Quercetin: Synergistic Interaction with Antibiotics against Colistin-Resistant Acinetobacter baumannii
Abstract
1. Introduction
2. Results
2.1. Antibacterial Susceptibility
2.2. Synergy Studies with QUE by Checkerboard
2.3. Time-Kill Studies
2.4. Measuring Cell Membrane Damage
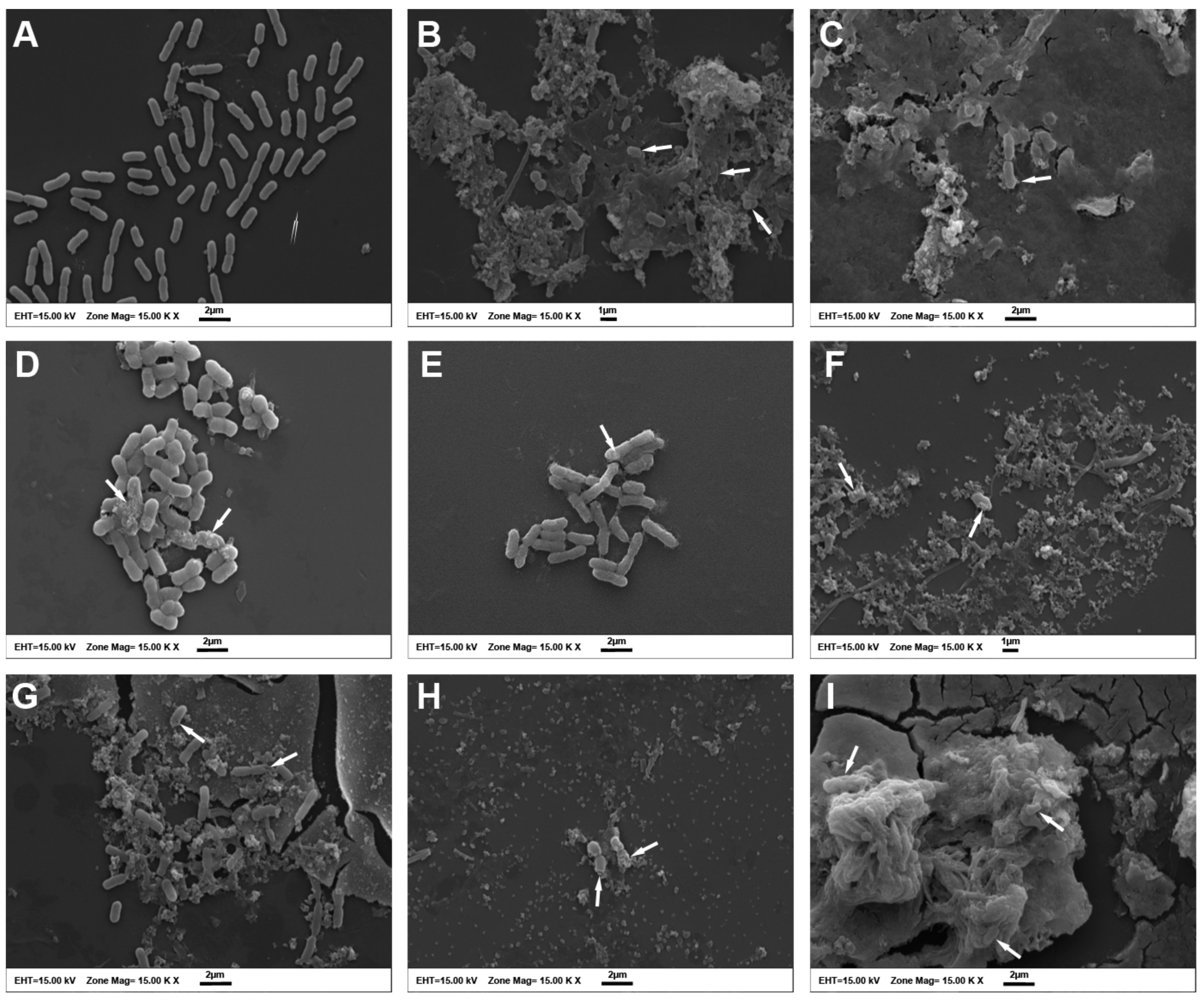
2.5. Scanning Electron Microscopy (SEM) Evaluation of Morphological Alterations
3. Discussion
4. Materials and Methods
4.1. Test Compounds
4.2. Bacterial Strains
4.3. MIC Determination
4.4. Checkerboard Synergy Test
4.5. Time-Kill Assay
4.6. Cytoplasmic Membrane Permeability Assay
4.7. Scanning Electron Microscopy
4.8. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Almasaudi, S.B. Acinetobacter spp. as Nosocomial Pathogens: Epidemiology and Resistance Features. Saudi J. Biol. Sci. 2018, 25, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Usmani, Y.; Ahmed, A.; Faizi, S.; Versiani, M.A.; Shamshad, S.; Khan, S.; Simjee, S.U. Antimicrobial and Biofilm Inhibiting Potential of an Amide Derivative [N-(2′, 4′-Dinitrophenyl)-3β-Hydroxyurs-12-En-28-Carbonamide] of Ursolic Acid by Modulating Membrane Potential and Quorum Sensing against Colistin Resistant Acinetobacter baumannii. Microb. Pathog. 2021, 157, 104997. [Google Scholar] [CrossRef] [PubMed]
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a Global Pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Rossolini, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the Antibiotic Resistance Crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef]
- Lee, J.-H. Perspectives towards Antibiotic Resistance: From Molecules to Population. J. Microbiol. 2019, 57, 181–184. [Google Scholar] [CrossRef]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of Antibiotic Resistance in Important Gram-Positive and Gram-Negative Pathogens and Novel Antibiotic Solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Howard, A.; O’Donoghue, M.; Feeney, A.; Sleator, R.D. Acinetobacter baumannii. Virulence 2012, 3, 243–250. [Google Scholar] [CrossRef]
- Rangel, K.; Chagas, T.P.G.; De-Simone, S.G. Acinetobacter baumannii Infections in Times of COVID-19 Pandemic. Pathogens 2021, 10, 1006. [Google Scholar] [CrossRef]
- Lin, M.-F. Antimicrobial Resistance in Acinetobacter baumannii: From Bench to Bedside. World J. Clin. Cases 2014, 2, 787. [Google Scholar] [CrossRef]
- Wong, D.; Nielsen, T.B.; Bonomo, R.A.; Pantapalangkoor, P.; Luna, B.; Spellberg, B. Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 2017, 30, 409–447. [Google Scholar] [CrossRef]
- Lee, C.-R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.-J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef]
- Qureshi, Z.A.; Hittle, L.E.; O’Hara, J.A.; Rivera, J.I.; Syed, A.; Shields, R.K.; Pasculle, A.W.; Ernst, R.K.; Doi, Y. Colistin-Resistant Acinetobacter baumannii: Beyond Carbapenem Resistance. Clin. Infect. Dis. 2015, 60, 1295–1303. [Google Scholar] [CrossRef]
- Lin, R.-D.; Chin, Y.-P.; Lee, M.-H. Antimicrobial Activity of Antibiotics in Combination with Natural Flavonoids against Clinical Extended-Spectrumβ-Lactamase (ESBL)-Producing Klebsiella pneumoniae. Phytother. Res. 2005, 19, 612–617. [Google Scholar] [CrossRef]
- Aleksic, V.; Knezevic, P. Antimicrobial and Antioxidative Activity of Extracts and Essential Oils of Myrtus communis L. Microbiol. Res. 2014, 169, 240–254. [Google Scholar] [CrossRef]
- Mutha, R.E.; Tatiya, A.U.; Surana, S.J. Flavonoids as Natural Phenolic Compounds and Their Role in Therapeutics: An Overview. Future J. Pharm. Sci. 2021, 7, 25. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Lamb, A.J. Recent Advances in Understanding the Antibacterial Properties of Flavonoids. Int. J. Antimicrob. Agents 2011, 38, 99–107. [Google Scholar] [CrossRef]
- Yang, D.; Wang, T.; Long, M.; Li, P. Quercetin: Its Main Pharmacological Activity and Potential Application in Clinical Medicine. Oxidative Med. Cell. Longev. 2020, 2020, 8825387. [Google Scholar] [CrossRef]
- Xu, D.; Hu, M.-J.; Wang, Y.-Q.; Cui, Y.-L. Antioxidant Activities of Quercetin and Its Complexes for Medicinal Application. Molecules 2019, 24, 1123. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.V.; Mistry, B.M.; Shinde, S.K.; Syed, R.; Singh, V.; Shin, H.-S. Therapeutic Potential of Quercetin as a Cardiovascular Agent. Eur. J. Med. Chem. 2018, 155, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Rauf, A.; Imran, M.; Khan, I.A.; ur-Rehman, M.-; Gilani, S.A.; Mehmood, Z.; Mubarak, M.S. Anticancer Potential of Quercetin: A Comprehensive Review. Phytother. Res. 2018, 32, 2109–2130. [Google Scholar] [CrossRef] [PubMed]
- Di Petrillo, A.; Orrù, G.; Fais, A.; Fantini, M.C. Quercetin and Its Derivates as Antiviral Potentials: A Comprehensive Review. Phytother. Res. 2022, 36, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.; Ullah, H.; Aschner, M.; Cheang, W.S.; Akkol, E.K. Neuroprotective Effects of Quercetin in Alzheimer’s Disease. Biomolecules 2019, 10, 59. [Google Scholar] [CrossRef]
- Lesjak, M.; Beara, I.; Simin, N.; Pintać, D.; Majkić, T.; Bekvalac, K.; Orčić, D.; Mimica-Dukić, N. Antioxidant and Anti-Inflammatory Activities of Quercetin and Its Derivatives. J. Funct. Foods 2018, 40, 68–75. [Google Scholar] [CrossRef]
- He, Z.; Zhang, X.; Song, Z.; Li, L.; Chang, H.; Li, S.; Zhou, W. Quercetin Inhibits Virulence Properties of Porphyromas gingivalis in Periodontal Disease. Sci. Rep. 2020, 10, 18313. [Google Scholar] [CrossRef]
- Roy, P.K.; Song, M.G.; Park, S.Y. The Inhibitory Effect of Quercetin on Biofilm Formation of Listeria monocytogenes Mixed Culture and Repression of Virulence. Antioxidants 2022, 11, 1733. [Google Scholar] [CrossRef]
- Wang, S.; Yao, J.; Zhou, B.; Yang, J.; Chaudry, M.T.; Wang, M.; Xiao, F.; Li, Y.; Yin, W. Bacteriostatic Effect of Quercetin as an Antibiotic Alternative In Vivo and Its Antibacterial Mechanism In Vitro. J. Food Prot. 2018, 81, 68–78. [Google Scholar] [CrossRef]
- Nguyen, T.L.A.; Bhattacharya, D. Antimicrobial Activity of Quercetin: An Approach to Its Mechanistic Principle. Molecules 2022, 27, 2494. [Google Scholar] [CrossRef]
- Vipin, C.; Saptami, K.; Fida, F.; Mujeeburahiman, M.; Rao, S.S.; Athmika; Arun, A.B.; Rekha, P.D. Potential Synergistic Activity of Quercetin with Antibiotics against Multidrug-Resistant Clinical Strains of Pseudomonas aeruginosa. PLoS ONE 2020, 15, e0241304. [Google Scholar] [CrossRef]
- Pal, A.; Tripathi, A. Quercetin Potentiates Meropenem Activity among Pathogenic Carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii. J. Appl. Microbiol. 2019, 127, 1038–1047. [Google Scholar] [CrossRef]
- Pal, A.; Tripathi, A. Demonstration of Bactericidal and Synergistic Activity of Quercetin with Meropenem among Pathogenic Carbapenem Resistant Escherichia coli and Klebsiella pneumoniae. Microb. Pathog. 2020, 143, 104120. [Google Scholar] [CrossRef]
- Betts, J.W.; Sharili, A.S.; Phee, L.M.; Wareham, D.W. In Vitro Activity of Epigallocatechin Gallate and Quercetin Alone and in Combination versus Clinical Isolates of Methicillin-Resistant Staphylococcus aureus. J. Nat. Prod. 2015, 78, 2145–2148. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial Resistance, Mechanisms and Its Clinical Significance. Dis. Mon. 2020, 66, 100971. [Google Scholar] [CrossRef]
- Reygaert, W.C. An Overview of the Antimicrobial Resistance Mechanisms of Bacteria. AIMS Microbiol. 2018, 4, 482–501. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- O’Neill, J. The Review on Antimicrobial Resistance. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. 2014. Available online: https://amr-review.org/Publications.html (accessed on 20 November 2022).
- Tiwari, P.; Khare, T.; Shriram, V.; Bae, H.; Kumar, V. Plant Synthetic Biology for Producing Potent Phyto-Antimicrobials to Combat Antimicrobial Resistance. Biotechnol. Adv. 2021, 48, 107729. [Google Scholar] [CrossRef]
- Siriwong, S.; Thumanu, K.; Hengpratom, T.; Eumkeb, G. Synergy and Mode of Action of Ceftazidime plus Quercetin or Luteolin on Streptococcus pyogenes. Evid.-Based Complement. Altern. Med. 2015, 2015, 759459. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, Y.; Liu, S.; Ye, D.; Chen, L.; Huang, N.; Zeng, W.; Liao, W.; Zhan, Y.; Zhou, T.; et al. Quercetin Rejuvenates Sensitization of Colistin-Resistant Escherichia coli and Klebsiella pneumoniae Clinical Isolates to Colistin. Front. Chem. 2021, 9, 795150. [Google Scholar] [CrossRef]
- Qu, S.; Dai, C.; Shen, Z.; Tang, Q.; Wang, H.; Zhai, B.; Zhao, L.; Hao, Z. Mechanism of Synergy Between Tetracycline and Quercetin Against Antibiotic Resistant Escherichia coli. Front. Microbiol. 2019, 10, 2536. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-López, R.; Solano-Gálvez, S.G.; Juárez Vignon-Whaley, J.J.; Abello Vaamonde, J.A.; Padró Alonzo, L.A.; Rivera Reséndiz, A.; Muleiro Álvarez, M.; Vega López, E.N.; Franyuti-Kelly, G.; Álvarez-Hernández, D.A.; et al. Acinetobacter baumannii Resistance: A Real Challenge for Clinicians. Antibiotics 2020, 9, 205. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, K.; Holley, R.A. Use of Natural Antimicrobials to Increase Antibiotic Susceptibility of Drug Resistant Bacteria. Int. J. Food Microbiol. 2010, 140, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Langeveld, W.T.; Veldhuizen, E.J.A.; Burt, S.A. Synergy between Essential Oil Components and Antibiotics: A Review. Crit. Rev. Microbiol. 2014, 40, 76–94. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI standard M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Moody, J. Synergism Testing: Broth Microdilution Checkerboard and Broth Macrodilution Methods. In Clinical Microbiology Procedures Handbook; Garcia, L., Ed.; ASM Press: Washington, DC, USA, 2010. [Google Scholar]
- Moody, J.; Knapp, C. Tests to Assess Bactericidal Activity. In Clinical Microbiology Procedures Handbook; Garcia, L., Ed.; ASM Press: Washington, DC, USA, 2010. [Google Scholar]
- Devi, K.P.; Sakthivel, R.; Nisha, S.A.; Suganthy, N.; Pandian, S.K. Eugenol Alters the Integrity of Cell Membrane and Acts against the Nosocomial Pathogen Proteus mirabilis. Arch. Pharm. Res. 2013, 36, 282–292. [Google Scholar] [CrossRef]
- Bendali, F.; Gaillard-Martinie, B.; Hebraud, M.; Sadoun, D. Kinetic of Production and Mode of Action of the Lactobacillus paracasei subsp. paracasei Anti-Listerial Bacteriocin, an Algerian Isolate. LWT-Food Sci. Technol. 2008, 41, 1784–1792. [Google Scholar] [CrossRef]
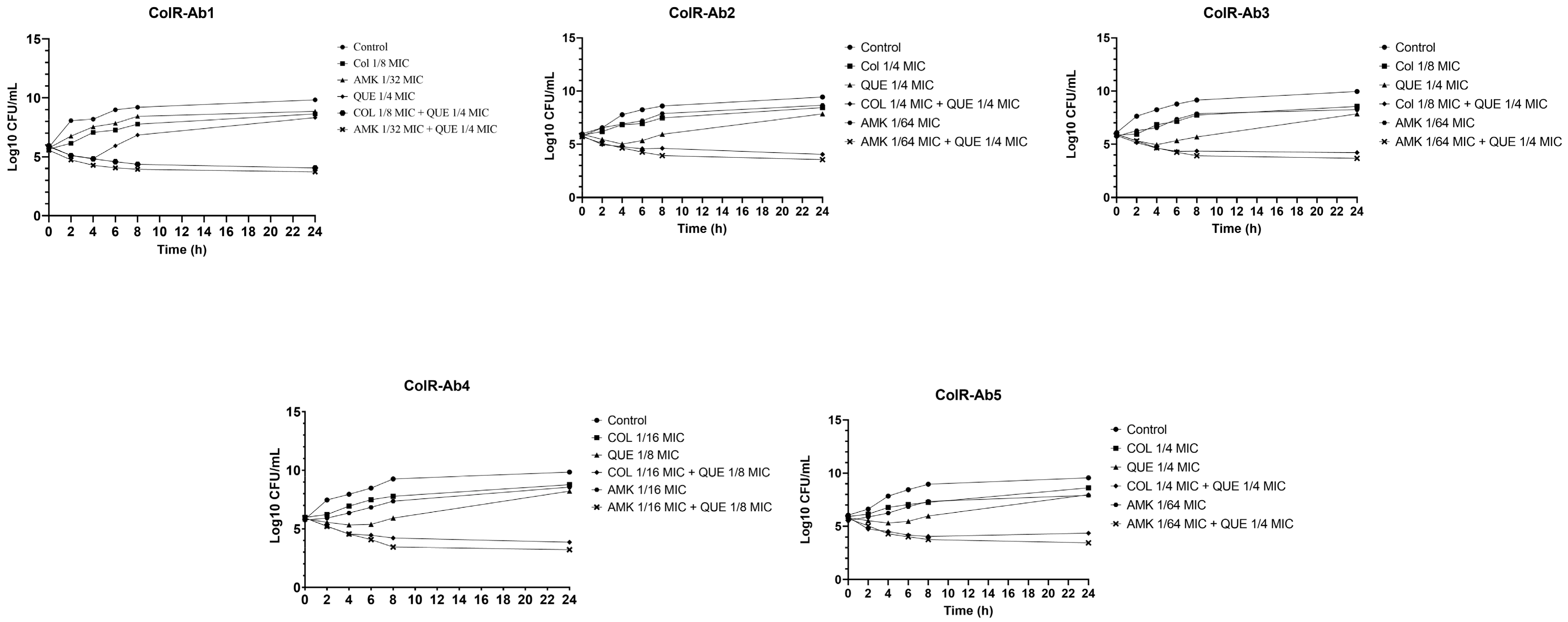

| Strains | MIC (µg/mL) | Mean FICI for QUE Combined With: | |||||
|---|---|---|---|---|---|---|---|
| QUE | COL | MEM | AMK | COL | MEM | AMK | |
| ColR-Ab1 | 128 | 8 | 64 | 8192 | 0.375 (S) | 0.515 (I) | 0.2825 (S) |
| ColR-Ab2 | 256 | 8 | 64 | 8192 | 0.5 (S) | 0.515 (I) | 0.2656 (S) |
| ColR-Ab3 | 256 | 32 | 32 | 8192 | 0.375 (S) | 0.53125 (I) | 0.2656 (S) |
| ColR-Ab4 | 256 | 32 | 64 | 8192 | 0.1875 (S) | 0.515 (I) | 0.1875 (S) |
| ColR-Ab5 | 256 | 8 | 64 | 16,384 | 0.5 (S) | 0.515 (I) | 0.2656 (S) |
| E. coli | 128 | 4 | ≤0.0625 | ≤0.0625 | - | - | - |
| Strains | MIC (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|
| AMK | CIP | GEN | IPM | LVX | MEM | SXT | COL * | |
| ColR-Ab1 | >32 | >1 | >4 | >8 | >8 | >8 | >8/152 | >4 |
| ColR-Ab2 | >32 | >1 | >4 | >8 | >8 | >8 | >8/152 | >4 |
| ColR-Ab3 | >32 | >1 | >4 | >8 | >8 | >8 | >8/152 | >4 |
| ColR-Ab4 | >32 | >1 | >4 | >8 | >8 | >8 | >8/152 | >4 |
| ColR-Ab5 | >32 | >1 | >4 | >8 | >8 | >8 | >8/152 | >4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odabaş Köse, E.; Koyuncu Özyurt, Ö.; Bilmen, S.; Er, H.; Kilit, C.; Aydemir, E. Quercetin: Synergistic Interaction with Antibiotics against Colistin-Resistant Acinetobacter baumannii. Antibiotics 2023, 12, 739. https://doi.org/10.3390/antibiotics12040739
Odabaş Köse E, Koyuncu Özyurt Ö, Bilmen S, Er H, Kilit C, Aydemir E. Quercetin: Synergistic Interaction with Antibiotics against Colistin-Resistant Acinetobacter baumannii. Antibiotics. 2023; 12(4):739. https://doi.org/10.3390/antibiotics12040739
Chicago/Turabian StyleOdabaş Köse, Elif, Özlem Koyuncu Özyurt, Süreyya Bilmen, Hakan Er, Cansu Kilit, and Esra Aydemir. 2023. "Quercetin: Synergistic Interaction with Antibiotics against Colistin-Resistant Acinetobacter baumannii" Antibiotics 12, no. 4: 739. https://doi.org/10.3390/antibiotics12040739
APA StyleOdabaş Köse, E., Koyuncu Özyurt, Ö., Bilmen, S., Er, H., Kilit, C., & Aydemir, E. (2023). Quercetin: Synergistic Interaction with Antibiotics against Colistin-Resistant Acinetobacter baumannii. Antibiotics, 12(4), 739. https://doi.org/10.3390/antibiotics12040739






