Abstract
Dairy foods are a staple in Lebanon, a low- and middle-income country that has been experiencing serious challenges to food safety and antimicrobial stewardship among other issues. The microbiological acceptability of dairy products has been of increasing concern. This is partially due to the failing economy and prolonged power outages that affect the quality of raw material and disrupt the dairy cold chain, respectively. Therefore, we assessed the microbiological acceptability of Akkawi, a popular white-brined cheese in Lebanon. For this purpose, we quantified the densities of Escherichia coli (a fecal indicator) and Staphylococcus aureus in cheeses collected from Lebanese retail stores. Additionally, we evaluated the antibiotic resistance profiles of the E. coli isolated from the cheese. E. coli and S. aureus were detected in 40 (80%) and 16 (32%) of the 50 cheese samples, respectively. Notably, 40 (80%) and 16 (32%) of the samples exceeded the maximum permissible limit of E. coli and S. aureus, respectively. A high percentage of the 118 E. coli isolated from the cheeses showed resistance to clinically and agriculturally important antibiotics, while 89 (75%) isolates were classified as multidrug-resistant (MDR). Given that Akkawi can be consumed without cooking, our findings highlight serious food safety and antimicrobial resistance problems that require immediate interventions.
1. Introduction
Dairy products are important components of a healthy and well-balanced diet [1]. White cheeses are particularly popular in many Mediterranean countries and are considered a nutritious staple food, providing proteins and micronutrients like calcium and phosphorous [2]. However, cheeses are high-risk foods that can be consumed without cooking, and contaminated cheeses can result in serious foodborne diseases. For example, several foodborne outbreaks were linked to cheese contaminated with Escherichia coli, Salmonella serovars, Listeria monocytogenes, and Staphylococcus aureus in Europe [3,4,5,6,7]. Additionally, several studies have reported the detection of E. coli, S. aureus, and L. monocytogenes in unpasteurized soft cheeses in the USA [2,8]. These contaminants pose a problem, because S. aureus is known to produce heat-stable enterotoxins that cause gastrointestinal disease [9], while L. monocytogenes is a major foodborne pathogen that is associated with severe health complications and deaths in pregnant women and immunocompromised individuals [10]. Similarly, pathogenic E. coli, such as Shiga toxin-producing E. coli O157:H7, can cause serious infections. Furthermore, it has been argued that foodborne pathogens are even more problematic in low- and middle-income countries that might suffer from under-developed public health systems [11].
The dairy market has a major agricultural and economic impact in Lebanon, being valued at $200 million USD and providing income for rural communities [12]. According to a report in 2016, 62,000 metric tons of dairy products are produced each year, with an average consumption of 24 kg of white cheese per capita in Lebanon [13]. Therefore, monitoring the safety of dairy products and ensuring the compliance of dairies are particularly important to stakeholders. However, Lebanon is currently facing major challenges to food safety due to weak governmental oversight, outdated national standards for microbiological and chemical contaminants, scarcity of baseline data, under-developed food safety monitoring systems, frail enforcement of food safety laws, and widespread pollution among others [14]. Due to these deficiencies, 42 outbreaks of foodborne disease, mainly associated with Salmonella infections, were reported in 2010, which is notable for a small country like Lebanon [15]. Additionally, the average number of reported food poisoning cases in Lebanon was 449 (±143) per year from 2012 to 2022 [16]. However, available data underestimate the reality of foodborne illnesses in Lebanon due to the lack of effective surveillance programs, and many foodborne pathogens, including Shigella spp., Campylobacter spp., L. monocytogenes, Cryptosporidium spp., norovirus, and hepatitis A virus among others, are not commonly investigated in clinical laboratories [17]. Furthermore, in the first ever nationwide study on food safety in Lebanon that was published in 2020, 28.7% of all tested foods were deemed microbiologically unsafe per the Lebanese ministry of public health [14]. The same report indicated that 28.3% of the dairy samples were microbiologically unacceptable, with bacterial pathogens such as L. monocytogenes being detected in cheeses. However, the prevalence of the pathogens and the contamination of different types of cheese were not thoroughly investigated. Given that Lebanon has been experiencing a protracted economic crisis, an influx of refugees (>1.5 million Syrian refugees), and shortages in medical supplies and hospital services [18,19,20], it is clear that foodborne illnesses will increase further, perpetuating the cycle of poverty and jeopardizing the well-being of the vulnerable, disenfranchised, and impoverished communities in Lebanon. Therefore, it is critical to assess the safety of important foods in Lebanon. However, only a limited number of studies have investigated the microbiological and chemical safety of cheese [14,21].
The problem of foodborne illness is aggravated by the rise of difficult-to-treat pathogens that acquire resistance to antimicrobial treatments [22]. Indeed, the extensive reliance on antibiotics in animal agriculture for the treatment and prevention of disease as well as for growth promotion has exacerbated the emergence of drug-resistant microorganisms [23]. The dissemination of multidrug-resistant (MDR) bacterial isolates in various food commodities, including milk and dairy products, is well documented in different countries [24,25,26,27]. Notably, the misuse of antimicrobials and the alarmingly high prevalence of drug resistance are also well-documented in the agricultural and environmental settings in Lebanon [28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47]. However, reports on the dissemination of antimicrobial resistance (AMR) in food commodities, especially dairy products, are scarce and these issues have not been thoroughly investigated in Lebanon [21,36]. Therefore, it was necessary to investigate the antimicrobial resistance profile of the bacterial contaminants in cheese in Lebanon.
The contamination of cheeses can occur during any step of processing, from milk production to cheese handling and storage. The densities of bacteria like E. coli and S. aureus are used as microbiological indicators to determine the acceptability and safety of cheese [9]. High bacterial densities of fecal indicators, like E. coli, also suggest the possible contamination of the food with enteric pathogens [48,49]. Additionally, the antibiotic resistance profiles of E. coli have been used as indicators for the emergence and spread of AMR in foods [50]. Taken together, we aimed to evaluate the microbiological safety of Akkawi, a highly popular white cheese, which we collected from the retail markets in Lebanon. Specifically, we quantified the prevalence and densities of E. coli and S. aureus in Akkawi cheese and compared them to the official cheese standards adopted by LIBNOR (Lebanese Standards Institute) [51,52]. Additionally, we assessed the antibiotic resistance phenotypes of the E. coli isolated from the cheese in order to evaluate the dissemination of AMR in this food product.
2. Results
2.1. Prevalence and Densities of E. coli and S. aureus
E. coli was detected in 40 (80%) of the 50 cheese samples. The average densities of E. coli ranged between 1 × 103 and 2 × 107 CFU/g. The highest average density of E. coli was in Akkawi samples [AA] at 2 × 107 CFU/g (Figure 1). S. aureus was detected in 16 (32%) of the cheese samples, with densities ranging between 1 × 102 and 2 × 105 CFU/g. The highest S. aureus density (2 x105 CFU/g) was detected in sample [SA] (Figure 1). There was no statistically significant (p value > 0.05) correlation between E. coli and S. aureus densities. The identities of the E. coli and S. aureus isolates selected from positive cheese samples were further confirmed by PCR analysis.
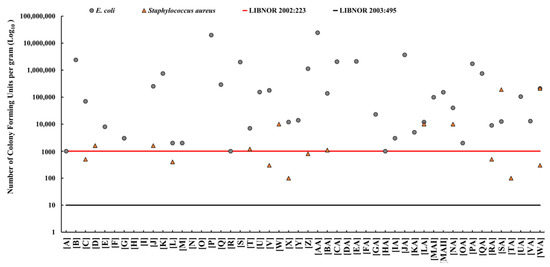
Figure 1.
The prevalence and densities (colony-forming units per gram; CFU/g) of Escherichia coli (grey dot) and Staphylococcus aureus (orange triangle) in Akkawi cheese in Lebanon. The red line indicates the maximum allowable limit for E. coli and S. aureus (1000 CFU/g) according to the Lebanese Standards Institution (LIBNOR, standard 2002:223). The black line indicates the permissible limit of E. coli and S. aureus (10 CFU/g) according to the LIBNOR standard 2003:495.
2.2. Evaluation of Cheese Accessibility by Comparing E. coli and S. aureus Densities to LIBNOR Standards
According to LIBNOR 2002:223, the maximum allowable limit of E. coli and S. aureus in cheese is 1000 CFU/g; however, this limit was decreased to 10 CFU/g in the updated LIBNOR 2003:495. Subsequently, using E. coli densities, 40 (80%) of the cheese samples were found to be microbiologically unacceptable (unsafe) and would be rejected. Furthermore, 8 (16%) of the cheese samples exceeded the maximum allowable limit of 1000 CFU/g of S. aureus based on LIBNOR (2002:223). Using the updated LIBNOR (2003:495) standard, the number of rejected samples increased to 16 (32%) for S. aureus and remained at 80% for E. coli (Figure 1).
2.3. Antimicrobial Susceptibility Profiles of E. coli Isolated from Cheese Samples
The 118 E. coli isolates were resistant to the control antibiotics PEN and ERY as expected. Furthermore, among the 118 isolates, resistance was noted against AMP (67% of the isolates), AMC (51%), FEP (19%), CTX (41%), LEX (67%), CFM (17%), DOR (39%), IPM (19%), MEM (37%), GEN (41%), KAN (36%), STR (58%), TET (40%), CIP (5%), NOR (4%), SXT (38%), and CHL (28%) (Figure 2 and Figure 3). Intermediate resistance was observed against AMP (9%), AMC (25%), FEP (49%), CTX (26%), CFM (18%), DOR (34%), IPM (35%), MEM (36%), GEN (14%), KAN (42%), STR (35%), TET (3%,), CIP (42%), NOR (3%), SXT (8%), and CHL (15%) (Figure 2 and Figure 3). Notably, 89 of the 118 isolates (75%) were classified as MDR, thus showing resistance to at least three different classes of antibiotics. Furthermore, 16 (14%), 11 (9%), and 15 (13%) of the 118 isolates were resistant to 8, 7, and 6 classes of antibiotics, respectively (Figure 3C). Of particular concern was the observation that 67 of the 118 isolates (56.7%) were resistant to carbapenems (DOR, IPM, and/or MEM) (Figure 2 and Table 1). Five distinct clusters with different AMR profiles were identified by using the hierarchical cluster analysis (HCL) method, highlighting the diversity of the AMR phenotypes of the isolates. Resistance to AMP-AMC-GEN-KAN-STR was prominent in clusters 1 and 5. Most of the isolates that resisted 5 and more classes of antibiotics were placed in clusters 1, 2, and 5. Isolates that were resistant to carbapenems (DOR-MEM) were found in clusters 2 and 4, while resistance to aminoglycosides (GEN-KAN-STR) was mainly found in clusters 1, 2, 4, and 5. Notably, clusters 2 and 5 showed high resistance to trimethoprim-sulfamethoxazole and chloramphenicol (SXT-CHL) (Figure 2).
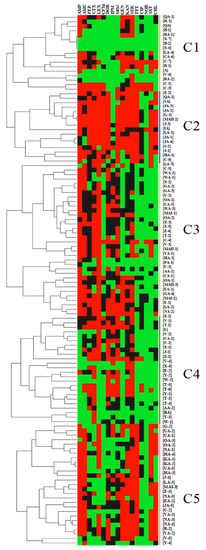
Figure 2.
Hierarchical clustering of 118 E. coli isolated from Akkawi cheese in Lebanon. The isolate labels (IDs) refer to the associated cheese sample and the isolate number (if more than one E. coli was isolated from the same cheese sample). For example, [B-1] indicates that E. coli isolate #1 that was retrieved from Akkawi sample B. Squares in red represent resistance while those in black and green indicate intermediate resistance and susceptibility, respectively. The clusters are represented by the letter C.
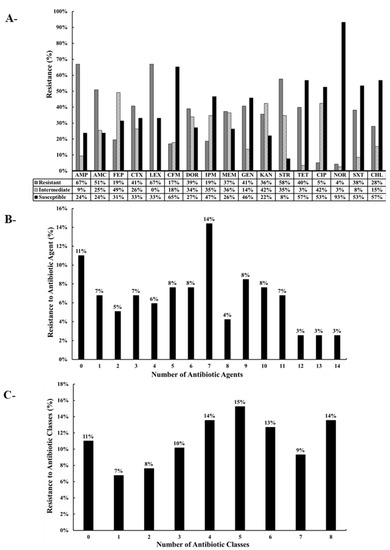
Figure 3.
(A) Percentage of antibiotic-resistant of E. coli recovered from Akkawi cheese in Lebanon. Ampicillin (AMP), amoxicillin + clavulanic acid (AMC), cefepime (FEP), cefotaxime (CTX), cephalexin (LEX), cefixime (CFM), doripenem (DOR), imipenem (IPM), meropenem (MEM), gentamicin (GEN), kanamycin (KAN), streptomycin (STR), tetracycline (TET), ciprofloxacin (CIP), norfloxacin (NOR), trimethoprim + sulfamethoxazole (SXT), and chloramphenicol (CHL). The antibiotics are arranged according to the order of antibiotics/classes listed in the CLSI guidelines. (B) Percentage of isolates (%) resistant to the different antibiotic agents. (C) Percentage of isolates (%) resistant to different antibiotic classes.

Table 1.
Antibiotic Resistance Profiles of 118 E. coli isolated from Akkawi cheese collected from Retail Stores in Beirut, Lebanon.
3. Discussion
Access to safe and nutritious food is necessary in order to maintain a healthy and productive community. Food safety is also a key to food security, which is especially critical in a country like Lebanon that has been experiencing increasing poverty due to an economic collapse. The latter has exacerbated pollution, the debilitation of infrastructure, and the proliferation of infectious diseases and antimicrobial resistance in the country [14,36,53]. Consequently, monitoring the safety of important foods is a critical need in Lebanon. Here, we focused on the safety of Akkawi, a white-brined cheese that is widely consumed (with or without cooking) in Lebanon and neighboring countries [54].
Our results revealed relatively high densities of E. coli and S. aureus in 80% and 32% of the cheese samples, respectively (Figure 1). We also found that 80% of the cheese samples would be rejected (deemed unacceptable for human consumption) based on the E. coli densities and LIBNOR standards (2002:223 and 2003:495), while up to 32% of the samples would be rejected based on S. aureus densities (Figure 1). Therefore, this study reported higher non-conformity rates in comparison to the limited studies available on dairy products in Lebanon [14,21]. Beyond the use of bacterial indicators, it is known that the enterotoxins secreted by S. aureus can cause gastrointestinal disease [9]. Given that a considerable number of people (~25% in some populations) carry S. aureus on their skin and/or in their nasal cavities, unhygienic practices while handling the cheese can lead to contamination [55,56]. This highlights the potential role of food handlers in the contamination of the cheese with important pathogens. Notably, it was estimated that S. aureus causes around 241,000 foodborne illnesses in the USA annually [55].
The high densities of E. coli, a fecal indicator, suggest the potential contamination of these cheeses with other enteric pathogens associated with fecal contamination [24,50]. Taken together, these results corroborated our previous analysis that showed that dairy foods, including cheeses, were at high risk of contamination with coliforms, L. monocytogenes, and E. coli [14]. Furthermore, fecal contamination can potentially occur at any stage of Akkawi cheese production. Although Akkawi is commonly produced from pasteurized milk, contamination can result from deficiencies in pasteurization as well as post-pasteurization processes that include hand-packing the cheese in draining hoops and curing it in brine solutions. Therefore, contamination can be attributed to the weak implementation of good hygienic practices among food handlers and food producers and/ or deficiencies in production and storage due to economic challenges that include unsustainable access to sources of electricity, clean water, and high-quality raw material.
Akkawi cheeses are not always sold in vacuum-sealed packages in Lebanon, especially those that are sourced from traditional and semi-modern dairies that usually provide the cheese in containers (buckets) filled with brine solution, and food handlers have to partition the cheese according to customers’ needs at the retail store. During sampling, we observed that the food handlers were wearing gloves, but that the gloves were used for extended periods of time and were not changed regularly. Moreover, the same cutting boards and knives were used to cut different cheese types and were not cleaned regularly, which increase the risk of cross-contamination. However, we found that even vacuum-sealed Akkawi cheese samples were contaminated with high densities of E. coli and S. aureus, which also suggested that the contamination might have occurred at the processing facilities prior to reaching the market, as explained earlier [25]. Beyond this, it is important to highlight that environmental pollution (e.g., contaminated water sources and pastures) can have major impacts on the safety and quality of dairy products. For example, a report in 2013 showed that the farming environment has a significant effect on the microbiological quality of milk [26], which would persist under deficient cheese production controls and processes. Taken together, strengthening food safety knowledge, endorsing the application of good personal hygiene, ensuring the availability of necessary factors (power, clean water and environment, and good quality raw material among others), establishing a sustainable contaminant-monitoring programs, and adherence to food safety standards are needed in order to maintain the safety of cheese and decrease the burden of foodborne illnesses.
The high prevalence of MDR E. coli in Akkawi cheese was not surprising (Figure 2 and Figure 3; Table 1), because several studies have reported a widespread antibiotic resistance in the Lebanese community, as well as in environmental and food matrices [29,34,35,37,38,39,46]. A recent study on dairy products in Lebanon screened 29 E. coli isolates that were found to be resistant to AMC (69% of isolates), CFM (11%), CHL (31%), and STR (100%), and all the tested isolates were multidrug-resistant [21]. However, in the aforementioned study, the number of E. coli isolates (n = 29) and antibiotics tested was very limited. In comparison, we tested 118 E. coli against 19 antibiotics that belonged to 9 different classes. In our study, the resistance against penicillins; AMP (67% of isolates), cephalosporins; FEP (19%,), CTX (41%), LEX (67%), CFM (17%), aminoglycosides; GEN (41%), KAN (36%), STR (58%), tetracyclines; TET (40%), and sulphonamides; SXT (38%) (Figure 3) was concerning but also expected, as these classes of antibiotics are heavily used in dairy cattle farming [57,58]. Our observations were further corroborated by a previous study that suggested that elevated levels of resistance against gentamicin can be attributed to the high use of this antibiotic in feed additives on Lebanese farms [59]. Additionally, another study conducted on dairy farms reported that aminoglycosides, which are normally used in clinical settings, are frequently used on animal farms in Lebanon [60]. Similarly, tetracycline was also reported to be extensively used in the Lebanese agricultural sector [60].
We noted that 56.7% of the E. coli isolates in our study were resistant to carbapenems, which is highly concerning. Carbapenems are considered last-resort antibiotics that are used as a salvage therapy to treat complicated MDR and extensively drug-resistant (XDR) Gram-negative infections [61]. When bacterial isolates develop resistance to carbapenems, colistin, one of the highest priority critically important antibiotics, is used as a viable treatment option [61]. However, the wide-emergence of resistance to colistin, due to the presence of the mobile colistin-resistant gene (mcr), has been well-document in Lebanon in various agricultural and environmental matrices [28,29,33,34,35,37,38,39,41,45,46]. The occurrence of resistance to critically important antibiotics in the food chain is a threat to public health and agriculture, heralding a rise in difficult-to-treat infections in both humans and farm animals. The high dissemination of antibiotic resistance in Lebanon has been attributed to the extensive reliance on and misuse of antibiotics in both animal farming and clinical settings [23]. It has been also established that there is a notable gap in knowledge about judicious antibiotic use among farmers in Lebanon [62]. For instance, dairy farmers believe that the use of antibiotics for prophylaxis and growth promotion is always beneficial, and that stopping the administration of antibiotics to their animals will lead to major economic losses [62]. Taken together, an immediate action plan is needed to address the dissemination of antibiotic resistance in Akkawi cheese and other food products.
4. Materials and Methods
4.1. Sample Collection
A total of 50 Akkawi cheese samples were aseptically collected from 16 major retail stores across Beirut, the capital of Lebanon, between September 2020 and October 2021. Forty-three cheese samples belonged to 9 brands, while 7 were unbranded. All samples were made from cows’ milk and collected from different lots. While Akkawi is normally made from pasteurized milk, this process was not evident in cases of unbranded cheese samples. The cheese samples were transported in a cooler with ice and processed immediately upon arrival at the laboratory (within 2 h of collection).
4.2. Sample Processing and Enumeration of E. coli and S. aureus
Each sample was aseptically processed by transferring 25 g of cheese to a sterile stomacher bag (Fisher Scientific, New Hampshire, USA) filled with 225 mL of buffered peptone water (BPW) (Oxoid, Hampshire, UK) [36]. The samples were then homogenized for 1 min at 250 rpms using a stomacher (Thomas Scientific, NJ, USA). The homogenates were 10-fold serially diluted (10−1, 10−2, 10−3, 10−4), and 100 µL of each dilution was plated in duplicate on the selective Chromogenic Tryptone Bile X-Glucuronic (TBX) agar (Oxoid, Hampshire, UK), and Baird-Parker (BPA) agar (Millipore-Sigma, Darmstadt, Germany) for the enumeration and isolation of E. coli and S. aureus, respectively. TBX plates were incubated at 37 °C for 24 h, and BPA plates were left for 48 h at 37 °C under aerobic conditions. After incubation, colonies that matched the diagnostic phenotypes of E. coli (Blue/Green colonies on TBX) and S. aureus (black shiny colonies with a halo on BPA) were counted, and the bacterial densities were reported as colony-forming units per gram of cheese (CFU/g) by averaging the counts from the duplicates.
The microbiological quality of the cheese (acceptability/rejection) was determined by comparing E. coli and S. aureus counts (CFU/g) to the two available standards published and adopted by LIBNOR (2002:223 and 2003:495). Specifically, standard (2002:223) indicates that the maximum permissible limit of E. coli and S. aureus in cheese is 1000 CFU/g [52]. Conversely, LIBNOR (2003:495) is more conservative, with a maximum allowable limit of 10 CFU/g [51]. E. coli and S. aureus isolates were purified, suspended in 1 mL Luria-Bertani (LB) broth (80%) (Oxoid, Hampshire, UK) with 0.5 mL glycerol (20%), and stored at −80 °C for further analysis [36].
4.3. Polymerase Chain Reaction (PCR) for the Confirmation of Identity of E. coli and S. aureus
Putative E. coli colonies were suspended in 100 µL of DNase-free water and placed in a water bath at 95 °C for 10 min to extract DNA [63]. The E. coli isolates were screened for a species-specific 16S rRNA gene fragment [64,65]. Briefly, PCR reactions (20 µL) were prepared, as reported elsewhere [38,39], and subjected to the following program: an initial denaturation at 95 °C followed by 38 cycles, each consisting of denaturation at 95 °C for 30 s, annealing at 58 °C for 45 s and extension at 72 °C for 1 min, followed by a final extension at 72 °C for 10 min [65]. The amplified PCR amplicons (544 bp) were visualized by gel electrophoresis. Reactions with DNase-free water instead of bacterial DNA were used as a negative control, while DNA extracted from E. coli DH5α was used as a positive control. Similarly, the PCR detection of a specific fragment of femB was used to confirm the identity of the S. aureus isolates [66]. However, the PCR program consisted of an initial denaturation at 95 °C for 4 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 45 s, and extension 72 °C for 3 min, which was followed by a final extension at 72 °C for 10 min. S. aureus ATCC® 6538 was used as a positive control.
4.4. Assessment of the Antibiotic Resistance Profiles of E. coli Using the Disk Diffusion Assay
The antibiotic resistance profiles of 118 E. coli isolates retrieved from the cheese (on average 3 E. coli, when available, were randomly selected from each positive cheese sample) were determined using the disk diffusion assay [67,68]. Pure E. coli colonies were suspended in Mueller–Hinton (MH) broth (Oxoid, Hampshire, UK). The optical density of the bacterial cultures was adjusted using the 0.5 McFarland standard and a spectrophotometer (Thermo Fisher Scientific, MA, USA). Thereafter, the cultures were spread on MH agar plates using swabs, as described in the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [68]. Nineteen different antibiotic disks (Oxoid, Hampshire, UK) belonging to 9 different classes were then added to the MH agar plates. The antibiotics included penicillins: ampicillin (AMP; 10 μg); β-lactam/β-lactamase inhibitor combinations: amoxicillin/clavulanic acid (AMC; 20 μg/10 μg); cephalosporins: cefepime (FEP; 30 μg), cefotaxime (CTX; 30 μg), cefalexin (LEX; 30 μg), and cefixime (CFM; 5 μg); carbapenems: doripenem (DOR; 10 μg), meropenem (MEM; 10 μg), and imipenem (IPM; 10 μg); aminoglycosides: gentamicin (GEN; 10 μg), kanamycin (KAN; 30 μg), and streptomycin (STR; 10 μg); tetracyclines: tetracycline (TET; 30 μg); quinolones and fluoroquinolones: ciprofloxacin (CIP; 5 μg) and norfloxacin (NOR; 10 μg); folate-pathway inhibitors: trimethoprim/sulfamethoxazole (SXT; 25 μg); and phenicols: chloramphenicol (CHL; 30 μg) [36,59]. The MH agar plates were then incubated at 37 °C for 18 h. Penicillin (PEN; 6 µg) and erythromycin (ERY; 15 µg) were used as controls [36,53]. Additionally, E. coli DH5α and E. coli ATCC® 25922 were used for quality control across the experiments. The diameters of the zones of inhibition around each antibiotic disk were measured and evaluated according to the CLSI and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) standards [68,69]. The breakpoints that were used to determine the susceptibility of the isolates to the different antibiotics are listed in the supplementary Table S1. Additionally, the AMR profiles were analyzed using the hierarchical clustering (HCL) approach. Briefly, isolates were represented in rows and antibiotics in columns, while the AMR profiles were coded as follows: with 0 indicating resistance, 0.5 representing intermediate resistance, and 1 corresponding to susceptibility. Hierarchical clustering was then conducted using Morpheus (https://software.broadinstitute.org/morpheus/, accessed on 15 December 2022) by setting the Pearson correlation as a distance metric and choosing the complete linkage method [70]. A graphical HCL analysis was produced (heat map), with green colors representing susceptibility (1), while black and red colors corresponded to intermediate resistance (0.5) and resistance (0), respectively.
4.5. Statistical Analysis
A simple linear regression analysis was conducted using SPSS 23.0 (IBM® SPSS® Statistics, NY, USA) to evaluate the correlation between the bacterial densities in the Akkawi cheese samples. Statistical tests were two-sided, with a type I error set at α = 0.05.
5. Conclusions
This study highlights the frequent occurrence of a fecal indicator, E. coli, and S. aureus in Akkawi cheese collected from retail stores across Beirut, Lebanon. Given the popularity of Akkawi cheese among the Lebanese population, the high contamination with these bacterial indicators raises public health concerns and highlights the need for robust food safety systems. Subsequently, microbial pathogens such as L. monocytogenes, which have been previously associated with cheese samples [14], must be fully investigated in order to identify and mitigate the potential sources of contamination. Furthermore, the occurrence of MDR E. coli in the cheese samples emphasizes a paramount need to strengthen antimicrobial stewardship, revise agricultural practices, and adopt policies to curb the spread of antibiotic resistance. The latter is not only a local problem, because AMR can spread across national borders, causing a wide threat to multiple stakeholders. Consequently, efforts must focus on decreasing the over reliance on antibiotics in both agriculture and medicine to preserve the efficacy of these highly important dugs, while seeking viable and sustainable alternatives that do not promote the spread of resistance.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12030610/s1, Table S1: The breakpoints that were used to interpret antibiotic susceptibility of the E. coli isolated from the cheese samples.
Author Contributions
Conceptualization, I.I.K.; methodology, I.I.K.; validation, I.I.K., N.D.H., J.W.H. and M.O.; formal analysis, I.I.K., N.D.H., J.W.H., M.O., K.E.-O., S.A.K. and I.T.; investigation, I.I.K., N.D.H., J.W.H., M.O., K.E.-O., S.A.K. and I.T.; resources, I.I.K.; data curation, I.I.K., N.D.H. and J.W.H.; writing—original draft preparation, I.I.K., N.D.H. and J.W.H.; writing—review and editing, I.I.K., J.W.H. and M.O; visualization, I.I.K.; supervision, I.I.K.; project administration, I.I.K.; funding acquisition; I.I.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by start-up funds from the Center for Food Safety, University of Georgia (UGA).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the relevant data have been included in this study.
Acknowledgments
M.O. is supported by the Atkinson Postdoctoral Fellowship (Cornell University).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Serra-Majem, L.; Román-Viñas, B.; Sanchez-Villegas, A.; Guasch-Ferré, M.; Corella, D.; La Vecchia, C. Benefits of the Mediterranean Diet: Epidemiological and Molecular Aspects. Mol. Asp. Med. 2019, 67, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Lee, H.; Lee, S.; Kim, S.; Yoon, Y. Cheese Microbial Risk Assessments—A Review. Asian-Australas. J. Anim. Sci. 2016, 29, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Ostyn, A.; De Buyser, M.L.; Guillier, F.; Groult, J.; Felix, B.; Salah, S.; Delmas, G.; Hennekinne, J.A. First Evidence of a Food Poisoning Outbreak due to Staphylococcal Enterotoxin Type E, France, 2009. Eurosurveill 2010, 15, 19528. [Google Scholar] [CrossRef]
- Napoleoni, M.; Villa, L.; Barco, L.; Busani, L.; Cibin, V.; Lucarelli, C.; Tiengo, A.; Dionisi, A.M.; Conti, F.; Da Silva Nunes, F.R.; et al. A Strong Evidence Outbreak of Salmonella Enteritidis in Central Italy Linked to the Consumption of Contaminated Raw Sheep Milk Cheese. Microorganisms 2021, 9, 2464. [Google Scholar] [CrossRef]
- Fretz, R.; Pichler, J.; Sagel, U.; Much, P.; Ruppitsch, W.; Pietzka, A.T.; Stöger, A.; Huhulescu, S.; Heuberger, S.; Appl, G.; et al. Update: Multinational Listeriosis Outbreak due to ‘Quargel’, a Sour Milk Curd Cheese, Caused by Two Different L. monocytogenes serotype 1/2a strains, 2009–2010. Eurosurveill 2010, 15, 19543. [Google Scholar] [CrossRef]
- Ung, A.; Baidjoe, A.Y.; Van Cauteren, D.; Fawal, N.; Fabre, L.; Guerrisi, C.; Danis, K.; Morand, A.; Donguy, M.P.; Lucas, E.; et al. Disentangling a Complex Nationwide Salmonella dublin Outbreak Associated with Raw-Milk Cheese Consumption, France, 2015 to 2016. Eurosurveill 2019, 24, 1700703. [Google Scholar] [CrossRef] [PubMed]
- Sarno, E.; Pezzutto, D.; Rossi, M.; Liebana, E.; Rizzi, V. A Review of Significant European Foodborne Outbreaks in the Last Decade. J. Food Prot. 2021, 84, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Rangel, J.M.; Sparling, P.H.; Crowe, C.; Griffin, P.M.; Swerdlow, D.L. Epidemiology of Escherichia coli O157:H7 Outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2005, 11, 603–609. [Google Scholar] [CrossRef] [PubMed]
- Kadariya, J.; Smith, T.C.; Thapaliya, D. Staphylococcus aureus and staphylococcal food-Borne Disease: An Ongoing Challenge in Public Health. Biomed Res. Int. 2014, 2014, 827965. [Google Scholar] [CrossRef]
- Jeffs, E.; Williman, J.; Brunton, C.; Gullam, J.; Walls, T. The Epidemiology of Listeriosis in Pregnant Women and Children in New Zealand from 1997 to 2016: An Observational Study. BMC Public Health 2020, 20, 116. [Google Scholar] [CrossRef]
- Yang, S.C.; Lin, C.H.; Aljuffali, I.A.; Fang, J.Y. Current Pathogenic Escherichia coli Foodborne Outbreak Cases and Therapy Development. Arch. Microbiol. 2017, 199, 811–825. [Google Scholar] [CrossRef]
- Daou, R.; Afif, C.; Joubrane, K.; Khabbaz, L.R.; Maroun, R.; Ismail, A.; Khoury, A.E. Occurrence of Aflatoxin M1 in Raw, Pasteurized, UHT Cows’ Milk, and Dairy Products in Lebanon. Food Control 2020, 111, 107055. [Google Scholar] [CrossRef]
- Daou, L.; El Azzi, D. Life Cycle Assessment Awareness: Impact on the Behaviour of Lebanese Consumers and Decision-Making. EuroMed J. Manag. 2021, 4, 19. [Google Scholar] [CrossRef]
- Kharroubi, S.; Nasser, N.A.; El-Harakeh, M.D.; Sulaiman, A.A.; Kassem, I.I. First Nation-Wide Analysis of Food Safety and Acceptability Data in Lebanon. Foods 2020, 9, 1717. [Google Scholar] [CrossRef]
- Fadlallah, S.M.; Shehab, M.; Cheaito, K.; Haidar-Ahmad, N.; El Hafi, B.; Saleh, M.; Nasser, Z.; El Hajj, R.; Ghosn, N.; Ammar, W.; et al. PulseNet Lebanon: An Overview of Its Activities, Outbreak Investigations, and Challenges. Foodborne Pathog. Dis. 2019, 16, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Lebanese Ministry of PublicHealth. Notifiable Communicable Diseases. Available online: https://www.moph.gov.lb/en/DynamicPages/index/8#/en/Pages/2/194/surveillance-data (accessed on 15 December 2022).
- Ikuta, K.S.; Swetschinski, L.R.; Robles Aguilar, G.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Davis Weaver, N.; Wool, E.E.; Han, C.; Gershberg Hayoon, A.; et al. Global Mortality Associated with 33 Bacterial Pathogens in 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- Kassem, I.I.; Osman, M. A brewing storm: The Impact of Economic Collapse on the Access to Antimicrobials in Lebanon. J. Glob. Antimicrob. Resist. 2022, 29, 313–315. [Google Scholar] [CrossRef]
- Osman, M.; Cummings, K.J.; El Omari, K.; Kassem, I.I. Catch-22: War, Refugees, COVID-19, and the Scourge of Antimicrobial Resistance. Front. Med. 2022, 9, 921921. [Google Scholar] [CrossRef]
- Osman, M.; Kasir, D.; Kassem, I.I.; Hamze, M. Shortage of Appropriate Diagnostics for Antimicrobial Resistance in Lebanese Clinical Settings: A Crisis Amplified by COVID-19 and Economic Collapse. J. Glob. Antimicrob. Resist. 2021, 27, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Kojok, H.E.; Khalil, M.; Hage, R.; Jammoul, R.; Jammoul, A.; Darra, N.E. Microbiological and Chemical Evaluation of Dairy Products Commercialized in the Lebanese Market. Vet. World 2022, 15, 2575–2586. [Google Scholar] [CrossRef]
- Waage, J.; Grace, D.; Fèvre, E.M.; McDermott, J.; Lines, J.; Wieland, B.; Naylor, N.R.; Hassell, J.M.; Chan, K. Changing Food Systems and Infectious Disease Risks in Low-Income and Middle-Income Countries. Lancet Planet. Health 2022, 6, e760–e768. [Google Scholar] [CrossRef]
- Kassem, I.I.; Hijazi, M.A.; Saab, R. On a Collision Course: The Availability and Use of Colistin-Containing Drugs in Human Therapeutics and Food-Animal Farming in Lebanon. J. Glob. Antimicrob. Resist. 2019, 16, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Hassani, S.; Moosavy, M.-H.; Gharajalar, S.N.; Khatibi, S.A.; Hajibemani, A.; Barabadi, Z. High Prevalence of Antibiotic Resistance in Pathogenic Foodborne Bacteria Isolated from Bovine Milk. Sci. Rep. 2022, 12, 3878. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Jay-Russell, M.; Lemay, D.G.; Mills, D.A. Reservoirs of Antimicrobial Resistance Genes in Retail Raw Milk. Microbiome 2020, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Mugoh, M.; Call, D.R.; Omulo, S. Antibiotic residues and antibiotic-resistant bacteria detected in milk marketed for human consumption in Kibera, Nairobi. PLoS ONE 2020, 15, e0233413. [Google Scholar] [CrossRef]
- Yao, J.; Gao, J.; Guo, J.; Wang, H.; Zhang, E.N.; Lin, Y.; Chen, Z.; Li, S.; Tao, S. Characterization of Bacteria and Antibiotic Resistance in Commercially Produced Cheeses Sold in China. J. Food Prot. 2022, 85, 484–493. [Google Scholar] [CrossRef]
- Kassem, I.I.; Assi, A.; Osman, M.; Mann, D.; Li, S.; Deng, X. Letter to the Editor: First Report of the Detection of the Plasmid-Borne Colistin Resistance Gene, mcr-1.26, in Multidrug-Resistant Escherichia coli Isolated from a Domesticated Pigeon. Microb. Drug Resist. 2022, 28, 821–823. [Google Scholar] [CrossRef]
- Abou Fayad, A.; El Azzi, M.; Sleiman, A.; Kassem, I.I.; Bawazeer, R.A.; Okdah, L.; Doumith, M.; Alghoribi, M.F.; Matar, G.M. Acquired Resistome and Plasmid Sequencing of mcr-1 carrying MDR Enterobacteriaceae from Poultry and Their Relationship to Sts Associated With Humans. JAC-Antimicrobial Resist. 2022, 4, dlab198. [Google Scholar] [CrossRef]
- Halimeh, F.B.; Rafei, R.; Diene, S.M.; Osman, M.; Kassem, I.I.; Jamal Akoum, R.; Moudani, W.; Hamze, M.; Rolain, J.-M. Genome Sequence of A Multidrug-Resistant Campylobacter coli Strain Isolated from a Newborn with Severe Diarrhea in Lebanon. Folia Microbiol. 2022, 67, 319–328. [Google Scholar] [CrossRef]
- Dabbousi, A.A.; Dabboussi, F.; Hamze, M.; Osman, M.; Kassem, I.I. The Emergence and Dissemination of Multidrug Resistant Pseudomonas aeruginosa in Lebanon: Current Status and Challenges during the Economic Crisis. Antibiotics 2022, 11, 687. [Google Scholar] [CrossRef]
- Kassem, I.I.; Hassan, J.; Esseili, M.A.; Mann, D.; Osman, M.; Li, S.; Deng, X. Draft Genome Sequences of Multidrug-Resistant Escherichia coli Isolated from River Water. Microbiol. Resour. Announc. 2022, 11, e00827-22. [Google Scholar] [CrossRef]
- Kassem, I.I.; Mann, D.; Li, S.; Deng, X. Draft Genome Sequences and Resistome Analysis of Multidrug-Resistant Mcr-1-Harbouring Escherichia coli Isolated from Pre-Harvest Poultry in Lebanon. J. Glob. Antimicrob. Resist. 2021, 25, 114–116. [Google Scholar] [CrossRef] [PubMed]
- Hassan, J.; Eddine, R.Z.; Mann, D.; Li, S.; Deng, X.; Saoud, I.P.; Kassem, I.I. The Mobile Colistin Resistance Gene, mcr-1.1, Is Carried On Incx4 Plasmids In Multidrug Resistant E. coli isolated From Rainbow Trout Aquaculture. Microorganisms 2020, 8, 1636. [Google Scholar] [CrossRef]
- Sourenian, T.; Mann, D.; Li, S.; Deng, X.; Jaafar, H.; Kassem, I.I. Dissemination of Multidrug-Resistant Escherichia Coli Harboring the Mobile Colistin Resistance Gene mcr-1.1 on Transmissible Plasmids in the Mediterranean Sea. J. Glob. Antimicrob. Resist. 2020, 22, 84–86. [Google Scholar] [CrossRef]
- Kassem, I.I.; Nasser, N.A.; Salibi, J. Prevalence and Loads of Fecal Pollution Indicators and the Antibiotic Resistance Phenotypes of Escherichia coli in Raw Minced Beef in Lebanon. Foods 2020, 9, 1543. [Google Scholar] [CrossRef] [PubMed]
- Alhaj Sulaiman, A.A.; Kassem, I.I. First Report of the Plasmid-Borne Colistin Resistance Gene (mcr-1) in Proteus mirabilis Isolated from Domestic and Sewer Waters in Syrian Refugee Camps. Travel Med. Infect. Dis. 2020, 33, 101482. [Google Scholar] [CrossRef] [PubMed]
- Hmede, Z.; Sulaiman, A.A.A.; Jaafar, H.; Kassem, I.I. Emergence of Plasmid-Borne Colistin Resistance Gene mcr-1 in Multidrug-Resistant Escherichia coli Isolated from Irrigation Water in Lebanon. Int. J. Antimicrob. Agents 2019, 54, 102–104. [Google Scholar] [CrossRef]
- Sulaiman, A.A.A.; Kassem, I.I. First Report on the Detection of the Plasmid-Borne Colistin Resistance Gene mcr-1 in Multi-Drug Resistant E. coli Isolated from Domestic and Sewer Waters in Syrian Refugee Camps in Lebanon. Travel Med. Infect. Dis. 2019, 30, 117–120. [Google Scholar] [CrossRef]
- Al-Mir, H.; Osman, M.; Drapeau, A.; Hamze, M.; Madec, J.-Y.; Haenni, M. Spread of ESC-, carbapenem- and colistin-resistant Escherichia coli clones and plasmids within and between food workers in Lebanon. J. Antimicrob. Chemother. 2021, 76, 3135–3143. [Google Scholar] [CrossRef] [PubMed]
- Al-Mir, H.; Osman, M.; Drapeau, A.; Hamze, M.; Madec, J.-Y.; Haenni, M. WGS Analysis of Clonal and Plasmidic Epidemiology of Colistin-Resistance Mediated by mcr Genes in the Poultry Sector in Lebanon. Front. Microbiol. 2021, 12, 624194. [Google Scholar] [CrossRef]
- Osman, M.; Al Bikai, A.; Rafei, R.; Mallat, H.; Dabboussi, F.; Hamze, M. Species Distribution and Antifungal Susceptibility Patterns of Clinical Candida Isolates in North Lebanon: A Pilot Cross-Sectional Multicentric Study. J. Med Mycol. 2020, 30, 100986. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.; Halimeh, F.B.; Rafei, R.; Mallat, H.; Tom, J.E.; Raad, E.B.; Diene, S.M.; Jamal, S.; Atrouni, A.A.; Dabboussi, F.; et al. Investigation of an XDR-Acinetobacter baumannii ST2 Outbreak in An Intensive Care Unit of a Lebanese Tertiary Care Hospital. Future Microbiol. 2020, 15, 1535–1542. [Google Scholar] [CrossRef]
- El Achkar, S.; Demanche, C.; Osman, M.; Rafei, R.; Ismail, M.B.; Yaacoub, H.; Pincon, C.; Duthoy, S.; De Matos, F.; Gaudin, C.; et al. Drug-Resistant Tuberculosis, Lebanon, 2016–2017. Emerg. Infect. Dis. 2019, 25, 564–568. [Google Scholar] [CrossRef]
- Al-Mir, H.; Osman, M.; Azar, N.; Madec, J.Y.; Hamze, M.; Haenni, M. Emergence of clinical mcr-1-positive Escherichia coli in Lebanon. J. Glob. Antimicrob. Resist. 2019, 19, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Hassan, J.; Mann, D.; Li, S.; Deng, X.; Kassem, I.I. Emergence of the Mobile Colistin Resistance Gene, mcr-1, in Multidrug-Resistant E. coli Isolated from the Fecal Matter of Toddlers in a Community. Antimicrob. Agents Chemother. 2021, 65, e00243-21. [Google Scholar] [CrossRef] [PubMed]
- Al Omari, S.; Al Mir, H.; Wrayde, S.; Merhabi, S.; Dhaybi, I.; Jamal, S.; Chahine, M.; Bayaa, R.; Tourba, F.; Tantawi, H.; et al. First Lebanese Antibiotic Awareness Week campaign: Knowledge, attitudes and practices towards antibiotics. J. Hosp. Infect. 2019, 101, 475–479. [Google Scholar] [CrossRef]
- Goddard, F.G.B.; Ban, R.; Barr, D.B.; Brown, J.; Cannon, J.; Colford, J.M., Jr.; Eisenberg, J.N.S.; Ercumen, A.; Petach, H.; Freeman, M.C.; et al. Measuring Environmental Exposure to Enteric Pathogens in Low-Income Settings: Review and Recommendations of an Interdisciplinary Working Group. Environ. Sci. Technol. 2020, 54, 11673–11691. [Google Scholar] [CrossRef]
- Holcomb, D.A.; Stewart, J.R. Microbial Indicators of Fecal Pollution: Recent Progress and Challenges in Assessing Water Quality. Curr. Environ. Health Rep. 2020, 7, 311–324. [Google Scholar] [CrossRef]
- Nyirabahizi, E.; Tyson, G.H.; Dessai, U.; Zhao, S.; Kabera, C.; Crarey, E.; Womack, N.; Crews, M.K.; Strain, E.; Tate, H. Evaluation of Escherichia coli as an Indicator for Antimicrobial Resistance in Salmonella Recovered from the Same Food or Animal Ceca Samples. Food Control 2020, 115, 107280. [Google Scholar] [CrossRef]
- Lebanese Specification Standard No. 495; LIBNOR. Akkawi Cheese. Lebanese Standards Institution: Beirut, Lebanon, 2003.
- Lebanese Specification Standard No. 223; LIBNOR. Akkawi Cheese. Lebanese Standards Institution: Beirut, Lebanon, 2002.
- Dagher, L.A.; Hassan, J.; Kharroubi, S.; Jaafar, H.; Kassem, I.I. Nationwide Assessment of Water Quality in Rivers across Lebanon by Quantifying Fecal Indicators Densities and Profiling Antibiotic Resistance of Escherichia coli. Antibiotics 2021, 10, 883. [Google Scholar] [CrossRef]
- Kamleh, R.; Olabi, A.; Toufeili, I.; Daroub, H.; Younis, T.; Ajib, R. The Effect of Partial Substitution of Nacl with Kcl on the Physicochemical, Microbiological and Sensory Properties of Akkawi Cheese. J. Sci. Food Agric. 2015, 95, 1940–1948. [Google Scholar] [CrossRef]
- Economou, V.; Gousia, P. Agriculture and Food Animals as a Source of Antimicrobial-Resistant Bacteria. Infect. Drug Resist. 2015, 8, 49. [Google Scholar] [CrossRef]
- Osman, M.; Kamal-Dine, K.; El Omari, K.; Rafei, R.; Dabboussi, F.; Hamze, M. Prevalence of Staphylococcus aureus methicil-lin-sensitive and Methicillin-Resistant Nasal Carriage in Food Handlers in Lebanon: A Potential Source of Transmission of Virulent Strains in the Community. Access Microbiol. 2019, 1, e000043. [Google Scholar] [CrossRef] [PubMed]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A Review of Antibiotic Use in Food Animals: Perspective, Policy, and Potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef]
- Dandachi, I.; Sokhn, E.S.; Dahdouh, E.A.; Azar, E.; El-Bazzal, B.; Rolain, J.-M.; Daoud, Z. Prevalence and Characterization of Multi-Drug-Resistant Gram-Negative Bacilli Isolated from Lebanese Poultry: A Nationwide Study. Front. Microbiol. 2018, 9, 550. [Google Scholar] [CrossRef] [PubMed]
- Zeina, K.; Fawwak, S. Quantification of Antibiotic Residues and Determination of Antimicrobial Resistance Profiles of Mi-croorganisms Isolated from Bovine Milk in Lebanon. Food Sci. Nutr. 2013, 4, 1. [Google Scholar]
- Jammoul, A.; El Darra, N. Evaluation of Antibiotics Residues in Chicken Meat Samples in Lebanon. Antibiotics 2019, 8, 69. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Dankar, I.; Hassan, H.; Serhan, M. Knowledge, attitudes, and perceptions of dairy farmers regarding antibiotic use: Lessons from a developing country. J. Dairy Sci. 2022, 105, 1519–1532. [Google Scholar] [CrossRef]
- Dashti, A.; Jadaon, M.; Abdulsamad, A.; Dashti, H. Heat Treatment of Bacteria: A Simple Method of DNA Extraction for Molecular Techniques. Kuwait Med. J. 2009, 41, 117–122. [Google Scholar]
- Hmede, Z.; Kassem, I.I. The Colistin Resistance Gene mcr-1 Is Prevalent in Commensal Escherichia coli Isolated from Preharvest Poultry in Lebanon. Antimicrob. Agents Chemother. 2018, 62, e01304-18. [Google Scholar] [CrossRef] [PubMed]
- Sabat, G.; Rose, P.; Hickey, W.J.; Harkin, J.M. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol. 2000, 66, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Jonas, D.; Speck, M.; Daschner, F.; Grundmann, H. Rapid PCR-Based Identification of Methicillin-Resistant Staphylococcus Aureus From Screening Swabs. Clin. Microbiol. 2002, 40, 1821–1823. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for In Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. EUCAST, Version 8.1. 2018. Available online: http://www.eucast.org (accessed on 10 December 2022).
- Erkmen, O.; Bozoglu, T.F. Food microbiology. In Principles into Practice; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).