Abstract
Antimicrobial resistance (AMR) is one of the world’s industrialized nations’ biggest issues. It has a significant influence on the ecosystem and negatively affects human health. The overuse of antibiotics in the healthcare and agri-food industries has historically been defined as a leading factor, although the use of antimicrobial-containing personal care products plays a significant role in the spread of AMR. Lotions, creams, shampoos, soaps, shower gels, toothpaste, fragrances, and other items are used for everyday grooming and hygiene. However, in addition to the primary ingredients, additives are included to help preserve the product by lowering its microbial load and provide disinfection properties. These same substances are released into the environment, escaping traditional wastewater treatment methods and remaining in ecosystems where they contact microbial communities and promote the spread of resistance. The study of antimicrobial compounds, which are often solely researched from a toxicological point of view, must be resumed considering the recent discoveries, to highlight their contribution to AMR. Parabens, triclocarban, and triclosan are among the most worrying chemicals. To investigate this issue, more effective models must be chosen. Among them, zebrafish is a crucial study system because it allows for the assessment of both the risks associated with exposure to these substances as well as environmental monitoring. Furthermore, artificial intelligence-based computer systems are useful in simplifying the handling of antibiotic resistance data and speeding up drug discovery processes.
1. Introduction
One of the key concerns for industry is the capacity to offer items that are risk-free for consumers while also guaranteeing the preservation of their chemical, physical, and microbiological properties. When it comes to producing edible goods or products used for personal hygiene and cleaning, additives play a significant role in regulating the bacterial load of the items. Without them, the material would quickly expire, and, more importantly, it would put the consumer at risk of unpredictable dangers. Antimicrobial agents are also added in the formulation of medical lotions, soap, creams, or sprays, used for everyday hygiene or the treatment of localized bacterial infections. Some of the most well-known chemicals include parabens, triclocarban, and triclosan. Starting in the mid-1920s, parabens (p-hydroxybenzoic acid esters) were commonly added to cosmetics, food products, and pharmaceuticals; their wide use was related to their low costs and good stability [1]. Triclocarban and triclosan can be found in several personal care products, such as toothpaste, detergents, shampoos, deodorants, and body washes [2]. These substances are considered contaminants of emerging concern because of their occurrence in outdoor and even indoor environments [3,4,5]. Their persistence in several environmental matrices is also the consequence of incomplete elimination by the traditional wastewater treatment process. For instance, analysis performed on effluent samples from wastewater treatment plants showed the presence of high paraben residuals. The most predominant form that was detected was methylparaben, which reached a concentration of 3830 ng/L in United States effluent before 2012 [6]. Parabens were also found in sediments and sludge samples that were collected in Japan and Korea [7].
Triclosan and triclocarban were also found at effluent discharge sites in the United States, Australia, Switzerland, Japan, and China, with concentrations ranging from 10.9 to 241 ng/L for triclosan and 23.9 to 342 ng/L for triclocarban, respectively [2,8,9,10,11,12]. The contamination is aggravated by these compounds’ low solubility, which causes them to accumulate and persist in surface waters and sediments. Surface sediments obtained from northern China’s coastal areas contained at least one of the primary paraben analogs, with values ranging from 1.37 to 24.2 ng/g (dry weight) [13]. The supplementary research involved sediment investigations from various Chinese rivers [14,15], as well as lakes and bays in America and Spain [7,16,17]. However, the issue of accumulation impacts not just terrestrial or aquatic ecosystems, but also the residential environment which serves as an extra storage location for harmful chemicals. An investigation of 80 samples of U.S. indoor dust gathered from residential dwellings and sporting facilities, for example, revealed the presence of antimicrobial compounds such as paraben esters (methyl, butyl, ethyl, benzyl paraben), triclocarban, and triclosan [18].
Several studies have demonstrated the role of these chemicals as possible endocrine disruptors [19,20]. Concerns have been expressed regarding how these substances may impair organism development, particularly in cases of indirect exposure, such as that experienced by pregnant women. Much research on zebrafish has revealed that the developmental processes of early life stages can be affected depending on the concentration examined [21,22,23]. The impacts on development have also been examined on human communities, and several findings have fueled worry [24,25,26].
Usage of Personal Care Products and the Spread of Infectious Diseases
Antimicrobial substances prevent the growth of germs through various mechanisms. The potential for choosing resistant strains indicates the dangers connected to the excessive use of these agents [27,28,29,30,31]. AMR, for example, is a concern in the treatment of Mycobacterium tuberculosis infections, since it limits the number of available antibiotics for its therapy [32]. The same goes for infections caused by Clostridium difficile, which continue to be a severe danger and burden for healthcare systems [33].
Antibiotic overuse in the treatment of infections, particularly in children, is historically linked to the issue of antimicrobial resistance (AMR) [34,35], but also the usage of personal care products plays a key role. These compounds are discharged into the environment, and due to their impact on microbial ecosystems, their persistence dictates when resistance phenomena start to occur. Due to their relatively high concentrations, these chemicals exert selective pressure on the microbial populations that inhabit both domestic and natural habitats [36].
In particular, the spread of more or less hazardous diseases has been encouraged by high cleanliness standards. The use of personal care products is directly tied to this paradox. As hands are a major source of bacterial and viral transmission, handwashing is critical in reducing the risk of infection, and the use of antibacterial soaps has been demonstrated in studies to help prevent cutaneous infections and treat skin lesions [37]. Correct hand cleansing may be effective in reducing bacterial illnesses such as gastrointestinal illness, but the additives contained in soaps may contribute to the spread of resistance. For instance, exposure to triclosan resulted in increased resistance to chloramphenicol and tetracycline in Salmonella enterica serovar Typhimurium [38] and Escherichia coli [39].
Many infectious disease symptoms were much more common in people with poor health or chronic problems who used antibacterial medications [40], signaling that more research is needed. The effects of antibiotic resistance on respiratory system health warrant special consideration [41]. Bacteria that are specifically selected by antimicrobial agents cause airway infections. Pseudomonas aeruginosa, for example, is responsible for lung infections and is resistant to triclosan at high concentrations [42]. Parabens are not effective against Staphylococcus aureus, the causative agent of staphylococcal pneumonia [43]. These are only a few of the hazards connected with utilizing antimicrobials that are detrimental both on their own and in combination with other antibiotics.
This review aims to look at any connections that might exist between using personal care products and developing AMR. We shall pay particular attention to three additives whose widespread use in a variety of preparations has prompted severe environmental concerns (parabens, triclocarban, and triclosan). Given the need to investigate these substances from a novel perspective, new experimental models must be developed to facilitate the collection of new data. Understanding these challenges is a critical step in creating awareness for the preservation of people and the environment.
2. Parabens, Triclocarban, and Triclosan: The Dark Side of Personal Care Products
Parabens, triclocarban, and triclosan are well-known chemicals found in a variety of products. Parabens are preservatives that are added to a variety of items to strengthen their stability over time, such as cosmetics, detergents, or lotions. They are derived from benzoic acid and comprise derivatives characterized by the different lengths of the side chain. Its excessive use has resulted in its release into the environment, where it has contaminated several matrices. Its presence in the environment also influenced microbial ecosystems, leading to the selection of resistant strains. Selvaraj and colleagues (2013) investigated the resistance of bacteria in sewage treatment plant effluents to parabens and discovered that S. aureus was the most resistant while P. aeruginosa is the least resistant. Butyl and ethyl parabens are often more toxic to bacteria [44]. These studies suggested that environmental paraben exposure could contribute to the development of resistance in pathogenic bacteria, leading to detrimental consequences for human health. Nonetheless, there is also concern that these chemicals, once swallowed, may interact with the microbial communities of the human mucous membranes. Even though there is relatively little research, investigations on rats yield fascinating outcomes. For example, low dose methylparaben exposure could influence bacterial composition changes in rats during their adolescence [45]. This finding is critical since gut dysbiosis may be one of the mechanisms permitting multidrug-resistant pathogenic bacteria to colonize the intestinal mucosa [46].
Triclocarban and triclosan are broad-spectrum antibacterial compounds that have found widespread use in a variety of cosmetic products including skin disinfectants and sanitizers. However, because of concerns about their impact on hormones and antibiotic resistance, regulatory bodies such as the FDA and the European Commission have prohibited the use of triclocarban and triclosan in some commodities. Despite this, they are still widely utilized in many consumer products, and once in the environment, these chemicals can be found in water, sediment, aquatic animals, dust, and even human bodies [47]. The presence of triclosan in sewage sludge makes it an important source of antibiotic resistance genes, and mutations in the enoyl-acyl carrier protein (ACP) reductase gene have been discovered in sensitive opportunistic bacteria exposed to triclosan [48]. Triclocarban exposure in sewage sludge can reduce antibiotic tolerance and promote the selection of cross-resistant and multidrug-resistant genes, helping ARGs and MGEs (antibiotic resistance genes and mobile genetic elements) propagate in bacterial communities [49,50].
To acquire a more complete picture, these compounds should not only be studied separately, but also in the context of combination toxicity [51,52]. The environment contains a mix of chemical contaminants, including pharmaceuticals and personal care products, industrial chemicals, nutrients derived from wastewater and manure, and dissolved organic matter. Wastewater treatment plants release these contaminants into rivers, and their quantities can vary depending on social and economic factors. For example, the COVID-19 pandemic has led to increased use of antibacterial substances and disinfectants, certainly impacting the environmental microbial communities [53].
The main properties and characteristics of parabens, triclocarban, and triclosan are summarized in Table 1. Even though they have long been considered safe products, studies have increased to date that warn of the potential risks [54,55,56,57,58,59], with the emergence of AMR being a major concern.

Table 1.
Main characteristics of parabens, triclocarban, and triclosan.
The Mechanisms of Antibiotic Resistance
Numerous studies have shown a connection between the usage of these compounds and the advent of AMR. For example, their impact on the microbial diversity of indoor environments was particularly well-expressed in a study by Fahimipour and colleagues (2018) [67]. Intriguingly, the study revealed that the abundance of microbial taxa with various resistance capacities is reflected in the presence of these chemicals in the household environment. They noticed the rise of phenotypes with cross-resistance to clarithromycin, ampicillin, or tetracycline [67]. These results supported the findings of Hartmann et al. (2016), who investigated the connection between the dust microbiome and the antibacterial substances triclosan, triclocarban, and paraben derivatives [68]. The analysis of dust resistome revealed a high abundance of diverse genes of resistance such as tet(W), blaSRT-1, and erm(B) [68]. They performed different analyses on 44 dust samples collected in an athletic and educational facility, showing a positive correlation between the concentrations of antimicrobial agents and the abundance of resistome (the collection of genes responsible for antibiotic resistance) [68].
The dust accumulated in homes or other buildings represents a suitable substrate for this kind of examination. It contains a sizable portion of biological components, such as microbes. Since indoor communities are strongly influenced by anthropogenic and domestic factors (temperature, humidity, diet, habits, and hygiene) [69], antimicrobial substances, dust quality, and an increase in antibiotic genes might be connected. Other surveys were carried out on dust samples from 63 Canadian houses, 10 private residences in the northwest of Spain, and 18 houses in Belgium, demonstrating that indoor dust may act as a sink of antimicrobial compounds, including the above-mentioned ones [70,71,72]. Dust contamination suggests that these substances may persist in the environment at home and continue to encounter people (especially children, who ingest them more easily).
In addition to the residential environment, these compounds continue to exist outdoors in many matrices, as was previously mentioned in the introduction. The main implication of their persistence is the selection of resistant bacterial communities. According to some studies, exposure to antimicrobials can favor the development of bacteria that are resistant to both biocides and antibiotics. This occurs because some of the genes that give resistance to biocides are found on the same genetic elements as those that provide resistance to antibiotics, and the use of biocides can encourage the spread of these genes among bacterial populations. For example, the presence of triclocarban and triclosan could influence the abundance of the tet(Q) gene, responsible for resistance to tetracycline, in aerobic-activated sludge microcosms [73]. The cross-resistance between triclosan and antibiotics in P. aeruginosa is another example. It had been proven that triclosan and antibiotic exposure might select for equivalent mutations that result in the expression of a multidrug efflux mechanism [74]. In addition, it has been demonstrated that exposure to environmentally relevant concentrations of triclosan could promote the conjugative transfer of multi-resistance genes across different bacterial genera, by the promotion of reactive oxygen species (ROS) generation [75].
Thus, it is evident that biocides may alter permeable bacterial cell walls, making it more challenging for antibiotics to kill germs by penetrating and entering the cells. In a 2005 study, Bredin and colleagues demonstrated that propylparaben can cause the release of potassium from E. coli cells via a permeabilizing impact, in which the porin OmpF appears to be involved [76]. As a result, this compound can select any Gram-negative bacteria with poor porin expression. Those same bacteria have been noted to exhibit a considerable level of β-lactam resistance [77]. In Figure 1, we summarize the main mechanisms of antimicrobial resistance mediated by the biocides reported above.
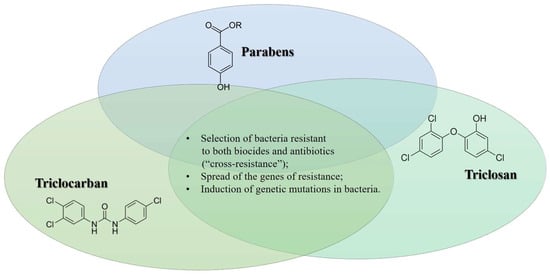
Figure 1.
Biocide and the onset of antimicrobial resistance: summary of the main mechanisms.
Research employing diverse models, ranging from cellular (or two-dimensional) to in vivo, has led to the understanding of resistance pathways, while epidemiological research has contributed significantly to our understanding of the trend and spread of AMR. It is critical to understand how antimicrobial resistance develops and spreads, as well as create measures to prevent and regulate its dissemination. This necessitates interdisciplinary study integrating microbiology, epidemiology, medicine, public health, and social sciences, among others. Through understanding antimicrobial resistance, we may create novel and effective antibiotics, adopt appropriate antibiotic use policies, and improve infection prevention and control strategies, ultimately leading to better health outcomes for individuals and communities.
3. Modeling and Tracking Antimicrobial Resistance: From Bi-Dimensional Models to Artificial Intelligence
The given examples highlight how AMR is an emerging worldwide health issue that is anticipated to get worse over time and eventually kill millions of people. According to the 2019 study on the worldwide burden of bacterial resistance, there were 4.95 million drug-resistant infection-related deaths, of which at least 1.27 million were directly attributable to drug resistance [78]. Therefore, the only factors that contribute to a greater death risk are ischemia and heart attacks. E. coli, S. aureus, K. pneumoniae, S. pneumoniae, and P. aeruginosa are a few of the key pathogens that are thought to be to blame for this rise in mortality.
However, the discovery of a significant link with antimicrobial compounds found in numerous widely used items makes it evident that more research into the mechanisms underlying antibiotic resistance is required, as well as the development of new tracking systems.
3.1. The Potentiality of the Zebrafish Model
Our understanding of the mechanisms underlying antibiotic resistance has surely been aided by the traditional method of researching how various chemicals affect bacterial cells directly [79,80,81,82]. The potential to research resistant infections using the zebrafish model is particularly intriguing because it already has many benefits and enables the investigation of numerous issues (Figure 2).
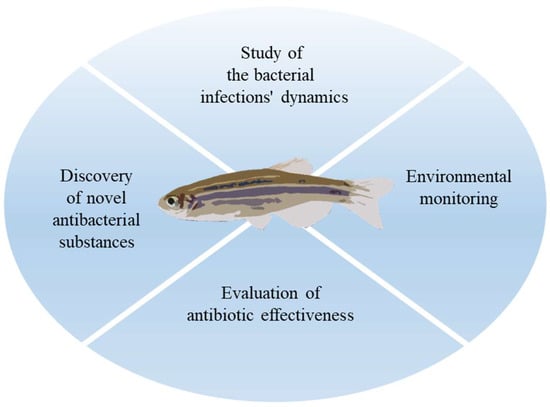
Figure 2.
The use of zebrafish in the study of AMR.
Given that it shares many metabolic, physiological, and developmental processes with humans, zebrafish can be utilized to research antibiotic resistance and the host’s response to bacterial infections [83]. New antibacterial substances can be tested for toxicity and effectiveness using zebrafish [84]. For instance, zebrafish can be exposed to various compound concentrations and several parameters such as survival, rate of development, and organ morphology could be monitored. Utilizing this method, one can find novel chemicals that are effective against particular strains of antibiotic-resistant bacteria [85].
The effects of antimicrobial drugs on development have been studied extensively in both embryonic and larval forms of zebrafish [21,23,86]. In this regard, it will undoubtedly make it possible to assess how antibiotic resistance impacts survival in infection cases and identify new targets to get around the problem of recurrent drug resistance. Zebrafish also serve as a model for environmental monitoring, making it feasible to simulate the effects of exposure to specific biocide doses while also allowing for the evaluation of the interaction with microbiota [87,88,89]. For instance, the study of bacterial species isolated from zebrafish itself raises intriguing questions, as demonstrated by a collection of 43 Aeromonas species that were isolated and recognized from distinct zebrafish specimens selected from various pet stores. The genes for antibiotic resistance have been recognized, and they have helped to establish the MDR (multidrug resistance) indexes [90].
The broad-spectrum antibiotic oxytetracycline (OTC) is an environmental polluter that is commonly used in aquaculture. The zebrafish model was utilized by Almeida and colleagues (2021) to examine the effects of chronic OTC exposure on the microbial population in the environment and the fish’s gut microbiota [91]. They discovered that OTC had a significant impact on the levels of particular bacteria after 5 days and 1 month of exposure [92]. The ability of an organism to recover at the microbiome level following chemical exposure is critical for assessing populations’ ability to recover from situations of intermittent or pulsed contamination [93].
Even more recently, the zebrafish model has proven to be especially useful in assessing how chlorine residues affect the spread of AMR. It is a quite relevant topic since the usage of chlorine-based disinfection products increased during the COVID-19 pandemic. The research showed that freshwater microbial communities can be disturbed by chlorine, even at modest levels, when it is introduced continuously [94]. This may lead to a decrease in their metabolic rate, an increase in potential pathogens, and an acceleration of the transmission of antibiotic resistance genes (ARGs). Zebrafish’s intestinal microbial population is also adversely impacted, which harms their growth and behavior. According to the study, microbial diversity and richness were reduced by exposure to 0.1 mg/L of chlorine, but they soon recovered. After 7 days of exposure, chlorine also had an impact on the majority of microbial species, but by 14 days, the effects had been reversed [94].
3.2. Artificial Intelligence against Antimicrobial Resistance
Traditional research offers a wealth of knowledge regarding the molecular pathways underlying antibiotic resistance, and even experiments carried out directly on sick animals give us a more comprehensive picture. However, new technologies play a significant part in monitoring antibiotic resistance. There may be several options to explore antibiotic resistance thanks to artificial intelligence (AI) [95]. AI may be used, for instance, to analyze vast volumes of data that are produced by studies on antibiotic resistance, spot trends, and make predictions about the spread of antibiotic resistance. AI can also be used to find new therapeutic targets and enhance the identification of new antibiotics. Large datasets of chemical compounds can be analyzed using machine learning algorithms to forecast their qualities, such as their capacity to combat a particular pathogenic bacterium or prevent antibiotic resistance [96]. AI can also help make the use of antibiotics more effective. For instance, patient response to a specific antibiotic can be predicted using machine learning algorithms, customizing treatment, and enhancing clinical efficacy [97]. AI enables health authorities to promptly implement preventive measures by predicting and tracking the spread of illnesses that are resistant to antibiotics in real-time [98]. AI can make a significant contribution to the fight against antibiotic resistance by helping with drug discovery, infection prevention and control, and patient-specific antibiotic treatment [99].
Machine learning, a subfield of artificial intelligence, has emerged as a viable way for dealing with this complexity by constructing algorithms capable of predicting outcomes with minimal human intervention. It can be used to predict resistance phenotypes from pathogen genomic data, to better understand antibiotic processes and medication development, and to aid in antimicrobial stewardship in clinical decision-making. While the area is still in its early stages, machine learning is expected to play a significant role in lowering the global burden of antimicrobial resistance [100]. By accelerating target identification, lead discovery, preclinical and clinical development, machine learning has the potential to revolutionize pharmaceutical drug discovery. Recent studies have shown that machine learning can be applied to the production of novel antimicrobials by learning small molecule structural features from existing screens in the context of antimicrobial development [101]. This advancement has included both novel screening procedures and new algorithms that increase algorithmic learning by utilizing chemical structure representations. Johnson et al. (2019), for example, used a genomic library and machine learning to uncover new therapeutic targets to establish a screening technique for discovering pharmacologic inhibitors of key genes in Mycobacterium tuberculosis [102].
4. Conclusions
Antibiotic resistance is a problem of global importance with repercussions on human health. Personal care products and, in particular, the antimicrobial additives they contain, are released into the environment and meet microbial communities, altering them and selecting resistant bacteria. These substances have also been detected in indoor environments, further increasing the concern about their persistence. We concentrated on three of these additives because they are found in many personal care products: parabens, triclosan, and triclocarban. Although legislation in this area has resulted in a reduction in the percentages of presence allowed in marketed products over time, they are still found in ecosystems due to their proclivity to persist. Therefore, it becomes necessary to research and examine how these compounds may affect microbial ecosystems and result in AMR. To accomplish this goal, new analytical approaches are also required to highlight the AMR processes and, more importantly, to find novel biological targets to solve the issue of resistance. We have concentrated on the use of zebrafish as an alternative to conventional models since it plays a crucial part in monitoring harmful compounds that are released into the environment. The monitoring of antibiotic resistance can also be aided by artificial intelligence and machine learning approaches which can enable the research of this issue through simulations and in silico models.
Author Contributions
Conceptualization, G.C. and C.M.; writing—original draft preparation, G.C. and C.M.; review and editing, E.B., M.P. and M.A.; figure preparation G.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Soni, M.G.; Carabin, I.G.; Burdock, G.A. Safety Assessment of Esters of P-Hydroxybenzoic Acid (Parabens). Food Chem. Toxicol. 2005, 43, 985–1015. [Google Scholar] [CrossRef]
- Halden, R.U.; Lindeman, A.E.; Aiello, A.E.; Andrews, D.; Arnold, W.A.; Fair, P.; Fuoco, R.E.; Geer, L.A.; Johnson, P.I.; Lohmann, R.; et al. The Florence Statement on Triclosan and Triclocarban. Environ. Health Perspect. 2017, 125, 064501. [Google Scholar] [CrossRef] [PubMed]
- Vimalkumar, K.; Seethappan, S.; Pugazhendhi, A. Fate of Triclocarban (TCC) in Aquatic and Terrestrial Systems and Human Exposure. Chemosphere 2019, 230, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, G.; Zhu, Q.; Liao, C. Occurrence of Parabens, Triclosan and Triclocarban in Paired Human Urine and Indoor Dust from Two Typical Cities in China and Its Implications for Human Exposure. Sci. Total Environ. 2021, 786, 147485. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Mortimer, M.; Cheng, H.; Sang, N.; Guo, L.-H. Parabens as Chemicals of Emerging Concern in the Environment and Humans: A Review. Sci. Total Environ. 2021, 778, 146150. [Google Scholar] [CrossRef]
- Loraine, G.A.; Pettigrove, M.E. Seasonal Variations in Concentrations of Pharmaceuticals and Personal Care Products in Drinking Water and Reclaimed Wastewater in Southern California. Environ. Sci. Technol. 2006, 40, 687–695. [Google Scholar] [CrossRef]
- Liao, C.; Lee, S.; Moon, H.-B.; Yamashita, N.; Kannan, K. Parabens in Sediment and Sewage Sludge from the United States, Japan, and Korea: Spatial Distribution and Temporal Trends. Environ. Sci. Technol. 2013, 47, 10895–10902. [Google Scholar] [CrossRef]
- Singer, H.; Müller, S.; Tixier, C.; Pillonel, L. Triclosan: Occurrence and Fate of a Widely Used Biocide in the Aquatic Environment: Field Measurements in Wastewater Treatment Plants, Surface Waters, and Lake Sediments. Environ. Sci. Technol. 2002, 36, 4998–5004. [Google Scholar] [CrossRef]
- Ying, G.-G.; Kookana, R.S. Triclosan in Wastewaters and Biosolids from Australian Wastewater Treatment Plants. Environ. Int. 2007, 33, 199–205. [Google Scholar] [CrossRef]
- Zhao, J.-L.; Ying, G.-G.; Liu, Y.-S.; Chen, F.; Yang, J.-F.; Wang, L. Occurrence and Risks of Triclosan and Triclocarban in the Pearl River System, South China: From Source to the Receiving Environment. J. Hazard. Mater. 2010, 179, 215–222. [Google Scholar] [CrossRef]
- Ying, G.-G.; Kookana, R.S.; Kolpin, D.W. Occurrence and Removal of Pharmaceutically Active Compounds in Sewage Treatment Plants with Different Technologies. J. Environ. Monit. 2009, 11, 1498–1505. [Google Scholar] [CrossRef]
- Nishi, I.; Kawakami, T.; Onodera, S. Monitoring of Triclosan in the Surface Water of the Tone Canal, Japan. Bull. Environ. Contam. Toxicol. 2008, 80, 163–166. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Shi, J.; Wang, X.; Zhu, Q.; Kannan, K. Occurrence and Distribution of Parabens and Bisphenols in Sediment from Northern Chinese Coastal Areas. Environ. Pollut. 2019, 253, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-H.; Feng, J.-L.; Xi, N.-N.; Sun, J.-H. Determination of Paraben and Its Metabolite in Sediments by Ultra Performance Liquid Chromatography-Triple Quadrupole Tandem Mass Spectrometry. Chin. J. Anal. Chem. 2018, 12, 260–264. [Google Scholar]
- Huang, C.; Wu, L.-H.; Liu, G.-Q.; Shi, L.; Guo, Y. Occurrence and Ecological Risk Assessment of Eight Endocrine-Disrupting Chemicals in Urban River Water and Sediments of South China. Arch. Environ. Contam. Toxicol. 2018, 75, 224–235. [Google Scholar] [CrossRef]
- Carmona, E.; Andreu, V.; Picó, Y. Occurrence of Acidic Pharmaceuticals and Personal Care Products in Turia River Basin: From Waste to Drinking Water. Sci. Total Environ. 2014, 484, 53–63. [Google Scholar] [CrossRef]
- Xue, X.; Xue, J.; Liu, W.; Adams, D.H.; Kannan, K. Trophic Magnification of Parabens and Their Metabolites in a Subtropical Marine Food Web. Environ. Sci. Technol. 2017, 51, 780–789. [Google Scholar] [CrossRef]
- Chen, J.; Hartmann, E.M.; Kline, J.; Van Den Wymelenberg, K.; Halden, R.U. Assessment of Human Exposure to Triclocarban, Triclosan and Five Parabens in U.S. Indoor Dust Using Dispersive Solid Phase Extraction Followed by Liquid Chromatography Tandem Mass Spectrometry. J. Hazard. Mater. 2018, 360, 623–630. [Google Scholar] [CrossRef]
- Kenda, M.; Karas, K.N.; Iida, M.; Kojima, H.; Sollner, D.M. Triclocarban, Triclosan, Bromochlorophene, Chlorophene, and Climbazole Effects on Nuclear Receptors: An in Silico and in Vitro Study. Environ. Health Perspect. 2020, 128, 107005. [Google Scholar] [CrossRef]
- Nowak, K.; Ratajczak-Wrona, W.; Górska, M.; Jabłońska, E. Parabens and Their Effects on the Endocrine System. Mol. Cell. Endocrinol. 2018, 474, 238–251. [Google Scholar] [CrossRef]
- Merola, C.; Lai, O.; Conte, A.; Crescenzo, G.; Torelli, T.; Alloro, M.; Perugini, M. Toxicological Assessment and Developmental Abnormalities Induced by Butylparaben and Ethylparaben Exposure in Zebrafish Early-Life Stages. Environ. Toxicol. Pharmacol. 2020, 80, 103504. [Google Scholar] [CrossRef] [PubMed]
- Iannetta, A.; Caioni, G.; Di Vito, V.; Benedetti, E.; Perugini, M.; Merola, C. Developmental Toxicity Induced by Triclosan Exposure in Zebrafish Embryos. Birth Defects Res. 2022, 114, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Caioni, G.; d’Angelo, M.; Panella, G.; Merola, C.; Cimini, A.; Amorena, M.; Benedetti, E.; Perugini, M. Environmentally Relevant Concentrations of Triclocarban Affect Morphological Traits and Melanogenesis in Zebrafish Larvae. Aquat. Toxicol. 2021, 236, 105842. [Google Scholar] [CrossRef] [PubMed]
- Pycke, B.F.G.; Geer, L.A.; Dalloul, M.; Abulafia, O.; Jenck, A.M.; Halden, R.U. Human Fetal Exposure to Triclosan and Triclocarban in an Urban Population from Brooklyn, New York. Environ. Sci. Technol. 2014, 48, 8831–8838. [Google Scholar] [CrossRef] [PubMed]
- Leppert, B.; Strunz, S.; Seiwert, B.; Schlittenbauer, L.; Schlichting, R.; Pfeiffer, C.; Röder, S.; Bauer, M.; Borte, M.; Stangl, G.I.; et al. Maternal Paraben Exposure Triggers Childhood Overweight Development. Nat. Commun. 2020, 11, 561. [Google Scholar] [CrossRef]
- Aker, A.M.; Ferguson, K.K.; Rosario, Z.Y.; Mukherjee, B.; Alshawabkeh, A.N.; Cordero, J.F.; Meeker, J.D. The Associations between Prenatal Exposure to Triclocarban, Phenols and Parabens with Gestational Age and Birth Weight in Northern Puerto Rico. Environ. Res. 2019, 169, 41–51. [Google Scholar] [CrossRef]
- El-Zubeir, I.; El Owni, O. Antimicrobial Resistance of Bacteria Associated with Raw Milk Contaminated by Chemical Preservatives. World J. Dairy Food Sci. 2009, 4, 65–69. [Google Scholar]
- Orús, P.; Gomez-Perez, L.; Leranoz, S.; Berlanga, M. Increasing Antibiotic Resistance in Preservative-Tolerant Bacterial Strains Isolated from Cosmetic Products. Int. Microbiol. 2015, 18, 51–59. [Google Scholar] [CrossRef]
- Romero, J.L.; Grande Burgos, M.J.; Pérez-Pulido, R.; Gálvez, A.; Lucas, R. Resistance to Antibiotics, Biocides, Preservatives and Metals in Bacteria Isolated from Seafoods: Co-Selection of Strains Resistant or Tolerant to Different Classes of Compounds. Front. Microbiol. 2017, 8, 1650. [Google Scholar] [CrossRef]
- Cheng, G.; Ning, J.; Ahmed, S.; Huang, J.; Ullah, R.; An, B.; Hao, H.; Dai, M.; Huang, L.; Wang, X.; et al. Selection and Dissemination of Antimicrobial Resistance in Agri-Food Production. Antimicrob. Resist. Infect. Control 2019, 8, 158. [Google Scholar] [CrossRef]
- Cen, T.; Zhang, X.; Xie, S.; Li, D. Preservatives Accelerate the Horizontal Transfer of Plasmid-Mediated Antimicrobial Resistance Genes via Differential Mechanisms. Environ. Int. 2020, 138, 105544. [Google Scholar] [CrossRef] [PubMed]
- Gygli, S.M.; Borrell, S.; Trauner, A.; Gagneux, S. Antimicrobial Resistance in Mycobacterium Tuberculosis: Mechanistic and Evolutionary Perspectives. FEMS Microbiol. Rev. 2017, 41, 354–373. [Google Scholar] [CrossRef] [PubMed]
- O’Grady, K.; Knight, D.R.; Riley, T.V. Antimicrobial Resistance in Clostridioides Difficile. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 2459–2478. [Google Scholar] [CrossRef]
- Habboush, Y.; Guzman, N. Antibiotic Resistance. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria—A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Yadav, S.; Jadeja, N.B.; Dafale, N.A.; Purohit, H.J.; Kapley, A. Pharmaceuticals and Personal Care Products Mediated Antimicrobial Resistance: Future Challenges. In Pharmaceuticals and Personal Care Products: Waste Management and Treatment Technology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 409–428. ISBN 978-0-12-816189-0. [Google Scholar]
- Larson, E. Hygiene of the Skin: When Is Clean Too Clean? Emerg. Infect. Dis. 2001, 7, 225–230. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Webber, M.A.; Jorgensen, F.; Woodward, M.J.; Piddock, L.J.V.; Humphrey, T.J. Prolonged Treatment of Salmonella Enterica Serovar Typhimurium with Commercial Disinfectants Selects for Multiple Antibiotic Resistance, Increased Efflux and Reduced Invasiveness. J. Antimicrob. Chemother. 2007, 60, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Braoudaki, M.; Hilton, A.C. Low Level of Cross-Resistance between Triclosan and Antibiotics in Escherichia coli K-12 and E. coli O55 Compared to E. coli O157. FEMS Microbiol. Lett. 2004, 235, 305–309. [Google Scholar] [CrossRef]
- Larson, E.L.; Lin, S.X.; Gomez-Pichardo, C.; Della-Latta, P. Effect of Antibacterial Home Cleaning and Handwashing Products on Infectious Disease Symptoms. Ann. Intern. Med. 2004, 140, 321–329. [Google Scholar] [CrossRef]
- Guitor, A.K.; Wright, G.D. Antimicrobial Resistance and Respiratory Infections. Chest 2018, 154, 1202–1212. [Google Scholar] [CrossRef]
- Chuanchuen, R.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. High-Level Triclosan Resistance in Pseudomonas Aeruginosa Is Solely a Result of Efflux. Am. J. Infect. Control 2003, 31, 124–127. [Google Scholar] [CrossRef]
- Bargiota, E.; Rico-Munoz, E.; Davidson, P.M. Lethal Effect of Methyl and Propyl Parabens as Related to Staphylococcus Aureus Lipid Composition. Int. J. Food Microbiol. 1987, 4, 257–266. [Google Scholar] [CrossRef]
- Selvaraj, K.K.; Sivakumar, S.; Sampath, S.; Shanmugam, G.; Sundaresan, U.; Ramaswamy, B.R. Paraben Resistance in Bacteria from Sewage Treatment Plant Effluents in India. Water Sci. Technol. 2013, 68, 2067–2073. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Raikhel, V.; Gopalakrishnan, K.; Fernandez-Hernandez, H.; Lambertini, L.; Manservisi, F.; Falcioni, L.; Bua, L.; Belpoggi, F.L.; Teitelbaum, S.; et al. Effect of Postnatal Low-Dose Exposure to Environmental Chemicals on the Gut Microbiome in a Rodent Model. Microbiome 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Matzaras, R.; Nikopoulou, A.; Protonotariou, E.; Christaki, E. Gut Microbiota Modulation and Prevention of Dysbiosis as an Alternative Approach to Antimicrobial Resistance: A Narrative Review. Yale J. Biol. Med. 2022, 95, 479–494. [Google Scholar]
- Zhang, D.; Lu, S. A Holistic Review on Triclosan and Triclocarban Exposure: Epidemiological Outcomes, Antibiotic Resistance, and Health Risk Assessment. Sci. Total Environ. 2023, 872, 162114. [Google Scholar] [CrossRef]
- Barrett, H.; Sun, J.; Gong, Y.; Yang, P.; Hao, C.; Verreault, J.; Zhang, Y.; Peng, H. Triclosan Is the Predominant Antibacterial Compound in Ontario Sewage Sludge. Environ. Sci. Technol. 2022, 56, 14923–14936. [Google Scholar] [CrossRef]
- Carey, D.E.; McNamara, P.J. Altered Antibiotic Tolerance in Anaerobic Digesters Acclimated to Triclosan or Triclocarban. Chemosphere 2016, 163, 22–26. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, J.; Wang, S.; Zhao, Y.; Dai, H.; Li, D.; Cui, Y.; Li, Z. Triclocarban Shifted the Microbial Communities and Promoted the Spread of Antibiotic Resistance Genes in Nitrifying Granular Sludge System. Bioresour. Technol. 2022, 347, 126429. [Google Scholar] [CrossRef]
- Wang, D.; Ning, Q.; Dong, J.; Brooks, B.W.; You, J. Predicting Mixture Toxicity and Antibiotic Resistance of Fluoroquinolones and Their Photodegradation Products in Escherichia coli. Environ. Pollut. 2020, 262, 114275. [Google Scholar] [CrossRef]
- Manaia, C.M.; Aga, D.S.; Cytryn, E.; Gaze, W.H.; Graham, D.W.; Guo, J.; Leonard, A.F.C.; Li, L.; Murray, A.K.; Nunes, O.C.; et al. The Complex Interplay Between Antibiotic Resistance and Pharmaceutical and Personal Care Products in the Environment. Environ. Toxicol. Chem. 2022. [Google Scholar] [CrossRef]
- Sharma, S.; Singh, A.; Banerjee, T. Antibacterial Agents Used in COVID-19: A Systematic Review and Meta-Analysis. Environ. Sustain. 2021, 4, 503–513. [Google Scholar] [CrossRef]
- Japanese Society of Chemotherapy; Antimicrobial Agents Safety Evaluation Standards Committee; Watanabe, A.; Tokue, Y.; Aoki, N.; Matsumoto, T.; Yanagihara, K.; Higa, F.; Tsuge, H.; Nagashima, M.; et al. Criteria for Safety Evaluation of Antimicrobial Agents. J. Infect. Chemother. 2011, 17, 139–147. [Google Scholar] [CrossRef]
- Kirchhof, M.G.; de Gannes, G.C. The Health Controversies of Parabens. Skin Ther. Lett. 2013, 18, 5–7. [Google Scholar]
- Weatherly, L.M.; Gosse, J.A. Triclosan Exposure, Transformation, and Human Health Effects. J. Toxicol. Environ. Health Part B 2017, 20, 447–469. [Google Scholar] [CrossRef] [PubMed]
- Musee, N. Environmental Risk Assessment of Triclosan and Triclocarban from Personal Care Products in South Africa. Environ. Pollut. 2018, 242, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Yun, H.; Liang, B.; Kong, D.; Li, X.; Wang, A. Fate, Risk and Removal of Triclocarban: A Critical Review. J. Hazard. Mater. 2020, 387, 121944. [Google Scholar] [CrossRef] [PubMed]
- Cherian, P.; Zhu, J.; Bergfeld, W.F.; Belsito, D.V.; Hill, R.A.; Klaassen, C.D.; Liebler, D.C.; Marks, J.G.; Shank, R.C.; Slaga, T.J.; et al. Amended Safety Assessment of Parabens as Used in Cosmetics. Int. J. Toxicol. 2020, 39, 5S–97S. [Google Scholar] [CrossRef]
- Lincho, J.; Martins, R.C.; Gomes, J. Paraben Compounds—Part I: An Overview of Their Characteristics, Detection, and Impacts. Appl. Sci. 2021, 11, 2307. [Google Scholar] [CrossRef]
- Nes, I.F.; Eklund, T. The Effect of Parabens on DNA, RNA and Protein Synthesis in Escherichia coli and Bacillus subtilis. J. Appl. Bacteriol. 1983, 54, 237–242. [Google Scholar] [CrossRef]
- Ma, Y.; Marquis, R.E. Irreversible Paraben Inhibition of Glycolysis by Streptococcus Mutans GS-5. Lett. Appl. Microbiol. 1996, 23, 329–333. [Google Scholar] [CrossRef]
- Freese, E.; Sheu, C.W.; Galliers, E. Function of Lipophilic Acids as Antimicrobial Food Additives. Nature 1973, 241, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Iacopetta, D.; Catalano, A.; Ceramella, J.; Saturnino, C.; Salvagno, L.; Ielo, I.; Drommi, D.; Scali, E.; Plutino, M.R.; Rosace, G.; et al. The Different Facets of Triclocarban: A Review. Molecules 2021, 26, 2811. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.-L.; Stingley, R.L.; Beland, F.A.; Harrouk, W.; Lumpkins, D.L.; Howard, P. Occurrence, Efficacy, Metabolism, and Toxicity of Triclosan. J. Environ. Sci. Health Part C 2010, 28, 147–171. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Yan, K.; Wallis, N.G.; Reed, S.; Moore, T.D.; Rittenhouse, S.F.; DeWolf, W.E., Jr.; Huang, J.; McDevitt, D.; Miller, W.H.; et al. Defining and Combating the Mechanisms of Triclosan Resistance in Clinical Isolates of Staphylococcus Aureus. Antimicrob. Agents Chemother. 2002, 46, 3343–3347. [Google Scholar] [CrossRef]
- Fahimipour, A.K.; Ben Mamaar, S.; McFarland, A.G.; Blaustein, R.A.; Chen, J.; Glawe, A.J.; Kline, J.; Green, J.L.; Halden, R.U.; Van Den Wymelenberg, K.; et al. Antimicrobial Chemicals Associate with Microbial Function and Antibiotic Resistance Indoors. mSystems 2018, 3, e00200-18. [Google Scholar] [CrossRef]
- Hartmann, E.M.; Hickey, R.; Hsu, T.; Betancourt Román, C.M.; Chen, J.; Schwager, R.; Kline, J.; Brown, G.Z.; Halden, R.U.; Huttenhower, C.; et al. Antimicrobial Chemicals Are Associated with Elevated Antibiotic Resistance Genes in the Indoor Dust Microbiome. Environ. Sci. Technol. 2016, 50, 9807–9815. [Google Scholar] [CrossRef]
- Thompson, J.R.; Argyraki, A.; Bashton, M.; Bramwell, L.; Crown, M.; Hursthouse, A.S.; Jabeen, K.; Marinho Reis, P.; Namdeo, A.; Nelson, A.; et al. Bacterial Diversity in House Dust: Characterization of a Core Indoor Microbiome. Front. Environ. Sci. 2021, 9, 533. [Google Scholar] [CrossRef]
- Geens, T.; Roosens, L.; Neels, H.; Covaci, A. Assessment of Human Exposure to Bisphenol-A, Triclosan and Tetrabromobisphenol-A through Indoor Dust Intake in Belgium. Chemosphere 2009, 76, 755–760. [Google Scholar] [CrossRef]
- Canosa, P.; Rodríguez, I.; Rubí, E.; Cela, R. Determination of Parabens and Triclosan in Indoor Dust Using Matrix Solid-Phase Dispersion and Gas Chromatography with Tandem Mass Spectrometry. Anal. Chem. 2007, 79, 1675–1681. [Google Scholar] [CrossRef]
- Fan, X.; Kubwabo, C.; Rasmussen, P.; Jones-Otazo, H. Simultaneous Quantitation of Parabens, Triclosan, and Methyl Triclosan in Indoor House Dust Using Solid Phase Extraction and Gas Chromatography-Mass Spectrometry. J. Environ. Monit. 2010, 12, 1891–1897. [Google Scholar] [CrossRef]
- Son, A.; Kennedy, I.M.; Scow, K.M.; Hristova, K.R. Quantitative Gene Monitoring of Microbial Tetracycline Resistance Using Magnetic Luminescent Nanoparticles. J. Environ. Monit. 2010, 12, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- Chuanchuen, R.; Beinlich, K.; Hoang, T.T.; Becher, A.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. Cross-Resistance between Triclosan and Antibiotics in Pseudomonas Aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Mutant Strain to Triclosan Selects NfxB Mutants Overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 2001, 45, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Wang, Y.; Li, J.; Mao, L.; Nguyen, S.H.; Duarte, T.; Coin, L.; Bond, P.; Yuan, Z.; Guo, J. Triclosan at Environmentally Relevant Concentrations Promotes Horizontal Transfer of Multidrug Resistance Genes within and across Bacterial Genera. Environ. Int. 2018, 121, 1217–1226. [Google Scholar] [CrossRef]
- Bredin, J.; Davin-Régli, A.; Pagès, J.-M. Propyl Paraben Induces Potassium Efflux in Escherichia coli. J. Antimicrob. Chemother. 2005, 55, 1013–1015. [Google Scholar] [CrossRef] [PubMed]
- Pagès, J.-M. Bacterial porin and antibiotic susceptibility. Med. Sci. 2004, 20, 346–351. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Levy, S.B. Active Efflux Mechanisms for Antimicrobial Resistance. Antimicrob. Agents Chemother. 1992, 36, 695–703. [Google Scholar] [CrossRef]
- Nikolaidis, I.; Favini-Stabile, S.; Dessen, A. Resistance to Antibiotics Targeted to the Bacterial Cell Wall. Protein Sci. 2014, 23, 243–259. [Google Scholar] [CrossRef]
- Willers, C.; Wentzel, J.F.; du Plessis, L.H.; Gouws, C.; Hamman, J.H. Efflux as a Mechanism of Antimicrobial Drug Resistance in Clinical Relevant Microorganisms: The Role of Efflux Inhibitors. Expert Opin. Ther. Targets 2017, 21, 23–36. [Google Scholar] [CrossRef]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Neely, M.N. The Zebrafish as a Model for Human Bacterial Infections. Methods Mol. Biol. 2017, 1535, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Rashidian, G.; Boldaji, J.T.; Rainis, S.; Prokić, M.D.; Faggio, C. Oregano (Origanum vulgare) Extract Enhances Zebrafish (Danio rerio) Growth Performance, Serum and Mucus Innate Immune Responses and Resistance against Aeromonas Hydrophila Challenge. Animals 2021, 11, 299. [Google Scholar] [CrossRef] [PubMed]
- Jijie, R.; Mihalache, G.; Balmus, I.-M.; Strungaru, S.-A.; Baltag, E.S.; Ciobica, A.; Nicoara, M.; Faggio, C. Zebrafish as a Screening Model to Study the Single and Joint Effects of Antibiotics. Pharmaceuticals 2021, 14, 578. [Google Scholar] [CrossRef] [PubMed]
- Merola, C.; Perugini, M.; Conte, A.; Angelozzi, G.; Bozzelli, M.; Amorena, M. Embryotoxicity of Methylparaben to Zebrafish (Danio rerio) Early-Life Stages. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 236, 108792. [Google Scholar] [CrossRef]
- Loftie-Eaton, W.; Crabtree, A.; Perry, D.; Millstein, J.; Baytosh, J.; Stalder, T.; Robison, B.D.; Forney, L.J.; Top, E.M. Contagious Antibiotic Resistance: Plasmid Transfer among Bacterial Residents of the Zebrafish Gut. Appl. Environ. Microbiol. 2021, 87, e02735-20. [Google Scholar] [CrossRef]
- Milanović, V.; Cardinali, F.; Aquilanti, L.; Maoloni, A.; Garofalo, C.; Zarantoniello, M.; Olivotto, I.; Riolo, P.; Ruschioni, S.; Isidoro, N.; et al. Quantitative Assessment of Transferable Antibiotic Resistance Genes in Zebrafish (Danio rerio) Fed Hermetia Illucens-Based Feed. Anim. Feed. Sci. Technol. 2021, 277, 114978. [Google Scholar] [CrossRef]
- Rasheed, S.; Fries, F.; Müller, R.; Herrmann, J. Zebrafish: An Attractive Model to Study Staphylococcus Aureus Infection and Its Use as a Drug Discovery Tool. Pharmaceuticals 2021, 14, 594. [Google Scholar] [CrossRef]
- Hossain, S.; Dahanayake, P.S.; De Silva, B.C.J.; Wickramanayake, M.V.K.S.; Wimalasena, S.H.M.P.; Heo, G.-J. Multidrug Resistant Aeromonas Spp. Isolated from Zebrafish (Danio rerio): Antibiogram, Antimicrobial Resistance Genes and Class 1 Integron Gene Cassettes. Lett. Appl. Microbiol. 2019, 68, 370–377. [Google Scholar] [CrossRef]
- Almeida, A.R.; Alves, M.; Domingues, I.; Henriques, I. The Impact of Antibiotic Exposure in Water and Zebrafish Gut Microbiomes: A 16S RRNA Gene-Based Metagenomic Analysis. Ecotoxicol. Environ. Saf. 2019, 186, 109771. [Google Scholar] [CrossRef]
- Almeida, A.R.; Tacão, M.; Soares, J.; Domingues, I.; Henriques, I. Tetracycline-Resistant Bacteria Selected from Water and Zebrafish after Antibiotic Exposure. Int. J. Environ. Res. Public Health 2021, 18, 3218. [Google Scholar] [CrossRef]
- Almeida, A.R.; Domingues, I.; Henriques, I. Zebrafish and Water Microbiome Recovery after Oxytetracycline Exposure. Environ. Pollut. 2021, 272, 116371. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Q.; Lu, T.; Zhang, J.; Sun, L.; Hu, B.; Hu, J.; Peñuelas, J.; Zhu, L.; Qian, H. Residual Chlorine Disrupts the Microbial Communities and Spreads Antibiotic Resistance in Freshwater. J. Hazard. Mater. 2022, 423, 127152. [Google Scholar] [CrossRef] [PubMed]
- Popa, S.L.; Pop, C.; Dita, M.O.; Brata, V.D.; Bolchis, R.; Czako, Z.; Saadani, M.M.; Ismaiel, A.; Dumitrascu, D.I.; Grad, S.; et al. Deep Learning and Antibiotic Resistance. Antibiotics 2022, 11, 1674. [Google Scholar] [CrossRef]
- Melo, M.C.R.; Maasch, J.R.M.A.; de la Fuente-Nunez, C. Accelerating Antibiotic Discovery through Artificial Intelligence. Commun. Biol. 2021, 4, 1050. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Maguire, F.; Tsang, K.K.; Gouliouris, T.; Peacock, S.J.; McAllister, T.A.; McArthur, A.G.; Beiko, R.G. Machine Learning for Antimicrobial Resistance Prediction: Current Practice, Limitations, and Clinical Perspective. Clin. Microbiol. Rev. 2022, 35, e0017921. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.-H.; Nguyen, N.Q.; Pham, H.T. A New Hope in the Fight Against Antimicrobial Resistance with Artificial Intelligence. IDR 2022, 15, 2685–2688. [Google Scholar] [CrossRef] [PubMed]
- Feretzakis, G.; Sakagianni, A.; Loupelis, E.; Kalles, D.; Skarmoutsou, N.; Martsoukou, M.; Christopoulos, C.; Lada, M.; Petropoulou, S.; Velentza, A.; et al. Machine Learning for Antibiotic Resistance Prediction: A Prototype Using Off-the-Shelf Techniques and Entry-Level Data to Guide Empiric Antimicrobial Therapy. Healthc. Inform. Res. 2021, 27, 214–221. [Google Scholar] [CrossRef]
- Anahtar, M.N.; Yang, J.H.; Kanjilal, S. Applications of Machine Learning to the Problem of Antimicrobial Resistance: An Emerging Model for Translational Research. J. Clin. Microbiol. 2021, 59, e01260-20. [Google Scholar] [CrossRef]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of Machine Learning in Drug Discovery and Development. Nat. Rev. Drug Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef]
- Johnson, E.O.; LaVerriere, E.; Office, E.; Stanley, M.; Meyer, E.; Kawate, T.; Gomez, J.E.; Audette, R.E.; Bandyopadhyay, N.; Betancourt, N.; et al. Large-Scale Chemical-Genetics Yields New M. Tuberculosis Inhibitor Classes. Nature 2019, 571, 72–78. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).