Performance of a Pilot-Scale Continuous Flow Ozone-Based Hospital Wastewater Treatment System
Abstract
1. Introduction
2. Materials and Methods
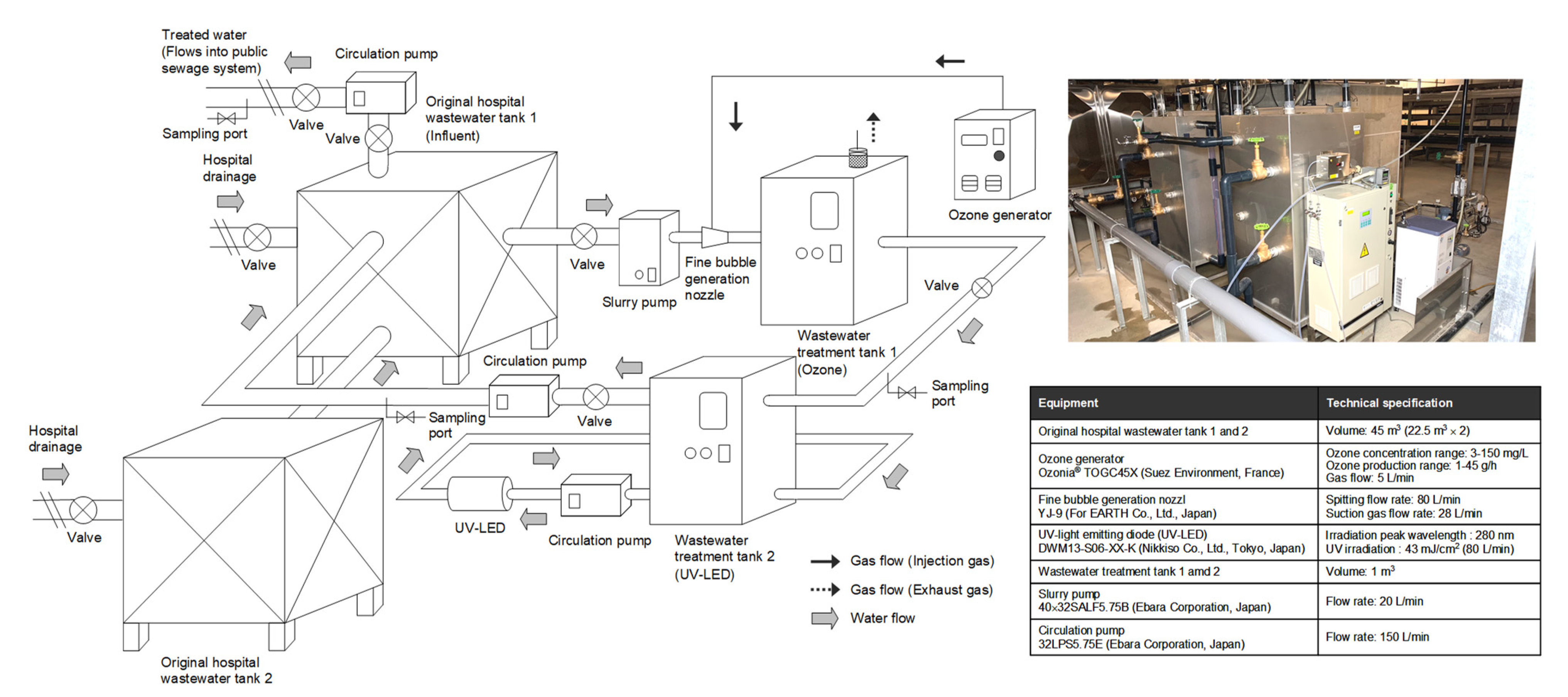
2.1. Hospital Wastewater Treatment Using Ozone Treatment Based on the Continuous Flow System
2.2. Viable Bacterial Counting of Wastewater Samples
2.3. Metagenomic DNA-Seq Analysis of Wastewater Samples
2.4. Analytical Procedures for Antimicrobials
3. Results
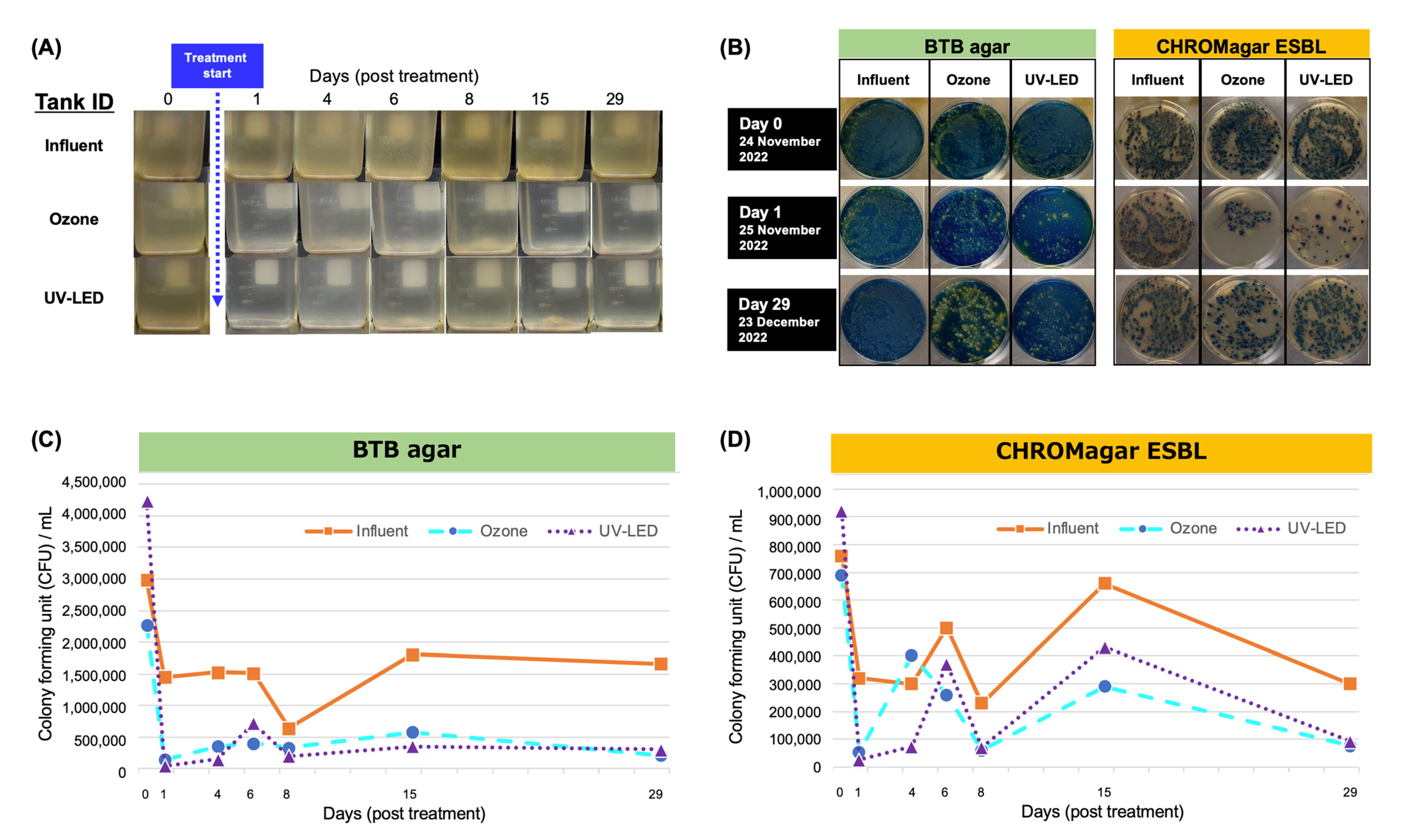
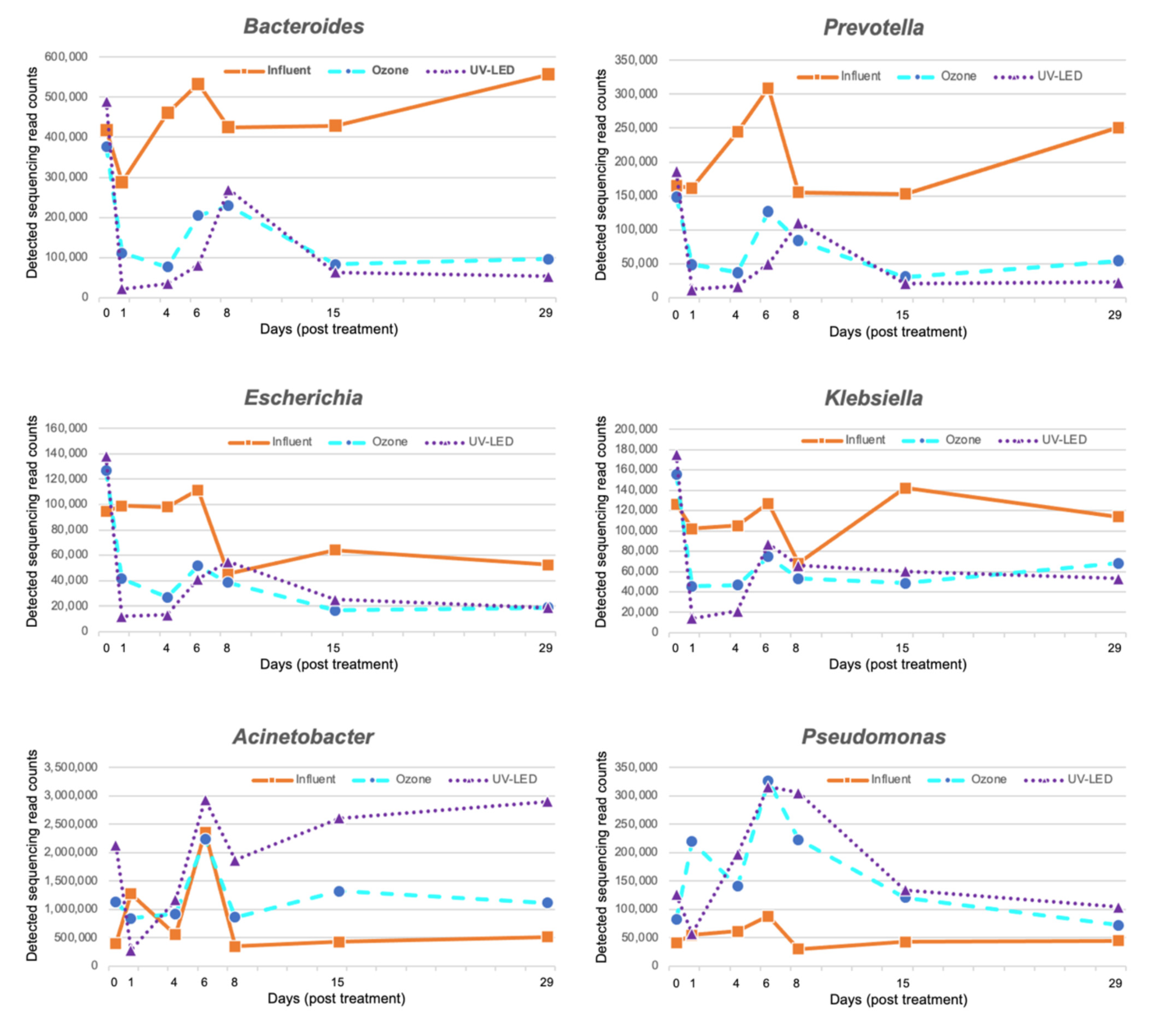
3.1. Proportion of Bacteria in Hospital Wastewater after Ozone Treatment
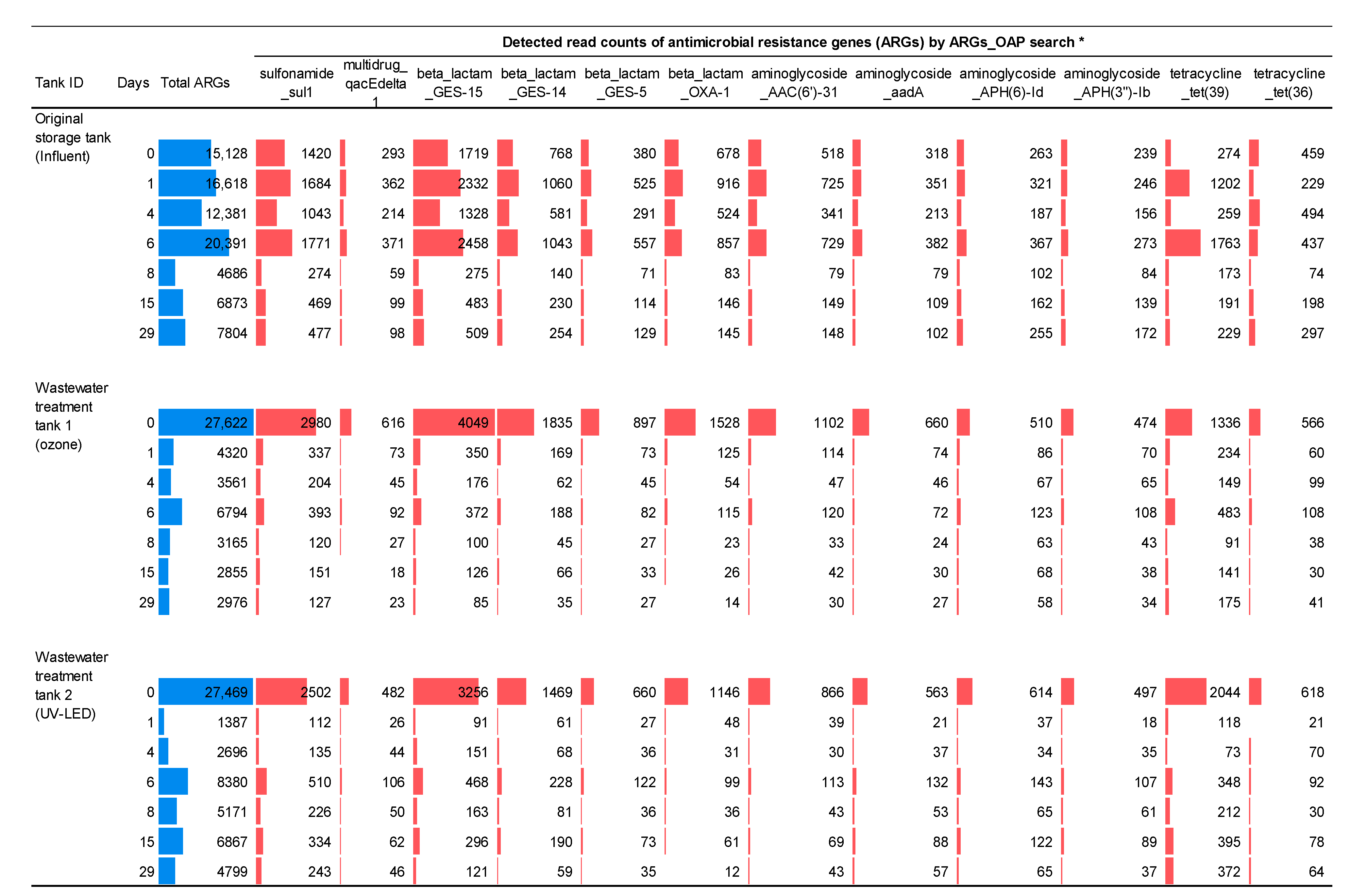
3.2. Resistome Analysis in Hospital Wastewater Subjected to Ozone Treatment
3.3. Removal of Antimicrobials by Ozone Treatment
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, S.; Zhang, C.; Li, F.; Hua, T.; Zhou, Q.; Ho, S.H. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: A critical review. J. Hazard. Mater. 2021, 411, 125148. [Google Scholar] [CrossRef] [PubMed]
- Baba, H.; Nishiyama, M.; Watanabe, T.; Kanamori, H. Review of antimicrobial resistance in wastewater in Japan: Current challenges and future perspectives. Antibiotics 2022, 11, 849. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, Y.; Li, L.; Liu, J.; Yan, X. Distribution, sources, and potential risks of antibiotic resistance genes in wastewater treatment plant: A review. Environ. Pollut. 2022, 310, 119870. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; pp. 1–232. [Google Scholar]
- US Department of Health and Human Services. Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States. 2019; p. 1. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 18 February 2023).
- Jim, O.N. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014, 1–16. [Google Scholar]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; pp. 1–19. [Google Scholar]
- National Action Plan on Antimicrobial Resistance (AMR) (2016–2020); The Government of Japan: Tokyo, Japan, 2016; pp. 1–69.
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems–A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Ezeuko, A.S.; Ojemaye, M.O.; Okoh, O.O.; Okoh, A.I. Technological advancement for eliminating antibiotic resistance genes from wastewater: A review of their mechanisms and progress. J. Environ. Chem. Eng. 2021, 9, 106183. [Google Scholar] [CrossRef]
- Cai, M.; Wang, Z.; Gu, H.; Dong, H.; Zhang, X.; Cui, N.; Zhou, L.; Chen, G.; Zou, G. Occurrence and temporal variation of antibiotics and antibiotic resistance genes in hospital inpatient department wastewater: Impacts of daily schedule of inpatients and wastewater treatment process. Chemosphere 2022, 292, 133405. [Google Scholar] [CrossRef]
- Pariente, M.I.; Segura, Y.; Álvarez-Torrellas, S.; Casas, J.A.; de Pedro, Z.M.; Diaz, E.; García, J.; López-Muñoz, M.J.; Marugán, J.; Mohedano, A.F.; et al. Critical review of technologies for the on-site treatment of hospital wastewater: From conventional to combined advanced processes. J. Environ. Manag. 2022, 320, 115769. [Google Scholar] [CrossRef]
- Ulvi, A.; Aydın, S.; Aydın, M.E. Fate of selected pharmaceuticals in hospital and municipal wastewater effluent: Occurrence, removal, and environmental risk assessment. Environ. Sci. Pollut. Res. 2022, 29, 75609–75625. [Google Scholar] [CrossRef]
- Cahill, N.; O’Connor, L.; Mahon, B.; Varley, Á.; McGrath, E.; Ryan, P.; Cormican, M.; Brehony, C.; Jolley, K.A.; Maiden, M.C.; et al. Hospital effluent: A reservoir for carbapenemase-producing Enterobacterales? Sci. Total Environ. 2019, 672, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Al Salah, D.M.M.; Ngweme, G.N.; Laffite, A.; Otamonga, J.-P.; Mulaji, C.; Poté, J. Hospital wastewaters: A reservoir and source of clinically relevant bacteria and antibiotic resistant genes dissemination in urban river under tropical conditions. Ecotoxicol. Environ. Saf. 2020, 200, 110767. [Google Scholar] [CrossRef] [PubMed]
- Gwenzi, W.; Musiyiwa, K.; Mangori, L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: A hotspot reservoir. J. Environ. Chem. Eng. 2020, 8, 102220. [Google Scholar] [CrossRef]
- Sekizuka, T.; Itokawa, K.; Tanaka, R.; Hashino, M.; Yatsu, K.; Kuroda, M. Metagenomic analysis of urban wastewater treatment plant effluents in tokyo. Infect. Drug Resist. 2022, 15, 4763–4777. [Google Scholar] [CrossRef] [PubMed]
- Sekizuka, T.; Tanaka, R.; Hashino, M.; Yatsu, K.; Kuroda, M. Comprehensive genome and plasmidome analysis of antimicrobial resistant bacteria in wastewater treatment plant effluent of Tokyo. Antibiotics 2022, 11, 1283. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Ye, J.; Yang, Q.; Hu, Y.; Zhang, T.; Jiang, L.; Munezero, S.; Lin, K.; Cui, C. Occurrence and removal of antibiotics, antibiotic resistance genes, and bacterial communities in hospital wastewater. Environ. Sci. Pollut. Res. 2021, 28, 57321–57333. [Google Scholar] [CrossRef]
- Azuma, T.; Katagiri, M.; Sekizuka, T.; Kuroda, M.; Watanabe, M. Inactivation of bacteria and residual antimicrobials in hospital wastewater by ozone treatment. Antibiotics 2022, 11, 862. [Google Scholar] [CrossRef]
- Weissbrodt, D.; Kovalova, L.; Ort, C.; Pazhepurackel, V.; Moser, R.; Hollender, J.; Siegrist, H.; McArdell, C.S. Mass flows of X-ray contrast media and cytostatics in hospital wastewater. Environ. Sci. Technol. 2009, 43, 4810–4817. [Google Scholar] [CrossRef]
- Santos, L.H.M.L.M.; Gros, M.; Rodriguez-Mozaz, S.; Delerue-Matos, C.; Pena, A.; Barceló, D.; Montenegro, M.C.B.S.M. Contribution of hospital effluents to the load of pharmaceuticals in urban wastewaters: Identification of ecologically relevant pharmaceuticals. Sci. Total Environ. 2013, 461–462, 302–316. [Google Scholar] [CrossRef]
- Aydin, S.; Aydin, M.E.; Ulvi, A.; Kilic, H. Antibiotics in hospital effluents: Occurrence, contribution to urban wastewater, removal in a wastewater treatment plant, and environmental risk assessment. Environ. Sci. Pollut. Res. 2019, 26, 544–558. [Google Scholar] [CrossRef]
- Azuma, T.; Otomo, K.; Kunitou, M.; Shimizu, M.; Hosomaru, K.; Mikata, S.; Ishida, M.; Hisamatsu, K.; Yunoki, A.; Mino, Y.; et al. ; et al. Environmental fate of pharmaceutical compounds and antimicrobial-resistant bacteria in hospital effluents, and contributions to pollutant loads in the surface waters in Japan. Sci. Total Environ. 2019, 657, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Afsa, S.; Hamden, K.; Lara Martin, P.A.; Mansour, H.B. Occurrence of 40 pharmaceutically active compounds in hospital and urban wastewaters and their contribution to mahdia coastal seawater contamination. Environ. Sci. Pollut. Res. 2020, 27, 1941–1955. [Google Scholar] [CrossRef] [PubMed]
- Harbarth, S.; Balkhy, H.H.; Goossens, H.; Jarlier, V.; Kluytmans, J.; Laxminarayan, R.; Saam, M.; Van Belkum, A.; Pittet, D. Antimicrobial resistance: One world, one fight! Antimicrob. Resist. Infect. Control 2015, 4, 49. [Google Scholar] [CrossRef]
- Noman, E.; Al-Gheethi, A.; Radin Mohamed, R.M.S.; Talip, B.; Al-Sahari, M.; Al-Shaibani, M. Quantitative microbiological risk assessment of complex microbial community in prawn farm wastewater and applicability of nanoparticles and probiotics for eliminating of antibiotic-resistant bacteria. J. Hazard. Mater. 2021, 419, 126418. [Google Scholar] [CrossRef] [PubMed]
- Schoen, M.E.; Jahne, M.A.; Garland, J.; Ramirez, L.; Lopatkin, A.J.; Hamilton, K.A. Quantitative microbial risk assessment of antimicrobial resistant and susceptible Staphylococcus aureus in reclaimed wastewaters. Environ. Sci. Technol. 2021, 55, 15246–15255. [Google Scholar] [CrossRef] [PubMed]
- Zainab, S.M.; Junaid, M.; Xu, N.; Malik, R.N. Antibiotics and antibiotic resistant genes (ATGs) in groundwater: A global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020, 187, 116455. [Google Scholar] [CrossRef]
- Anand, U.; Reddy, B.; Singh, V.K.; Singh, A.K.; Kesari, K.K.; Tripathi, P.; Kumar, P.; Tripathi, V.; Simal-Gandara, J. Potential environmental and human health risks caused by antibiotic-resistant bacteria (ARB), antibiotic resistance genes (ARGs) and emerging contaminants (ECS) from municipal solid waste (MSW) landfill. Antibiotics 2021, 10, 374. [Google Scholar] [CrossRef]
- Booton, R.D.; Meeyai, A.; Alhusein, N.; Buller, H.; Feil, E.; Lambert, H.; Mongkolsuk, S.; Pitchforth, E.; Reyher, K.K.; Sakcamduang, W.; et al. ; et al. One health drivers of antibacterial resistance: Quantifying the relative impacts of human, animal and environmental use and transmission. One Health 2021, 12, 100220. [Google Scholar] [CrossRef]
- González-Plaza, J.J.; Blau, K.; Milaković, M.; Jurina, T.; Smalla, K.; Udiković-Kolić, N. Antibiotic-manufacturing sites are hot-spots for the release and spread of antibiotic resistance genes and mobile genetic elements in receiving aquatic environments. Environ. Int. 2019, 130, 104735. [Google Scholar] [CrossRef]
- Menz, J.; Olsson, O.; Kümmerer, K. Antibiotic residues in livestock manure: Does the eu risk assessment sufficiently protect against microbial toxicity and selection of resistant bacteria in the environment? J. Hazard. Mater. 2019, 379, 120807. [Google Scholar] [CrossRef]
- Hossain, A.; Habibullah-Al-Mamun, M.; Nagano, I.; Masunaga, S.; Kitazawa, D.; Matsuda, H. Antibiotics, antibiotic-resistant bacteria, and resistance genes in aquaculture: Risks, current concern, and future thinking. Environ. Sci. Pollut. Res. 2022, 29, 11054–11075. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhou, P.; Shimabuku, K.K.; Fang, X.; Li, S.; Lee, Y.; Dodd, M.C. Degradation and deactivation of bacterial antibiotic resistance genes during exposure to free chlorine, monochloramine, chlorine dioxide, ozone, ultraviolet light, and hydroxyl radical. Environ. Sci. Technol. 2019, 53, 2013–2026. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A. Hospital Wastewater Treatment: Global Scenario and Case Studies; IWA Publishing: London, UK, 2022. [Google Scholar]
- Parida, V.K.; Sikarwar, D.; Majumder, A.; Gupta, A.K. An assessment of hospital wastewater and biomedical waste generation, existing legislations, risk assessment, treatment processes, and scenario during COVID-19. J. Environ. Manag. 2022, 308, 114609. [Google Scholar] [CrossRef] [PubMed]
- Anthony, E.T.; Ojemaye, M.O.; Okoh, O.O.; Okoh, A.I. A critical review on the occurrence of resistomes in the environment and their removal from wastewater using apposite treatment technologies: Limitations, successes and future improvement. Environ. Pollut. 2020, 263, 113791. [Google Scholar] [CrossRef]
- Mahaney, A.P.; Franklin, R.B. Persistence of wastewater-associated antibiotic resistant bacteria in river microcosms. Sci. Total Environ. 2022, 819, 153099. [Google Scholar] [CrossRef]
- Ahmed, S.; Khan, F.S.A.; Mubarak, N.M.; Khalid, M.; Tan, Y.H.; Mazari, S.A.; Karri, R.R.; Abdullah, E.C. Emerging pollutants and their removal using visible-light responsive photocatalysis– A comprehensive review. J. Environ. Chem. Eng. 2021, 9, 106643. [Google Scholar] [CrossRef]
- Ao, X.; Eloranta, J.; Huang, C.H.; Santoro, D.; Sun, W.; Lu, Z.; Li, C. Peracetic acid-based advanced oxidation processes for decontamination and disinfection of water: A review. Water Res. 2021, 188, 116479. [Google Scholar] [CrossRef]
- Divyapriya, G.; Singh, S.; Martínez-Huitle, C.A.; Scaria, J.; Karim, A.V.; Nidheesh, P.V. Treatment of real wastewater by photoelectrochemical methods: An overview. Chemosphere 2021, 276, 130188. [Google Scholar] [CrossRef]
- Asghar, A.; Lutze, H.V.; Tuerk, J.; Schmidt, T.C. Influence of water matrix on the degradation of organic micropollutants by ozone based processes: A review on oxidant scavenging mechanism. J. Hazard. Mater. 2022, 429, 128189. [Google Scholar] [CrossRef]
- Zhou, S.; Marcelino, K.R.; Wongkiew, S.; Sun, L.; Guo, W.; Khanal, S.K.; Lu, H. Untapped potential: Applying microbubble and nanobubble technology in water and wastewater treatment and ecological restoration. ACS ES&T Eng. 2022, 2, 1558–1573. [Google Scholar]
- Loeb, B.L. Forty years of advances in ozone technology. A review of ozone: Science & engineering. Ozone Sci. Eng. 2018, 40, 3–20. [Google Scholar]
- Rekhate, C.V.; Srivastava, J.K. Recent advances in ozone-based advanced oxidation processes for treatment of wastewater- a review. Chem. Eng. J. Adv. 2020, 3, 100031. [Google Scholar] [CrossRef]
- Lim, S.; Shi, J.L.; von Gunten, U.; McCurry, D.L. Ozonation of organic compounds in water and wastewater: A critical review. Water Res. 2022, 213, 118053. [Google Scholar] [CrossRef]
- Aleksić, S.; Žgajnar Gotvajn, A.; Premzl, K.; Kolar, M.; Turk, S.Š. Ozonation of amoxicillin and ciprofloxacin in model hospital wastewater to increase biotreatability. Antibiotics 2021, 10, 1407. [Google Scholar] [CrossRef] [PubMed]
- Azuma, T.; Hayashi, T. Disinfection of antibiotic-resistant bacteria in sewage and hospital effluent by ozonation. Ozone Sci. Eng. 2021, 43, 413–426. [Google Scholar] [CrossRef]
- Czekalski, N.; Imminger, S.; Salhi, E.; Veljkovic, M.; Kleffel, K.; Drissner, D.; Hammes, F.; Bürgmann, H.; von Gunten, U. Inactivation of antibiotic resistant bacteria and resistance genes by ozone: From laboratory experiments to full-scale wastewater treatment. Environ. Sci. Technol. 2016, 50, 11862–11871. [Google Scholar] [CrossRef]
- Basturk, I.; Varank, G.; Murat-Hocaoglu, S.; Yazici-Guvenc, S.; Oktem-Olgun, E.E.; Canli, O. Characterization and treatment of medical laboratory wastewater by ozonation: Optimization of toxicity removal by central composite design. Ozone Sci. Eng. 2021, 43, 213–227. [Google Scholar] [CrossRef]
- Walsh, T.R. A one-health approach to antimicrobial resistance. Nat. Microbiol. 2018, 3, 854–855. [Google Scholar] [CrossRef]
- Miłobedzka, A.; Ferreira, C.; Vaz-Moreira, I.; Calderón-Franco, D.; Gorecki, A.; Purkrtova, S.; Jan, B.; Dziewit, L.; Singleton, C.M.; Nielsen, P.H.; et al. ; et al. Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the One-Health Cycle. J. Hazard. Mater. 2022, 424, 127407. [Google Scholar] [CrossRef]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Antibiotic resistance: Moving from individual health norms to social norms in one health and global health. Front. Microbiol. 2020, 11, 01914. [Google Scholar] [CrossRef]
- Lépesová, K.; Olejníková, P.; Mackuľak, T.; Cverenkárová, K.; Krahulcová, M.; Bírošová, L. Hospital wastewater—Important source of multidrug resistant coliform bacteria with ESBL-production. Int. J. Environ. Res. Public Health 2020, 17, 7827. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Ke, M.; Zhang, Q.; Zhang, F.; Lu, T.; Sun, L.; Qian, H. Response of microbial antibiotic resistance to pesticides: An emerging health threat. Sci. Total Environ. 2022, 850, 158057. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzfathy, M.; Malayeri, A.H.; Mohseni, M.; Taghipour, F. Uv-led fluence determination by numerical method for microbial inactivation studies. J.Photochem. Photobiol. A Chem. 2020, 392, 112406. [Google Scholar] [CrossRef]
- Shimoda, H.; Matsuda, J.; Iwasaki, T.; Hayasaka, D. Efficacy of 265-nm ultraviolet light in inactivating infectious SARS-CoV-2. J. Photochem. Photobiol. 2021, 7, 100050. [Google Scholar] [CrossRef]
- Biancullo, F.; Moreira, N.F.F.; Ribeiro, A.R.; Manaia, C.M.; Faria, J.L.; Nunes, O.C.; Castro-Silva, S.M.; Silva, A.M.T. Heterogeneous photocatalysis using UVA-LEDs for the removal of antibiotics and antibiotic resistant bacteria from urban wastewater treatment plant effluents. Chem. Eng. J. 2019, 367, 304–313. [Google Scholar] [CrossRef]
- Gerchman, Y.; Mamane, H.; Friedman, N.; Mandelboim, M. Uv-led disinfection of coronavirus: Wavelength effect. J. Photochem. Photobiol. B Biol. 2020, 212, 112044. [Google Scholar] [CrossRef]
- Wan, Q.; Cao, R.; Wen, G.; Xu, X.; Xia, Y.; Wu, G.; Li, Y.; Wang, J.; Xu, H.; Lin, Y.; et al. Efficacy of UV-LED based advanced disinfection processes in the inactivation of waterborne fungal spores: Kinetics, photoreactivation, mechanism and energy requirements. Sci. Total Environ. 2022, 803, 150107. [Google Scholar] [CrossRef]
- Dong, C.; Fang, W.; Yi, Q.; Zhang, J. A comprehensive review on reactive oxygen species (ROS) in advanced oxidation processes (AOPs). Chemosphere 2022, 308, 136205. [Google Scholar] [CrossRef]
- Lozano, I.; Pérez-Guzmán, C.J.; Mora, A.; Mahlknecht, J.; Aguilar, C.L.; Cervantes-Avilés, P. Pharmaceuticals and personal care products in water streams: Occurrence, detection, and removal by electrochemical advanced oxidation processes. Sci. Total Environ. 2022, 827, 154348. [Google Scholar] [CrossRef]
- Saravanan, A.; Deivayanai, V.C.; Kumar, P.S.; Rangasamy, G.; Hemavathy, R.V.; Harshana, T.; Gayathri, N.; Alagumalai, K. A detailed review on advanced oxidation process in treatment of wastewater: Mechanism, challenges and future outlook. Chemosphere 2022, 308, 136524. [Google Scholar] [CrossRef]
- Yuan, T.; Pian, Y. Hospital wastewater as hotspots for pathogenic microorganisms spread into aquatic environment: A review. Front. Environ. Sci. 2023, 10, 1734. [Google Scholar] [CrossRef]
- Zhu, L.; Yuan, L.; Shuai, X.Y.; Lin, Z.J.; Sun, Y.J.; Zhou, Z.C.; Meng, L.X.; Ju, F.; Chen, H. Deciphering basic and key traits of antibiotic resistome in influent and effluent of hospital wastewater treatment systems. Water Res. 2023, 231, 119614. [Google Scholar] [CrossRef] [PubMed]
- Yamada, S.; Amano, T.; Minagawa, H. A study for distribution of microbubbles and effects of oxygen supplying into water. Trans. Jpn. Soc. Mech. Eng. B 2005, 71, 1301–1306. [Google Scholar] [CrossRef]
- Hashimoto, K.; Kubota, N.; Okuda, T.; Nakai, S.; Nishijima, W.; Motoshige, H. Reduction of ozone dosage by using ozone in ultrafine bubbles to reduce sludge volume. Chemosphere 2021, 274, 129922. [Google Scholar] [CrossRef]
- Zheng, J.; Su, C.; Zhou, J.; Xu, L.; Qian, Y.; Chen, H. Effects and mechanisms of ultraviolet, chlorination, and ozone disinfection on antibiotic resistance genes in secondary effluents of municipal wastewater treatment plants. Chem. Eng. J. 2017, 317, 309–316. [Google Scholar] [CrossRef]
- Dunkin, N.; Weng, S.; Coulter, C.G.; Jacangelo, J.G.; Schwab, K.J. Impacts of virus processing on human norovirus gi and gii persistence during disinfection of municipal secondary wastewater effluent. Water Res. 2018, 134, 1–12. [Google Scholar] [CrossRef]
- Takeuchi, F.; Sekizuka, T.; Yamashita, A.; Ogasawara, Y.; Mizuta, K.; Kuroda, M. Mepic, metagenomic pathogen identification for clinical specimens. Jpn. J. Infect. Dis. 2014, 67, 62–65. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. Megan community edition—Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Yin, X.; Jiang, X.T.; Chai, B.; Li, L.; Yang, Y.; Cole, J.R.; Tiedje, J.M.; Zhang, T. Args-oap v2.0 with an expanded sarg database and hidden markov models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes. Bioinformatics 2018, 34, 2263–2270. [Google Scholar] [CrossRef]
- Yin, X.; Zheng, X.; Li, L.; Zhang, A.N.; Jiang, X.T.; Zhang, T. ARGs-OAP v3.0: Antibiotic-resistance gene database curation and analysis pipeline optimization. Engineering 2022, in press. [Google Scholar] [CrossRef]
- Tran, N.H.; Reinhard, M.; Gin, K.Y.H. Occurrence and fate of emerging contaminants in municipal wastewater treatment plants from different geographical regions-A review. Water Res. 2018, 133, 182–207. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Health Labour and Welfare (Japan). Ministry of Health Labour and Welfare, Japan. Japan Nosocomial Infections Surveillance (JANIS), Nosocomial Infections Surveillance for Drug-Resistant Bacteria. Available online: https://janis.mhlw.go.jp/english/index.asp (accessed on 24 March 2023).
- Ministry of Health Labour and Welfare (Japan). Annual Report on Statistics of Production by Pharmaceutical Industry in 2021. Available online: https://www.mhlw.go.jp/topics/yakuji/2021/nenpo/index.html (accessed on 24 March 2023).
- Prasse, C.; Schlüsener, M.P.; Schulz, R.; Ternes, T.A. Antiviral drugs in wastewater and surface waters: A new pharmaceutical class of environmental relevance? Environ. Sci. Technol. 2010, 44, 1728–1735. [Google Scholar] [CrossRef]
- Azuma, T.; Ishiuchi, H.; Inoyama, T.; Teranishi, Y.; Yamaoka, M.; Sato, T.; Mino, Y. Occurrence and fate of selected anticancer, antimicrobial, and psychotropic pharmaceuticals in an urban river in a subcatchment of the Yodo River basin, Japan. Environ. Sci. Pollut. Res. 2015, 22, 18676–18686. [Google Scholar] [CrossRef] [PubMed]
- Petrović, M.; Škrbić, B.; Živančev, J.; Ferrando-Climent, L.; Barcelo, D. Determination of 81 pharmaceutical drugs by high performance liquid chromatography coupled to mass spectrometry with hybrid triple quadrupole–linear ion trap in different types of water in Serbia. Sci. Total Environ. 2014, 468–469, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Schlüsener, M.P.; Hardenbicker, P.; Nilson, E.; Schulz, M.; Viergutz, C.; Ternes, T.A. Occurrence of venlafaxine, other antidepressants and selected metabolites in the rhine catchment in the face of climate change. Environ. Pollut. 2015, 196, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Shukla, P.; Giri, B.S.; Chowdhary, P.; Chandra, R.; Gupta, P.; Pandey, A. Prevalence and hazardous impact of pharmaceutical and personal care products and antibiotics in environment: A review on emerging contaminants. Environ. Res. 2021, 194, 110664. [Google Scholar] [CrossRef]
- Azuma, T.; Arima, N.; Tsukada, A.; Hirami, S.; Matsuoka, R.; Moriwake, R.; Ishiuchi, H.; Inoyama, T.; Teranishi, Y.; Yamaoka, M.; et al. ; et al. Detection of pharmaceuticals and phytochemicals together with their metabolites in hospital effluents in Japan, and their contribution to sewage treatment plant influents. Sci. Total Environ. 2016, 548–549, 189–197. [Google Scholar] [CrossRef]
- Oliveira, T.S.; Murphy, M.; Mendola, N.; Wong, V.; Carlson, D.; Waring, L. Characterization of pharmaceuticals and personal care products in hospital effluent and waste water influent/effluent by direct-injection LC-MS-MS. Sci. Total Environ. 2015, 518–519, 459–478. [Google Scholar] [CrossRef]
- Singh, R.R.; Angeles, L.F.; Butryn, D.M.; Metch, J.W.; Garner, E.; Vikesland, P.J.; Aga, D.S. Towards a harmonized method for the global reconnaissance of multi-class antimicrobials and other pharmaceuticals in wastewater and receiving surface waters. Environ. Int. 2019, 124, 361–369. [Google Scholar] [CrossRef]
- Kharel, S.; Stapf, M.; Miehe, U.; Ekblad, M.; Cimbritz, M.; Falås, P.; Nilsson, J.; Sehlén, R.; Bester, K. Ozone dose dependent formation and removal of ozonation products of pharmaceuticals in pilot and full-scale municipal wastewater treatment plants. Sci. Total Environ. 2020, 731, 139064. [Google Scholar] [CrossRef]
- Issaka, E.; Amu-Darko, J.N.-O.; Yakubu, S.; Fapohunda, F.O.; Ali, N.; Bilal, M. Advanced catalytic ozonation for degradation of pharmaceutical pollutants―A review. Chemosphere 2022, 289, 133208. [Google Scholar] [CrossRef]
- Iakovides, I.C.; Michael-Kordatou, I.; Moreira, N.F.F.; Ribeiro, A.R.; Fernandes, T.; Pereira, M.F.R.; Nunes, O.C.; Manaia, C.M.; Silva, A.M.T.; Fatta-Kassinos, D. Continuous ozonation of urban wastewater: Removal of antibiotics, antibiotic-resistant Escherichia coli and antibiotic resistance genes and phytotoxicity. Water Res. 2019, 159, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Chávez, A.M.; Beltrán, F.J.; López, J.; Javier Rivas, F.; Álvarez, P.M. On the importance of reactions in the proximity of the gas–water interface: Application to direct ozone reactions of antibiotics in water. Chem. Eng. J. 2023, 458, 141408. [Google Scholar] [CrossRef]
- Ge, L.; Na, G.; Zhang, S.; Li, K.; Zhang, P.; Ren, H.; Yao, Z. New insights into the aquatic photochemistry of fluoroquinolone antibiotics: Direct photodegradation, hydroxyl-radical oxidation, and antibacterial activity changes. Sci. Total Environ. 2015, 527–528, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lv, K.; Deng, C.; Yu, Z.; Shi, J.; Johnson, A.C. Persistence and migration of tetracycline, sulfonamide, fluoroquinolone, and macrolide antibiotics in streams using a simulated hydrodynamic system. Environ. Pollut. 2019, 252, 1532–1538. [Google Scholar] [CrossRef]
- Lima, L.M.; Silva, B.N.M.d.; Barbosa, G.; Barreiro, E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112829. [Google Scholar] [CrossRef]
- Robles-Jimenez, L.E.; Aranda-Aguirre, E.; Castelan-Ortega, O.A.; Shettino-Bermudez, B.S.; Ortiz-Salinas, R.; Miranda, M.; Li, X.; Angeles-Hernandez, J.C.; Vargas-Bello-Pérez, E.; Gonzalez-Ronquillo, M. Worldwide traceability of antibiotic residues from livestock in wastewater and soil: A systematic review. Animals 2022, 12, 60. [Google Scholar] [CrossRef]
- Vione, D.; Feitosa-Felizzola, J.; Minero, C.; Chiron, S. Phototransformation of selected human-used macrolides in surface water: Kinetics, model predictions and degradation pathways. Water Res. 2009, 43, 1959–1967. [Google Scholar] [CrossRef]
- Carvalho, I.T.; Santos, L. Antibiotics in the aquatic environments: A review of the european scenario. Environ. Int. 2016, 94, 736–757. [Google Scholar] [CrossRef]
- Izadi, E.; Afshan, G.; Patel, R.P.; Rao, V.M.; Liew, K.B.; Meor Mohd Affandi, M.M.R.; Kifli, N.; Suleiman, A.; Lee, K.S.; Sarker, M.M.R.; et al. ; et al. Levofloxacin: Insights into antibiotic resistance and product quality. Front. Pharmacol. 2019, 10, 00881. [Google Scholar] [CrossRef]
- Norte, T.H.d.O.; Marcelino, R.B.P.; Medeiros, F.H.A.; Moreira, R.P.L.; Amorim, C.C.; Lago, R.M. Ozone oxidation of β-lactam antibiotic molecules and toxicity decrease in aqueous solution and industrial wastewaters heavily contaminated. Ozone Sci. Eng. 2018, 40, 385–391. [Google Scholar] [CrossRef]
- Tufail, A.; Price, W.E.; Mohseni, M.; Pramanik, B.K.; Hai, F.I. A critical review of advanced oxidation processes for emerging trace organic contaminant degradation: Mechanisms, factors, degradation products, and effluent toxicity. J. Water Proc. Eng. 2021, 40, 101778. [Google Scholar] [CrossRef]
- Zilberman, A.; Gozlan, I.; Avisar, D. Pharmaceutical transformation products formed by ozonation—Does degradation occur? Molecules 2023, 28, 1227. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xi, H.; Xu, L.; Jin, M.; Zhao, W.; Liu, H. Ecotoxicological effects, environmental fate and risks of pharmaceutical and personal care products in the water environment: A review. Sci. Total Environ. 2021, 788, 147819. [Google Scholar] [CrossRef]
- Li, J.; Li, W.; Liu, K.; Guo, Y.; Ding, C.; Han, J.; Li, P. Global review of macrolide antibiotics in the aquatic environment: Sources, occurrence, fate, ecotoxicity, and risk assessment. J. Hazard. Mater. 2022, 439, 129628. [Google Scholar] [CrossRef]
- Magdeburg, A.; Stalter, D.; Oehlmann, J. Whole effluent toxicity assessment at a wastewater treatment plant upgraded with a full-scale post-ozonation using aquatic key species. Chemosphere 2012, 88, 1008–1014. [Google Scholar] [CrossRef]
- Díaz-Garduño, B.; Pintado-Herrera, M.G.; Biel-Maeso, M.; Rueda-Márquez, J.J.; Lara-Martín, P.A.; Perales, J.A.; Manzano, M.A.; Garrido-Pérez, C.; Martín-Díaz, M.L. Environmental risk assessment of effluents as a whole emerging contaminant: Efficiency of alternative tertiary treatments for wastewater depuration. Water Res. 2017, 119, 136–149. [Google Scholar] [CrossRef]
- Tamura, I.; Yasuda, Y.; Kagota, K.; Yoneda, S.; Nakada, N.; Kumar, V.; Kameda, Y.; Kimura, K.; Tatarazako, N.; Yamamoto, H. Contribution of pharmaceuticals and personal care products (PPCPs) to whole toxicity of water samples collected in effluent-dominated urban streams. Ecotoxicol. Environ. Safe. 2017, 144, 338–350. [Google Scholar] [CrossRef]
- Ike, I.A.; Karanfil, T.; Cho, J.; Hur, J. Oxidation byproducts from the degradation of dissolved organic matter by advanced oxidation processes—A critical review. Water Res. 2019, 164, 114929. [Google Scholar] [CrossRef]
- Tufail, A.; Price, W.E.; Hai, F.I. A critical review on advanced oxidation processes for the removal of trace organic contaminants: A voyage from individual to integrated processes. Chemosphere 2020, 260, 127460. [Google Scholar] [CrossRef]
- Castellano-Hinojosa, A.; Gallardo-Altamirano, M.J.; González-López, J.; González-Martínez, A. Anticancer drugs in wastewater and natural environments: A review on their occurrence, environmental persistence, treatment, and ecological risks. J. Hazard. Mater. 2023, 447, 130818. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Pathak, V.; Tripathi, D.M.; Tripathi, B.D. Application of ozone based treatments of secondary effluents. Bioresour. Technol. 2011, 102, 2481–2486. [Google Scholar] [CrossRef] [PubMed]
- Foroughi, M.; Khiadani, M.; Kakhki, S.; Kholghi, V.; Naderi, K.; Yektay, S. Effect of ozonation-based disinfection methods on the removal of antibiotic resistant bacteria and resistance genes (ARB/ARGs) in water and wastewater treatment: A systematic review. Sci. Total Environ. 2022, 811, 151404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ma, X.; Luo, L.; Hu, N.; Duan, J.; Tang, Z.; Zhong, R.; Li, Y. The prevalence and characterization of extended-spectrum β-lactamase- and carbapenemase-producing bacteria from hospital sewage, treated effluents and receiving rivers. Int. J. Environ. Res. Public Health 2020, 17, 1183. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Vambol, V.; Vambol, S.; Bolibrukh, B.; Sillanpaa, M.; Changani, F.; Esrafili, A.; Yousefi, M. Hospital effluent guidelines and legislation scenario around the globe: A critical review. J. Environ. Chem. Eng. 2021, 9, 105874. [Google Scholar] [CrossRef]
- Verlicchi, P. Trends, new insights and perspectives in the treatment of hospital effluents. Curr. Opin. Environ. Sci. Health 2021, 19, 100217. [Google Scholar] [CrossRef]
- Ajala, O.J.; Tijani, J.O.; Salau, R.B.; Abdulkareem, A.S.; Aremu, O.S. A review of emerging micro-pollutants in hospital wastewater: Environmental fate and remediation options. Results Eng. 2022, 16, 100671. [Google Scholar] [CrossRef]
- Bian, J.; Wang, H.; Ding, H.; Song, Y.; Zhang, X.; Tang, X.; Zhong, Y.; Zhao, C. Unveiling the dynamics of antibiotic resistome, bacterial communities, and metals from the feces of patients in a typical hospital wastewater treatment system. Sci. Total Environ. 2023, 858, 159907. [Google Scholar] [CrossRef]
- Perveen, S.; Pablos, C.; Reynolds, K.; Stanley, S.; Marugán, J. Growth and prevalence of antibiotic-resistant bacteria in microplastic biofilm from wastewater treatment plant effluents. Sci. Total Environ. 2023, 856, 159024. [Google Scholar] [CrossRef]
- Korichi, W.; Ibrahimi, M.; Loqman, S.; Ouhdouch, Y.; Younes, K.; Lemée, L. Assessment of actinobacteria use in the elimination of multidrug-resistant bacteria of Ibn Tofail hospital wastewater (Marrakesh, Morocco): A chemometric data analysis approach. Environ. Sci. Pollut. Res. 2021, 28, 26840–26848. [Google Scholar] [CrossRef]
- Kovalova, L.; Siegrist, H.; von Gunten, U.; Eugster, J.; Hagenbuch, M.; Wittmer, A.; Moser, R.; McArdell, C.S. Elimination of micropollutants during post-treatment of hospital wastewater with powdered activated carbon, ozone, and UV. Environ. Sci. Technol. 2013, 47, 7899–7908. [Google Scholar] [CrossRef] [PubMed]
- Hiller, C.X.; Hübner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J.E. Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Mousazadeh, M.; Kabdaşlı, I.; Khademi, S.; Sandoval, M.A.; Moussavi, S.P.; Malekdar, F.; Gilhotra, V.; Hashemi, M.; Dehghani, M.H. A critical review on the existing wastewater treatment methods in the COVID-19 era: What is the potential of advanced oxidation processes in combatting viral especially SARS-CoV-2? J. Water Proc. Eng. 2022, 49, 103077. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azuma, T.; Katagiri, M.; Sasaki, N.; Kuroda, M.; Watanabe, M. Performance of a Pilot-Scale Continuous Flow Ozone-Based Hospital Wastewater Treatment System. Antibiotics 2023, 12, 932. https://doi.org/10.3390/antibiotics12050932
Azuma T, Katagiri M, Sasaki N, Kuroda M, Watanabe M. Performance of a Pilot-Scale Continuous Flow Ozone-Based Hospital Wastewater Treatment System. Antibiotics. 2023; 12(5):932. https://doi.org/10.3390/antibiotics12050932
Chicago/Turabian StyleAzuma, Takashi, Miwa Katagiri, Naobumi Sasaki, Makoto Kuroda, and Manabu Watanabe. 2023. "Performance of a Pilot-Scale Continuous Flow Ozone-Based Hospital Wastewater Treatment System" Antibiotics 12, no. 5: 932. https://doi.org/10.3390/antibiotics12050932
APA StyleAzuma, T., Katagiri, M., Sasaki, N., Kuroda, M., & Watanabe, M. (2023). Performance of a Pilot-Scale Continuous Flow Ozone-Based Hospital Wastewater Treatment System. Antibiotics, 12(5), 932. https://doi.org/10.3390/antibiotics12050932








