Using an Antibiogram Profile to Improve Infection Control and Rational Antimicrobial Therapy in an Urban Hospital in The Gambia, Strategies and Lessons for Low- and Middle-Income Countries
Abstract
1. Introduction
2. Results
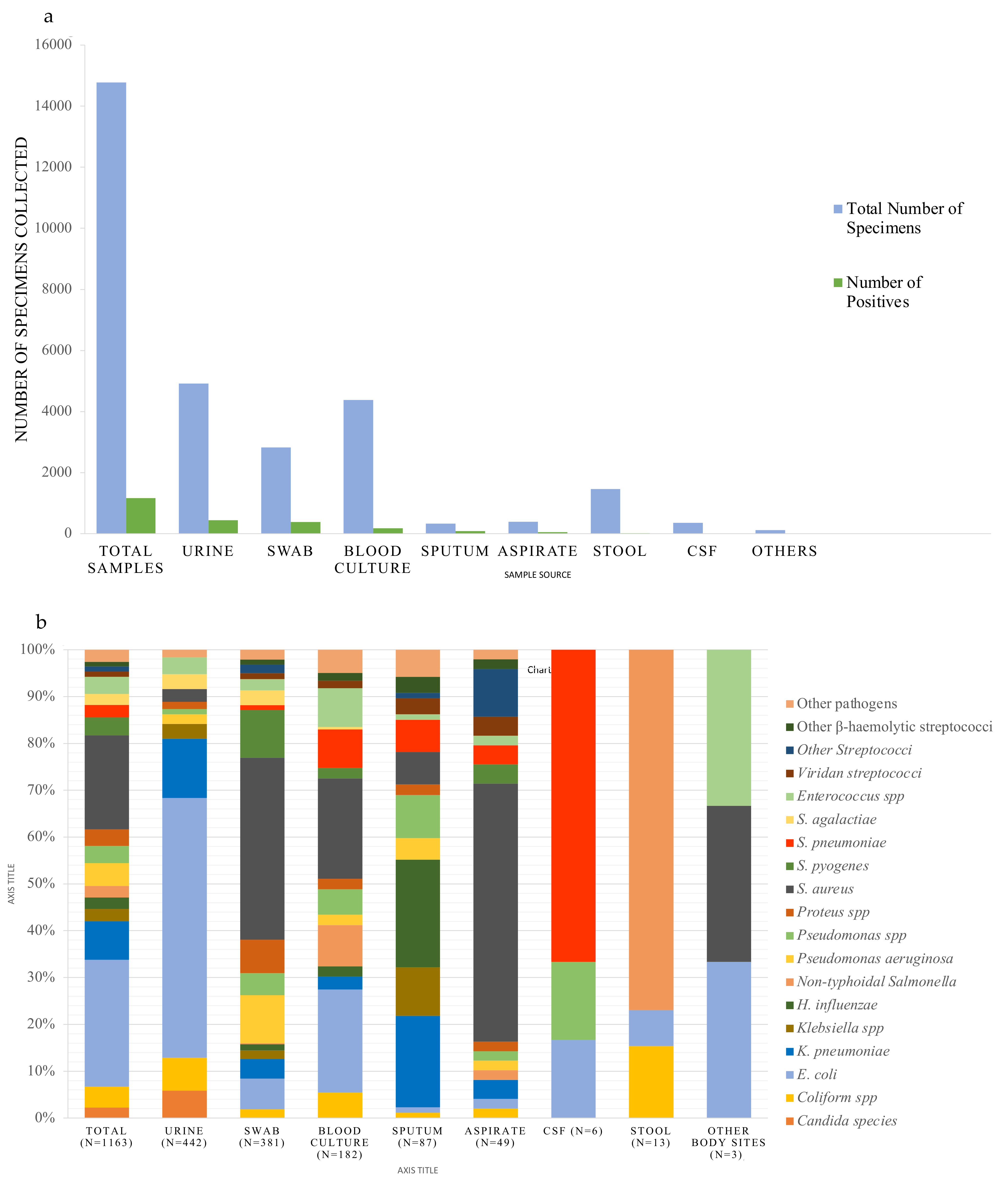
2.1. Samples and Pathogen Distribution
2.2. Susceptibility Profiles of Pathogens
3. Discussion
4. Materials and Methods
4.1. Study Design and Setting
4.2. Microbiological Procedures
4.3. Analysis
4.4. Ethical Review and Approval
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Strategy for Containment of Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2001; p. 105.
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 6736. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2017; pp. 1–28.
- Holloway, K.; Mathai, E.; Gray, A. Surveillance of antimicrobial resistance in resource-constrained settings–experience from five pilot projects. Trop. Med. Int. Health 2010, 16, 368–374. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021; WHO: Geneva, Switzerland, 2021.
- Elton, L.; Thomason, M.J.; Tembo, J.; Velavan, T.P.; Pallerla, S.R.; Arruda, L.B.; Vairo, F.; Montaldo, C.; Ntoumi, F.; Hamid, M.M.A.; et al. Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrob. Resist. Infect. Control. 2020, 9, 145. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Model List of Essential Medicines; WHO: Geneva, Switzerland, 2021.
- Handler, J.F.; Stelling, J. Analysis and presentation of cumulative antibiograms: A new consensus guideline from the Clinical and Laboratory Standards Institute. Clin. Infect. Diseases. 2007, 44, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Ahmend, I.; Zafar, H.; Lakhnana, N.; Ishtiaq, S.; Tauseef, K.; Zahid, M.; Kazmi, A. Hospital Antibiogram: A Necessity in Monitoring Sensitivity of Isolates and Rationale Use of Antibiotics. Br. Microbiol. Res. J. 2016, 13, 1–8. [Google Scholar] [CrossRef]
- Joshi, S. Hospital antibiogram: A necessity. Indian J. Med. Microbiol. 2010, 28, 277–280. [Google Scholar] [CrossRef]
- Avdic, E.; Carroll, K.C. The role of the microbiology laboratory in antimicrobial stewardship programs. Infect. Dis. Clin. N. Am. 2014, 28, 215–235. [Google Scholar] [CrossRef]
- El-Azizi, M.; Mushtaq, A.; Drake, C.; Lawhorn, J.; Barenfanger, J.; Verhulst, S.; Khardori, N. Evaluating antibiograms to monitor drug resistance. Emerg. Infect. Dis. 2005, 11, 1301–1302. [Google Scholar] [CrossRef]
- Ambretti, S.; Gagliotti, C.; Luzzaro, F.; Malacarne, P.; Pan, A.; Pieretti, B.; Tascini, C.; Sarti, M.; CoSIAS-AMCLI, C. Reporting epidemiology of antibiotic resistance. Microbiol. Med. 2015, 30. [Google Scholar] [CrossRef]
- Archibald, L.K.; Reller, L.B. Clinical microbiology in developing countries. Emerg. Infect. Dis. 2001, 7, 302–305. [Google Scholar] [CrossRef]
- Petti, C.A.; Polage, C.R.; Quinn, T.C.; Ronald, A.R.; Sande, M.A. Laboratory medicine in Africa: A barrier to effective health care. Clin. Infect. Dis. 2006, 42, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Iwu, C.D.; Patrick, S.M. An insight into the implementation of the global action plan on antimicrobial resistance in the WHO African region: A roadmap for action. Int. J. Antimicrob. Agents 2021, 58, 106411. [Google Scholar] [CrossRef] [PubMed]
- Truong, W.R.; Hidayat, L.; Bolaris, M.A.; Nguyen, L.; Yamaki, J. The antibiogram: Key considerations for its development and utilization. JAC Antimicrob. Resist. 2021, 3, dlab060. [Google Scholar] [CrossRef] [PubMed]
- Clinical Laboratory Standards Institute. M39-A4 Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data; Approved Guideline–Fourth Edition; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2014. [Google Scholar]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global Trends in Antimicrobial Resistance in Animals in Low- and Middle-income Countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef] [PubMed]
- Lacy, M.K.; Klutman, N.E.; Horvat, R.T.; Zapantis, A. Antibiograms: New NCCLS guidelines, development, and clinical application. Hosp. Pharm. 2004, 39, 542–553. [Google Scholar] [CrossRef]
- Hill, P.C.; Onyeama, C.O.; Ikumapayi, U.N.A.; Secka, O.; Ameyaw, S.; Simmonds, N.; Donkor, S.A.; Howie, S.R.; Tapgun, M.; Corrah, T.; et al. Bacteraemia in patients admitted to an urban hospital in West Africa. BMC Infect. Dis. 2007, 7, 2–10. [Google Scholar] [CrossRef]
- Darboe, S.; Okomo, U.; Muhammad, A.K.; Ceesay, B.; Jallow, M.; Usuf, E.; Tweed, S.; Akpalu, E.; Kwambana-Adams, B.; Kariuki, S.; et al. Community-acquired Invasive Bacterial Disease in Urban Gambia, 2005–2015: A Hospital-based Surveillance. Clin. Infect. Dis. 2019, 69, 105–113. [Google Scholar] [CrossRef]
- Laupland, K.B.; Gregson, D.B.; Flemons, W.W.; Hawkins, D.; Ross, T.; Church, D.L. Burden of community-onset bloodstream infection: A population-based assessment. Epidemiol. Infect. 2007, 135, 1037–1042. [Google Scholar] [CrossRef]
- Laupland, K.B.; Lyytikäinen, O.; Søgaard, M.; Kennedy, K.J.; Knudsen, J.D.; Ostergaard, C.; Galbraith, J.C.; Valiquette, L.; Jacobsson, G.; Collignon, P.; et al. The changing epidemiology of Staphylococcus aureus bloodstream infection: A multinational population-based surveillance study. Clin. Microbiol. Infect. 2013, 19, 465–471. [Google Scholar] [CrossRef]
- Mackenzie, G.A.; Hill, P.C.; Jeffries, D.J.; Hossain, I.; Uchendu, U.; Ameh, D.; Ndiaye, M.; Adeyemi, O.; Pathirana, J.; Olatunji, Y.; et al. Effect of the introduction of pneumococcal conjugate vaccination on invasive pneumococcal disease in The Gambia: A population-based surveillance study. Lancet Infect. Dis. 2016, 16, 703–711. [Google Scholar] [CrossRef]
- Cutts, F.T.; Zaman, S.M.A.; Enwere, G.; Jaffar, S.; Levine, O.S.; Okoko, J.B.; Oluwalana, C.; Vaughan, A.; Obaro, S.K.; Leach, A.; et al. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: Randomised, double-blind, placebo-controlled trial. Lancet 2005, 365, 1139–1146. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, G.A.; Hill, P.C.; Sahito, S.M.; Jeffries, D.J.; Hossain, I.; Bottomley, C.; Uchendu, U.; Ameh, D.; Ndiaye, M.; Osuorah, C.D.; et al. Impact of the introduction of pneumococcal conjugate vaccination on pneumonia in The Gambia: Population-based surveillance and case-control studies. Lancet Infect. Dis. 2017, 17, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Ikumapayi, U.N.; Antonio, M.; Sonne-Hansen, J.; Biney, E.; Enwere, G.; Okoko, B.; Oluwalana, C.; Vaughan, A.; Zaman, S.M.A.; Greenwood, B.M.; et al. Molecular Epidemiology of Community-Acquired Invasive Non-Typhoidal Salmonella among Children Aged 2 29 Months in Rural Gambia and Discovery of a New Serovar, Salmonella Enterica Dingiri. J. Med. Microbiol. 2007, 56, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Darboe, S.; Bradbury, R.S.; Phelan, J.; Kanteh, A.; Muhammad, A.-K.; Worwui, A.; Yang, S.; Nwakanma, D.; Pe-rez-Sepulveda, B.; Kariuki, S.; et al. Genomic diversity and antimicrobial resistance among non-typhoidal Salmonella associated with human disease in The Gambia. Microb. Genom. 2022, 8, 000785. [Google Scholar] [CrossRef] [PubMed]
- Kanteh, A.; Sesay, A.K.; Alikhan, N.-F.; Ikumapayi, U.N.; Salaudeen, R.; Manneh, J.; Olatunji, Y.; Page, A.J.; Macken-zie, G. Invasive atypical non-typhoidal Salmonella serovars in The Gambia. Microb. Genom. 2021, 7, 000677. [Google Scholar] [CrossRef]
- Su, T.-Y.; Lee, M.-H.; Huang, C.-T.; Liu, T.-P.; Lu, J.-J. The clinical impact of patients with bloodstream infection with different groups of Viridans group streptococci by using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). Medicine 2018, 97, e13607. [Google Scholar] [CrossRef]
- Erdem, I.; Kara Ali, R.; Ardic, E.; Elbasan Omar, S.; Mutlu, R.; Topkaya, A.E. Community-acquired lower urinary tract infections: Etiology, antimicrobial resistance, and treatment results in female patients. J. Glob. Infect. Dis. 2018, 10, 129–132. [Google Scholar] [CrossRef]
- Armitage, E.P.; Senghore, E.; Darboe, S.; Barry, M.; Camara, J.; Bah, S.; Marks, M.; Cerami, C.; Roca, A.; Antonio, M.; et al. High burden and seasonal variation of paediatric scabies and pyoderma prevalence in the Gambia: A cross-sectional study. PLoS Negl. Trop. Dis. 2019, 13, e0007801. [Google Scholar] [CrossRef]
- Uwaezuoke, S.N.; Ndu, I.K. The prevalence and risk of urinary tract infection in malnourished children: A systematic review and meta-Analysis. BMC Pediatr. 2019, 19, 261. [Google Scholar] [CrossRef]
- Donkor, E.S.; Horlortu, P.Z. Community acquired urinary tract infections among adults in Accra, Ghana. Infect. Drug Resist. 2019, 12, 2059–2067. [Google Scholar] [CrossRef]
- Darboe, S.; Dobreniecki, S.; Jarju, S.; Jallow, M.; Mohammed, N.I.; Wathuo, M.; Ceesay, B.; Tweed, S.; Basu Roy, R.; Okomo, U.; et al. Prevalence of Panton-Valentine Leukocidin (PVL) and Antimicrobial Resistance in Community-Acquired Clinical Staphylococcus aureus in an Urban Gambian Hospital: A 11-year period retrospective pilot study. Front. Cell Infect. Microbiol. 2019, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, B.T.; Ashley, E.A.; Ongarello, S.; Havumaki, J.; Wijegoonewardena, M.; González, I.J.; Dittrich, S. Antimicrobial resistance in Africa: A systematic review. BMC Infect. Dis. 2017, 17, 616. [Google Scholar] [CrossRef] [PubMed]
- Chaw, P.S.; Schlinkmann, K.M.; Raupach-Rosin, H.; Karch, A.; Pletz, M.W.; Huebner, J.; Nyan, O.; Mikolajczyk, R. Antibiotic use on paediatric inpatients in a teaching hospital in the Gambia, a retrospective study. Antimicrob. Resist. Infect. Control. 2018, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chaw, P.S.; Schlinkmann, K.M.; Raupach-Rosin, H.; Karch, A.; Pletz, M.W.; Huebner, J.; Mikolajczyk, R. Knowledge, attitude and practice of Gambian health practitioners towards antibiotic prescribing and microbiological testing: A crosssectional survey. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Review, T.G.C. The Gambia Country Review: Key Data. In The Gambia Country Review; International Monetary Fund: Washington, DC, USA, 2009. [Google Scholar]
- Ceesay, S.J.; Casals-Pascual, C.; Nwakanma, D.C.; Walther, M.; Gomez-Escobar, N.; Fulford, A.J.C.; Takem, E.N.; Nogaro, S.; Bojang, K.A.; Corrah, T.; et al. Continued decline of malaria in The Gambia with implications for elimination. PLoS ONE 2010, 5, e12242. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. CLSI Document M100-S25; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2015. [Google Scholar]

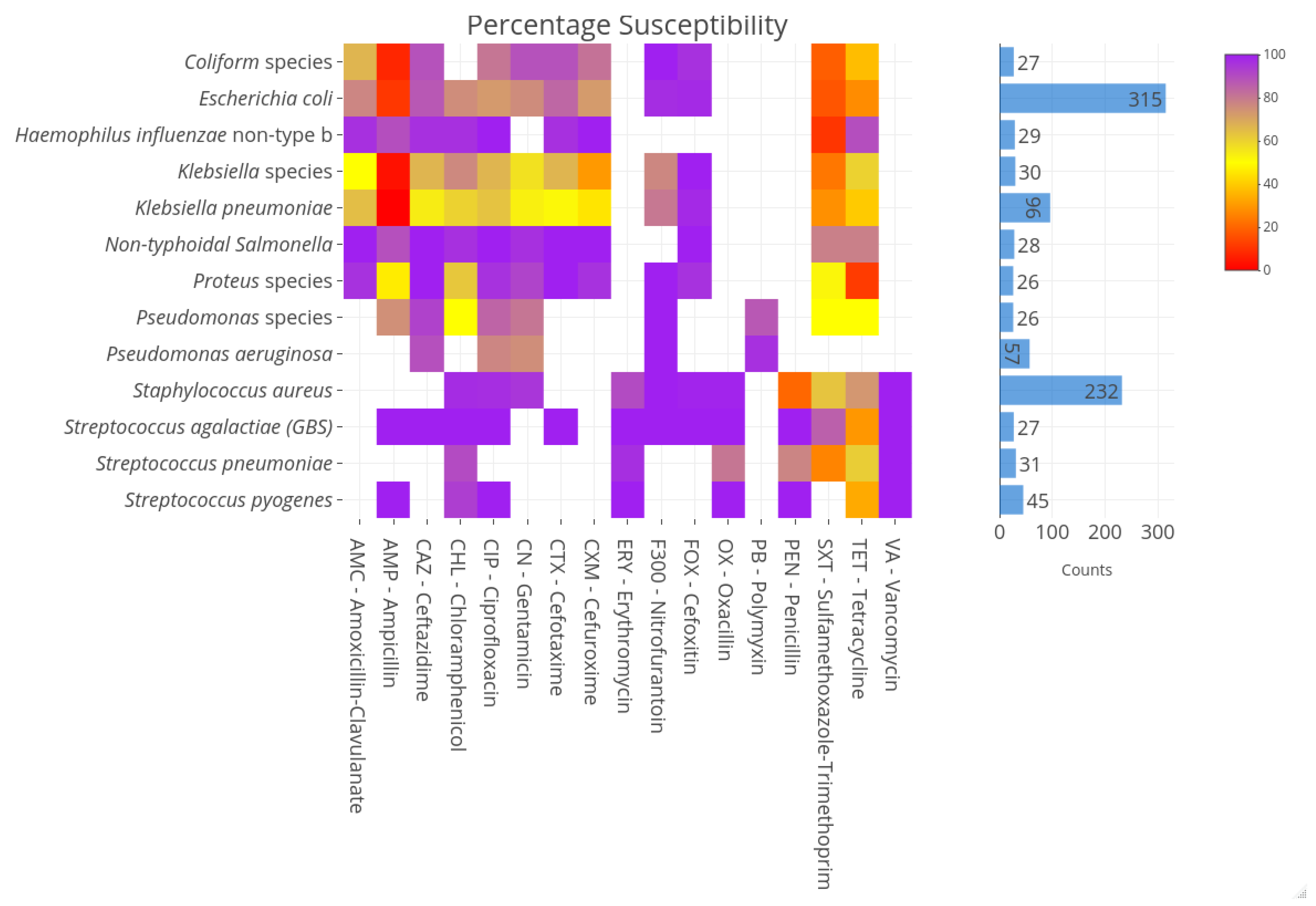
| Organism | Number | % Susceptibility | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-negatives | PEN | AMP | SXT | CN | CHL | TET | CIP | CXM | ERY | PB | OX | FOX | CTX | CAZ | VA | AMC | |
| E. coli | 40 | Na | 13 | 15 | 83 | 85 | 38 | 85 | 38 | Na | Na | Na | 97 | 90 | 88 | Na | 80 |
| NTS | 16 * | Na | 100 | 88 | 94 | 100 | 88 | 100 | 100 | Na | Na | Na | 100 | 100 | 100 | Na | 100 |
| Pseudomonas species | 10 * | Na | Na | 22 | 90 | 22 | 63 | 89 | 50 | Na | Na | Na | NA | Na | 100 | Na | Na |
| Gram-positives | |||||||||||||||||
| S. aureus | 39 | 13 | Na | 69 | 97 | 97 | 74 | 100 | Na | 87 | Na | 100 | 100 | Na | Na | 100 | Na |
| S. pneumoniae | 15 * | 80 | Na | 7 | Na | 93 | 67 | Na | Na | 93 | Na | 80 | Na | Na | Na | 100 | Na |
| Enterococcus species | 15 * | 92 | 92 | Na | Na | 83 | 58 | 83 | Na | 58 | Na | Na | Na | Na | Na | 100 | Na |
| Organism | Number | % Susceptibility | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-negatives | PEN | AMP | OB | OX | SXT | CN | TET | F300 | CIP | ERY | CXM | CAZ | CTX | FOX | AMC | PB | VA | |
| E coli | 245 | Na | 11 | Na | Na | 16 | 72 | 26 | 97 | 69 | Na | 78 | 88 | 84 | 98 | 77 | Na | Na |
| K. pneumoniae | 56 | Na | 0 | Na | Na | 16 | 46 | 33 | 80 | 59 | Na | 29 | 41 | 40 | 96 | 54 | Na | Na |
| Coliform species | 31 | Na | 7 | Na | Na | 23 | 87 | 38 | 100 | 83 | Na | 74 | 80 | 87 | 87 | 61 | Na | Na |
| Klebsiella species | 14 * | Na | 0 | Na | Na | 7 | 36 | 57 | 79 | 57 | Na | 36 | 50 | 50 | 100 | 29 | Na | Na |
| Gram-positives | ||||||||||||||||||
| Candida species | 26 * | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na | Na |
| Enterococcus species | 16 * | 94 | 88 | Na | Na | Na | Na | 13 | 94 | 75 | 81 | Na | Na | Na | Na | Na | Na | 94 |
| S. agalactiae | 14 * | 100 | 100 | Na | Na | 86 | Na | 7 | 100 | 100 | 100 | Na | Na | Na | Na | Na | Na | 100 |
| S. aureus | 12 * | 58 | Na | 100 | 100 | 58 | 100 | 92 | 100 | 100 | 100 | NA | Na | Na | 100 | Na | Na | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darboe, S.; Mirasol, R.; Adejuyigbe, B.; Muhammad, A.K.; Nadjm, B.; De St. Maurice, A.; Dogan, T.L.; Ceesay, B.; Umukoro, S.; Okomo, U.; et al. Using an Antibiogram Profile to Improve Infection Control and Rational Antimicrobial Therapy in an Urban Hospital in The Gambia, Strategies and Lessons for Low- and Middle-Income Countries. Antibiotics 2023, 12, 790. https://doi.org/10.3390/antibiotics12040790
Darboe S, Mirasol R, Adejuyigbe B, Muhammad AK, Nadjm B, De St. Maurice A, Dogan TL, Ceesay B, Umukoro S, Okomo U, et al. Using an Antibiogram Profile to Improve Infection Control and Rational Antimicrobial Therapy in an Urban Hospital in The Gambia, Strategies and Lessons for Low- and Middle-Income Countries. Antibiotics. 2023; 12(4):790. https://doi.org/10.3390/antibiotics12040790
Chicago/Turabian StyleDarboe, Saffiatou, Ruel Mirasol, Babapelumi Adejuyigbe, Abdul Khalie Muhammad, Behzad Nadjm, Annabelle De St. Maurice, Tiffany L. Dogan, Buntung Ceesay, Solomon Umukoro, Uduak Okomo, and et al. 2023. "Using an Antibiogram Profile to Improve Infection Control and Rational Antimicrobial Therapy in an Urban Hospital in The Gambia, Strategies and Lessons for Low- and Middle-Income Countries" Antibiotics 12, no. 4: 790. https://doi.org/10.3390/antibiotics12040790
APA StyleDarboe, S., Mirasol, R., Adejuyigbe, B., Muhammad, A. K., Nadjm, B., De St. Maurice, A., Dogan, T. L., Ceesay, B., Umukoro, S., Okomo, U., Nwakanma, D., Roca, A., Secka, O., Forrest, K., & Garner, O. B. (2023). Using an Antibiogram Profile to Improve Infection Control and Rational Antimicrobial Therapy in an Urban Hospital in The Gambia, Strategies and Lessons for Low- and Middle-Income Countries. Antibiotics, 12(4), 790. https://doi.org/10.3390/antibiotics12040790








