli and Klebsiella Infections in the United States: A Systematic Literature Review. Infect. Control. Hosp. Epidemiol. 2017, 38, 1209–1215. [Google Scholar] [CrossRef]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America 2022 Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2022, 75, 187–212. [Google Scholar] [CrossRef]
- Harris, P.N.A.; Tambyah, P.A.; Lye, D.C.; Mo, Y.; Lee, T.H.; Yilmaz, M.; Alenazi, T.H.; Arabi, Y.; Falcone, M.; Bassetti, M.; et al. Effect of Piperacillin-Tazobactam vs Meropenem on 30-Day Mortality for Patients With E coli or Klebsiella pneumoniae Bloodstream Infection and Ceftriaxone Resistance: A Randomized Clinical Trial. JAMA 2018, 320, 984–994. [Google Scholar] [CrossRef]
- Cain, S.E.; Kohn, J.; Bookstaver, P.B.; Albrecht, H.; Al-Hasan, M.N. Stratification of the impact of inappropriate empirical antimicrobial therapy for Gram-negative bloodstream infections by predicted prognosis. Antimicrob. Agents Chemother. 2015, 59, 245–250. [Google Scholar] [CrossRef]
- Battle, S.E.; Bookstaver, P.B.; Justo, J.A.; Kohn, J.; Albrecht, H.; Al-Hasan, M.N. Association between inappropriate empirical antimicrobial therapy and hospital length of stay in Gram-negative bloodstream infections: Stratification by prognosis. J. Antimicrob. Chemother. 2017, 72, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Madrid-Morales, J.; Sharma, A.; Reveles, K.; Velez-Mejia, C.; Hopkins, T.; Yang, L.; Walter, E.; Cadena, J. Validation of Available Extended-Spectrum-Beta-Lactamase Clinical Scoring Models in Predicting Drug Resistance in Patients with Enteric Gram-Negative Bacteremia Treated at South Texas Veterans Health Care System. Antimicrob. Agents Chemother. 2021, 65, e02562-20. [Google Scholar] [CrossRef]
- Augustine, M.R.; Testerman, T.L.; Justo, J.A.; Bookstaver, P.B.; Kohn, J.; Albrecht, H.; Al-Hasan, M.N. Clinical Risk Score for Prediction of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Bloodstream Isolates. Infect. Control. Hosp. Epidemiol. 2017, 38, 266–272. [Google Scholar] [CrossRef]
- Kumar, A. Early Antimicrobial Therapy in Severe Sepsis and Septic Shock. Curr. Infect. Dis. Rep. 2010, 12, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Bookstaver, P.B.; Nimmich, E.B.; Smith, T.J.; Justo, J.A.; Kohn, J.; Hammer, K.L.; Troficanto, C.; Albrecht, H.A.; Al-Hasan, M.N. Cumulative Effect of an Antimicrobial Stewardship and Rapid Diagnostic Testing Bundle on Early Streamlining of Antimicrobial Therapy in Gram-Negative Bloodstream Infections. Antimicrob. Agents Chemother. 2017, 61, e00189-17. [Google Scholar] [CrossRef]
- Cortazzo, V.; D’inzeo, T.; Giordano, L.; Menchinelli, G.; Liotti, F.M.; Fiori, B.; De Maio, F.; Luzzaro, F.; Sanguinetti, M.; Posteraro, B.; et al. Comparing BioFire FilmArray BCID2 and BCID Panels for Direct Detection of Bacterial Pathogens and Antimicrobial Resistance Genes from Positive Blood Cultures. J. Clin. Microbiol. 2021, 59, e03163-20. [Google Scholar] [CrossRef]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef]
- Chen, L.F.; Freeman, J.T.; Nicholson, B.; Keiger, A.; Lancaster, S.; Joyce, M.; Woods, C.W.; Cook, E.; Adcock, L.; Louis, S.; et al. Widespread dissemination of CTX-M-15 genotype extended-spectrum-β-lactamase-producing enterobacteriaceae among patients presenting to community hospitals in the southeastern United States. Antimicrob. Agents Chemother. 2014, 58, 1200–1202. [Google Scholar] [CrossRef] [PubMed]
- Sparks, R.; Balgahom, R.; Janto, C.; Polkinghorne, A.; Branley, J. Evaluation of the BioFire Blood Culture Identification 2 panel and impact on patient management and antimicrobial stewardship. Pathology 2021, 53, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Sze, D.T.T.; Lau, C.C.Y.; Chan, T.M.; Ma, E.S.K.; Tang, B.S.F. Comparison of novel rapid diagnostic of blood culture identification and antimicrobial susceptibility testing by Accelerate Pheno system and BioFire FilmArray Blood Culture Identification and BioFire FilmArray Blood Culture Identification 2 panels. BMC Microbiol. 2021, 21, 350. [Google Scholar] [CrossRef]
- Comini, S.; Bianco, G.; Boattini, M.; Banche, G.; Ricciardelli, G.; Allizond, V.; Cavallo, R.; Costa, C. Evaluation of a diagnostic algorithm for rapid identification of Gram-negative species and detection of extended-spectrum β-lactamase and carbapenemase directly from blood cultures. J. Antimicrob. Chemother. 2022, 77, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- Boattini, M.; Bianco, G.; Comini, S.; Iannaccone, M.; Casale, R.; Cavallo, R.; Nordmann, P.; Costa, C. Direct detection of extended-spectrum-β-lactamase-producers in Enterobacterales from blood cultures: A comparative analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Cerrudo, V.; Cortés-Cuevas, J.L.; García-Fernández, S.; Morosini, M.I.; Cantón, R.; Sánchez-Díaz, A.M. Usefulness of EUCAST rapid antibiotic susceptibility breakpoints and screening cut-off values directly from blood cultures for the inference of β-lactam resistance mechanisms in Enterobacterales. JAC Antimicrob. Resist. 2023, 5, dlad017. [Google Scholar] [CrossRef] [PubMed]


| Variable | Total (n = 146) | Pre-Intervention (n = 45) | Post-Intervention (n = 101) | p-Value |
|---|---|---|---|---|
| Median age (IQR) in years | 66 (51–75) | 68 (50–76) | 63 (51–75) | 0.27 |
| Male sex | 85 (58) | 24 (53) | 61 (60) | 0.42 |
| Hospital-onset BSI | 37 (25) | 8 (18) | 29 (29) | 0.16 |
| Residence at SNF | 33 (23) | 12 (27) | 21 (21) | 0.43 |
| Urinary source of BSI | 86 (59) | 26 (58) | 60 (59) | 0.85 |
| Diabetes mellitus | 63 (43) | 17 (38) | 46 (46) | 0.38 |
| End-stage renal disease | 13 (9) | 5 (11) | 8 (8) | 0.53 |
| Liver cirrhosis | 4 (3) | 3 (7) | 1 (1) | 0.09 |
| Cancer | 24 (16) | 9 (20) | 15 (15) | 0.44 |
| Immune compromised | 18 (12) | 4 (9) | 14 (14) | 0.40 |
| ICU admission | 43 (29) | 14 (31) | 29 (29) | 0.77 |
| Antibiotic | Pre-Intervention Period (n = 45) | Post-Intervention Period (n = 101) | ||
|---|---|---|---|---|
| Before Gram Stain | After Gram Stain | Before Gram Stain | After Gram Stain | |
| Ampicillin/sulbactam | 0 (0) | 0 (0) | 3 (3) | 0 (0) |
| Ceftriaxone | 9 (20) | 8 (18) | 28 (28) | 5 (5) |
| Cefepime | 4 (9) | 3 (7) | 29 (29) | 14 (14) |
| Piperacillin/tazobactam | 14 (31) | 12 (27) | 26 (26) | 12 (12) |
| Ertapenem | 3 (7) | 7 (16) | 7 (7) | 52 (52) |
| Meropenem | 8 (18) | 9 (20) | 6 (6) | 14 (14) |
| Fluoroquinolones * | 6 (13) | 3 (7) | 1 (1) | 4 (4) |
| Gentamicin | 1 (2) | 3 (7) | 1 (1) | 0 (0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haimerl, B.J.; Encinas, R.; Justo, J.A.; Kohn, J.; Bookstaver, P.B.; Winders, H.R.; Al-Hasan, M.N. Optimization of Empirical Antimicrobial Therapy in Enterobacterales Bloodstream Infection Using the Extended-Spectrum Beta-Lactamase Prediction Score. Antibiotics 2023, 12, 1003. https://doi.org/10.3390/antibiotics12061003
Haimerl BJ, Encinas R, Justo JA, Kohn J, Bookstaver PB, Winders HR, Al-Hasan MN. Optimization of Empirical Antimicrobial Therapy in Enterobacterales Bloodstream Infection Using the Extended-Spectrum Beta-Lactamase Prediction Score. Antibiotics. 2023; 12(6):1003. https://doi.org/10.3390/antibiotics12061003
Chicago/Turabian StyleHaimerl, Brian J., Rodrigo Encinas, Julie Ann Justo, Joseph Kohn, P. Brandon Bookstaver, Hana Rac Winders, and Majdi N. Al-Hasan. 2023. "Optimization of Empirical Antimicrobial Therapy in Enterobacterales Bloodstream Infection Using the Extended-Spectrum Beta-Lactamase Prediction Score" Antibiotics 12, no. 6: 1003. https://doi.org/10.3390/antibiotics12061003
APA StyleHaimerl, B. J., Encinas, R., Justo, J. A., Kohn, J., Bookstaver, P. B., Winders, H. R., & Al-Hasan, M. N. (2023). Optimization of Empirical Antimicrobial Therapy in Enterobacterales Bloodstream Infection Using the Extended-Spectrum Beta-Lactamase Prediction Score. Antibiotics, 12(6), 1003. https://doi.org/10.3390/antibiotics12061003









