Serving Two Masters: Effect of Escherichia coli Dual Resistance on Antibiotic Susceptibility
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Evolution Experiment
2.3. Bacteriophage Resistance Assay
2.4. Phenotypic Assays: 24 h Minimum Inhibitory Concentration Growth
2.5. T7 Bacteriophage Resistance Assay
2.6. Dual Resistance Assay: Excess Iron (III) and Escherichia Phage T7
2.7. Genomic Analysis
2.8. Statistics
3. Results
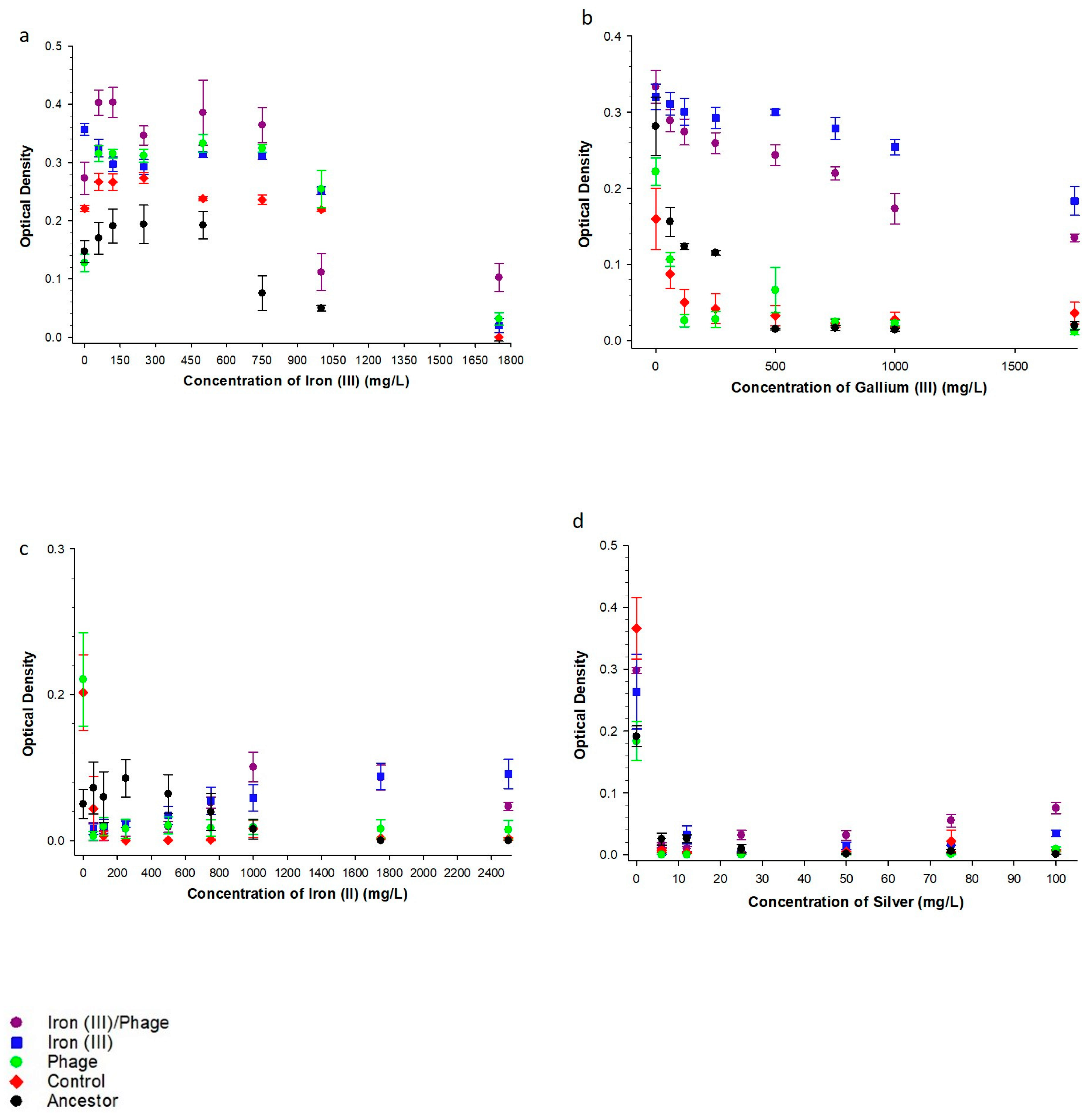
3.1. The Effect of Sequential Iron (III), Bacteriophage Selection on Metal Resistance
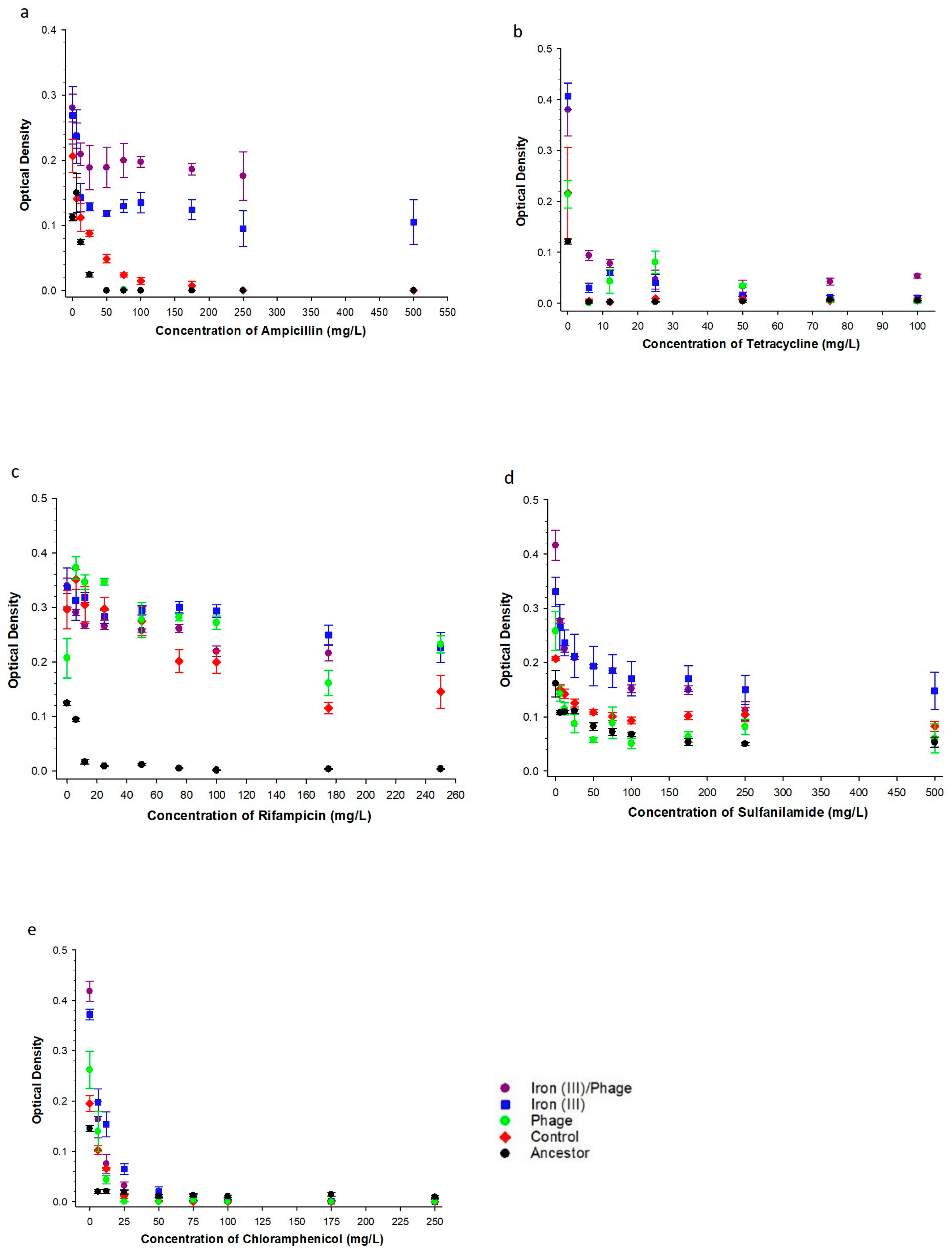
3.2. The Effect of Sequential Iron (III), Bacteriophage Selection on Antibiotic Resistance
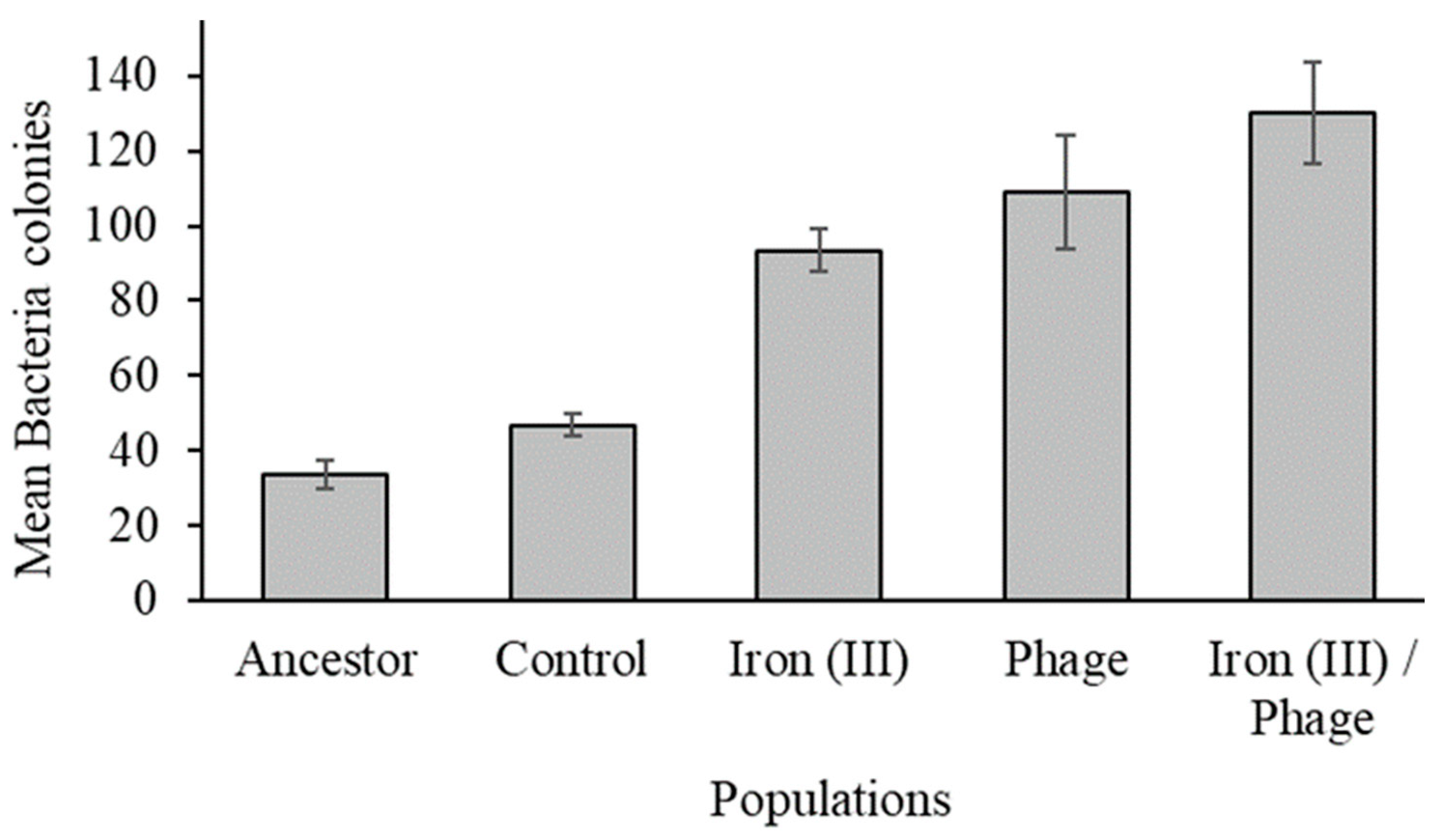
3.3. The Effect of Sequential Selection on Bacteriophage Resistance
3.4. Whole Genome Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aminov, R.I.; Mackie, R.I. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 2007, 271, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Bin Zaman, S.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus 2017, 9, 1403. [Google Scholar] [CrossRef] [PubMed]
- Sugden, R.; Kelly, R.; Davies, S. Combatting antimicrobial resistance globally. Nat. Microbiol. 2016, 1, 16187. [Google Scholar] [CrossRef]
- Graves, J.L. Antimicrobial Nanomaterials: Principles and Applications; Elseveir: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Sulakvelidze, A.; Alavidze, Z.; Morris, J.G., Jr. Bacteriophage therapy. Antimicrob. Agents Chemother. 2001, 45, 649–659. [Google Scholar] [CrossRef]
- Carrillo, C.L.; Atterbury, R.J.; El-Shibiny, A.; Connerton, P.L.; Dillon, E.; Scott, A.; Connerton, I.F. Bacteriophage Therapy To Reduce Campylobacter jejuni Colonization of Broiler Chickens. Appl. Environ. Microbiol. 2005, 71, 6554–6563. [Google Scholar] [CrossRef]
- Borie, C.; Albala, I.; Sánchez, P.; Sánchez, M.L.; Ramírez, S.; Navarro, C.; Morales, M.A.; Retamales, J.; Robeson, J. Bacteri-ophage treatment reduces Salmonella colonization of infected chickens. Avian Dis. 2008, 52, 64–67. [Google Scholar] [CrossRef]
- Huff, W.E.; Huff, G.R.; Rath, N.C.; Balog, J.M.; Donoghue, A.M. Prevention of Escherichia coli infection in broiler chickens with a bacteriophage aerosol spray. Poult. Sci. 2002, 81, 1486–1491. [Google Scholar] [CrossRef] [PubMed]
- Saez, A.C.; Zhang, J.; Rostagno, M.H.; Ebner, P.D. Direct Feeding of Microencapsulated Bacteriophages to ReduceSalmonel-laColonization in Pigs. Foodborne Pathog. Dis. 2011, 8, 1269–1274. [Google Scholar] [CrossRef]
- Graves, J.L.; Ewunkem, A.J.; Thomas, M.D.; Han, J.; Rhinehardt, K.L.; Boyd, S.; Edmondson, R.; Jeffers-Francis, L.; Harrison, S.H. Experimental Evolution of Metal Resistance in Bacteria. In Evolution in Action—Past, Present and Future. Genetic and Evolutionary Computation; Banzhaf, W., Cheng, B.H.C., Deb, K., Holekamp, K.E., Lenski, R.E., Ofria, C., Pennock, R.T., Punch, W.F., Whittaker, D.J., Eds.; Springer: Cham, Switzerland, 2020; pp. 91–106. [Google Scholar]
- Lee, C.; Kim, J.Y.; Lee, W.I.; Nelson, K.L.; Yoon, J.; Sedlak, D.L. Bactericidal Effect of Zero-Valent Iron Nanoparticles on Escherichia coli. Environ. Sci. Technol. 2008, 42, 4927–4933. [Google Scholar] [CrossRef]
- Ansari, S.A.; Oves, M.; Satar, R.; Khan, A.; Ahmad, S.I.; Alam Jafri, M.; Zaidi, S.K.; Alqahtani, M.H. Antibacterial activity of iron oxide nanoparticles synthesized by co-precipitation technology against Bacillus cereus and Klebsiella pneumoniae. Pjct 2017, 19, 110–115. [Google Scholar] [CrossRef]
- Seo, S.W.; Kim, D.; Latif, H.; O’Brien, E.J.; Szubin, R.; Palsson, B.O. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 2014, 5, 4910. [Google Scholar] [CrossRef]
- Nunoshiba, T.; Hidalgo, E.; Amabile Cuevas, C.F.; Demple, B. Two-stage control of an oxidative stress regulon: The Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 1992, 174, 6054–6060. [Google Scholar] [CrossRef] [PubMed]
- Ewunkem, J.A.; Rodgers, M.; Campbell, D.; Staley, C.; Subedi, K.; Boyd, S.; Graves, J.L. Experimental evolution of magnetite resistance in Escherichia coli. Nanomaterials 2021, 11, 790. [Google Scholar] [CrossRef]
- Zheng, M.; Åslund, F.; Storz, G. Activation of the OxyR Transcription Factor by Reversible Disulfide Bond Formation. Science 1998, 279, 1718–1722. [Google Scholar] [CrossRef]
- Graves, J.L., Jr.; Ewunkem, A.J.; Ward, J.; Staley, C.; Thomas, M.D.; Rhinehardt, K.L.; Han, J.; Harrison, S.H. Experimental evolution of gallium (III) resistance in Escherichia coli. Evol. Med. Public Health 2019, 2019, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.D.; Ewunkem, A.J.; Boyd, S.; Williams, D.K.; Moore, A.; Rhinehardt, K.L.; Beveren, E.; Yang, B.; Tapia, A.; Han, J.; et al. Too much of a good thing: Adaption to iron (II) intoxication in Escherichia coli. Evol. Med. Public Health 2021, 9, 53–67. [Google Scholar] [CrossRef]
- Conibear, T.C.; Collins, S.L.; JS, W. Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS ONE 2009, 14, 6289. [Google Scholar] [CrossRef]
- Tyerman, J.G.; Ponciano, J.M.; Joyce, P.; Forney, L.J.; Harmon, L.J. The evolution of antibiotic susceptibility and resistance during the formation of Escherichia colibiofilms in the absence of antibiotics. BMC Evol. Biol. 2013, 13, 22. [Google Scholar] [CrossRef]
- Wrande, M.; Roth, J.R.; Hughes, D. Accumulation of mutants in “aging” bacterial colonies is due to growth under selection, not stress-induced mutagenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 11863–11868. [Google Scholar] [CrossRef]
- Gu, H.; Lee, S.W.; Carnicelli, J.; Jiang, Z.; Ren, D. Antibiotic Susceptibility of Escherichia coli Cells during Early-Stage Biofilm Formation. J. Bacteriol. 2019, 201, 34. [Google Scholar] [CrossRef] [PubMed]
- Ferenci, T. Trade-off Mechanisms Shaping the Diversity of Bacteria. Trends Microbiol. 2015, 24, 209–223. [Google Scholar] [CrossRef]
- Yokoyama, M.; Stevens, E.; Laabei, M.; Bacon, L.; Heesom, K.; Bayliss, S.; Ooi, N.; O’Neill, A.J.; Murray, E.; Williams, P. Epistasis analysis uncovers hidden antibiotic resistance-associated fitness costs hampering the evolution of MRSA. Genome Biol. 2018, 19, 94. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, 002. [Google Scholar] [CrossRef]
- Heinrichs, D.E.; Yethon, J.A.; Whitfield, C. Molecular basis for structural diversity in the core regions of the lipopolysaccha-rides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 1998, 30, 221–232. [Google Scholar] [CrossRef]
- Riley, M.; Abe, T.; Arnaud, M.B.; Berlyn, M.K.; Blattner, F.R.; Chaudhuri, R.R.; Glasner, J.D.; Horiuchi, T.; Keseler, I.M.; Kosuge, T.; et al. Escherichia coli K-12: A cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006, 34, 1–9. [Google Scholar] [CrossRef]
- Hall, B.G.; Acar, H.; Nandipati, A.; Barlow, M. Growth Rates Made Easy. Mol. Biol. Evol. 2013, 31, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48, 5–16. [Google Scholar] [CrossRef]
- Hershberg, R. Antibiotic-Independent Adaptive Effects of Antibiotic Resistance Mutations. Trends Genet. 2017, 33, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Fard, R.M.N.; Heuzenroeder, M.W.; Barton, M.D. Antimicrobial and heavy metal resistance in commensal enterococci isolated from pigs. Veter. Microbiol. 2011, 148, 276–282. [Google Scholar] [CrossRef]
- Vahjen, W.; Pietruszyńska, D.; Starke, I.C.; Zentek, J. High dietary zinc supplementation increases the occurrence of tetracy-cline and sulfonamide resistance genes in the intestine of weaned pigs. Gut Pathog. 2015, 7, 23. [Google Scholar] [CrossRef]
- Lenski, R.E. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 2017, 11, 2181–2194. [Google Scholar] [CrossRef]
- Kedziora, A.; Gerasymchuk, Y.; Sroka, E.; Bugla-Płoskońska, G.; Doroszkiewicz, W.; Rybak, Z.; Hreniak, D.C.; Wilgusz, R.; Strek, A.W. Use of the materials based on partially reduced graphene-oxide with silver nanoparticle as bacteriostatic and bac-tericidal agent. Polym. Med 2014, 43, 129–134. [Google Scholar]
- Berardi, A.E.; Esfeld, K.; Jäggi, L.; Mandel, T.; Cannarozzi, G.M.; Kuhlemeier, C. Complex evolution of novel red floral color in Petunia. Plant Cell 2021, 33, 2273–2295. [Google Scholar] [CrossRef]
- Reynolds, M.G. Compensatory Evolution in Rifampin-Resistant Escherichia coli. Genetics 2000, 156, 1471–1481. [Google Scholar] [CrossRef]
- Cutugno, L.; Mc Cafferty, J.; Pané-Farré, J.; O’Byrne, C.; Boyd, A. rpoB mutations conferring rifampicin-resistance affect growth, stress response and motility in Vibrio vulnificus. Microbiology 2020, 166, 1160–1170. [Google Scholar] [CrossRef] [PubMed]
- Ibanez-Ruiz, M.; Robbe-Saule, V.; Hermant, D.; Labrude, S.; Norel, F. Identification of RpoS (sigma(S))-regulated genes in Salmonella enterica serovar typhimurium. J. Bacteriol. 2000, 182, 5749–5756. [Google Scholar] [CrossRef] [PubMed]
- Figueira, R.; Brown, D.R.; Ferreira, D.; Eldridge, M.J.G.; Burchell, L.; Pan, Z.; Helaine, S.; Wigneshweraraj, S. Adaptation to sustained nitrogen starvation by Escherichia coli requires the eukaryote-like serine/threonine kinase YeaG. Sci. Rep. 2015, 5, 17524. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Polen, T.; Heuveling, J.; Wendisch, V.F.; Hengge, R. Genome-Wide Analysis of the General Stress Response Network in Escherichia coli: σ S -Dependent Genes, Promoters, and Sigma Factor Selectivity. J. Bacteriol. 2005, 187, 1591–1603. [Google Scholar] [CrossRef]
- Hengge-Aronis, R. Signal transduction and regulatory mechanisms involved in control of the σS subunit of RNA polymerase in Escherichia coli. Microbiol. Mol. Biol. Rev. 2002, 66, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Al Mamun, A.A.M.; Lombardo, M.; Shee, C.; Lisewski, A.M.; Gonzalez, C.; Lin, D.; Nehring, R.B.; Saint-Ruf, C.; Gibson, J.L.; Frisch, R.L. Identity and Function of a Large Gene Network Underlying Mutagenic Repair of DNA Breaks. Science 2012, 338, 1344–1348. [Google Scholar] [CrossRef] [PubMed]
- Beher, M.G.; Schnaitman, A.C. Regulation of the OmpA outer membrane protein of Escherichia coli. J. Bacteriol. 1981, 147, 972–985. [Google Scholar] [CrossRef]
- Reyes-Cortés, R.; Martínez-Peñafiel, E.; Martínez-Pérez, F.; Garza, M.; Kameyama, L. A novel strategy to isolate cell-envelope mutants resistant to phage infection: Bacteriophage mEp213 requires lipopolysaccharides in addition to FhuA to enter Escherichia coli K-12. Microbiology 2012, 158, 3063–3071. [Google Scholar] [CrossRef]
- Hantke, K. Compilation of Escherichia coli K-12 outer membrane phage receptors—Their function and some historical remarks. FEMS Microbiol. Lett. 2020, 367, 013. [Google Scholar] [CrossRef]
- Kadrmas, J.L.; Raetz, C.R.H. Enzymatic Synthesis of Lipopolysaccharide in Escherichia coli. Purification and properties of heptosyltransferase i. J. Biol. Chem. 1998, 273, 2799–2807. [Google Scholar] [CrossRef]
- Guisbert, E.; Rhodius, V.A.; Ahuja, N.; Witkin, E.; Gross, C.A. Hfq Modulates the σE-Mediated Envelope Stress Response and the σ32-Mediated Cytoplasmic Stress Response in Escherichia coli. J Bacteriol 2007, 189, 1963–1973. [Google Scholar] [CrossRef]
- Meberg, B.M.; Sailer, F.C.; Nelson, D.E.; Young, K.D. Reconstruction of Escherichia coli mrcA (PBP 1a) Mutants Lacking Mul-tiple Combinations of Penicillin Binding Proteins. J Bacteriol 2001, 183, 6148–6149. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A class of chaperone-like ATPases associated with the assem-bly, operation, and disassembly of protein complexes. Genome Res 1999, 9, 27–43. [Google Scholar] [CrossRef]
- Weichart, D.; Querfurth, N.; Dreger, M.; Hengge-Aronis, R. Global Role for ClpP-Containing Proteases in Stationary-Phase Adaptation of Escherichia coli. J Bacteriol 2003, 185, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Portalier, R.C.; Stoeber, F.R.L. NAD-oxydoréductase d’Escherichia coli K12. Purification, propriétés et indi-vidualité [D-altronate: NAD-oxidoreductase in Escherichia coli K12. Purification, properties, and specificity. Eur. J. Biochem. 1972, 26, 50–61. [Google Scholar] [CrossRef]
- Cotte-Pattat, N.; Robert-Baudouy, J. Isolation of fusions between the lac genes and several genes of the exu regulon: Analysis of their regulation, determination of the transcription direction of the uxaC-uxaA operon, in Escherichia coli K-12. Mol.Genet.Genom 1981, 182, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Rungrassamee, W.; Liu, X.; Pomposiello, P.J. Activation of glucose transport under oxidative stress in Escherichia coli. Arch Microbiol 2008, 190, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Ferrières, L.; Aslam, S.N.; Cooper, R.M.; Clarke, D.J. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 2007, 153, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, M.; Franchini, A.; Egli, T.; Belkin, S. Induction of the yjbEFGH operon is regulated by growth rate and oxygen con-centration. Arch Microbiol 2007, 189, 219–226. [Google Scholar] [CrossRef]
- Hirakawa, H.; Nishino, K.; Yamada, J.; Hirata, T.; Yamaguchi, A. Lactam resistance modulated by the overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J Antimicrob Chemother 2003, 52, 576–582. [Google Scholar] [CrossRef] [PubMed]

| (a) | ||||
| Substance | Range Tested | Population | Concentration | Interaction |
| Iron (III)/Phage > Controls | ||||
| Iron (III) | 100–1750 mg/L | F = 10.88, p = 0.002 | F = 133.21, p = <0.001 | F = 4.67, p = <0.001 |
| Iron (II) | 100–5000 mg/L | F = 40.58, p = <0.001 | F = 1.84, p = 0.090 | F = 3.34, p = 0.004 |
| Gallium (III) | 100–1750 mg/L | F = 653.99, p = <0.001 | F = 27.88, p = <0.001 | F = 15.72, p = <0.001 |
| Silver (I) | 5–100 mg/L | F = 157.74, p = <0.001 | F = 42.87, p = <0.001 | F = 43.99, p = <0.001 |
| Ampicillin | 20–500 mg/L | F = 82.44, p = <0.001 | F = 9.62, p = <0.001 | F = 1.31, p = 0.258 |
| Tetracycline | 5–100 mg/L | F = 60.13, p = <0.001 | F = 33.82, p = <0.001 | F = 21.11, p = <0.001 |
| Chloramphenicol | 5–250 mg/L | F = 39.69, p = 0.000 | F = 42.62, p = 0.000 | F = 6.71, p = 0.000 |
| Sulfonamide | 10–500 mg/L | F = 514.31, p = 0.000 | F = 49.23, p = 0.000 | F = 11.39, p = 0.000 |
| Rifampicin | 5–250 mg/L | F = 8.08, p = 0.006 | F = 73.18, p = 0.000 | F = 3.39, p = 0.003 |
| Iron (III)/Phage > Ancestor | ||||
| Iron (III) | 100–1750 mg/L | F = 150.54, p = 0.000 | F = 25.78, p = 0.000 | F = 5.56, p = 0.000 |
| Iron (II) | 100–5000 mg/L | F = 1.63, p = 0.207 | F = 0.24, p = 0.980 | F = 4.80, p = 0.000 |
| Gallium (III) | 100–1750 mg/L | F = 854.93, p = 0.000 | F = 69.20, p = 0.000 | F = 14.33, p = 0.000 |
| Silver (I) | 5–100 mg/L | F = 62.23, p = 0.000 | F = 10.41, p = 0.000 | F = 13.99, p = 0.000 |
| Ampicillin | 20–500 mg/L | F = 374.13, p = 0.000 | F = 7.71, p = 0.000 | F = 1.75, p = 0.108 |
| Tetracycline | 5–100 mg/L | F = 87.07, p = 0.000 | F = 9.98, p = 0.000 | F = 7.27, p = 0.000 |
| Chloramphenicol | 5–250 mg/L | F = 40.02, p = 0.000 | F = 42.51, p = 0.000 | F = 6.66, p = 0.000 |
| Sulfonamide | 10–500 mg/L | F = 1215.90, p = 0.000 | F = 62.81, p = 0.000 | F = 14.15, p = 0.000 |
| Rifampicin | 5–250 mg/L | F = 2762.29, p = 0.000 | F = 52.99, p = 0.000 | F = 51.72, p = 0.000 |
| Iron (III)/Phage > Iron (III) resistant | ||||
| Iron (III) | 100–1750 mg/L | F = 52.29, p = <0.001 | F = 82.59, p = <0.001 | F = 1.25, p = 0.283 |
| Ampicillin | 20–500 mg/L | F = 37.37, p = <0.001 | F = 3.31, p = 0.004 | F = 0.635, p = 0.475 |
| Tetracycline | 5–100 mg/L | F = 28.23, p = <0.001 | F = 24.51, p = <0.001 | F = 1.17, p = 0.335 |
| Iron (III)/Phage = Iron (III) resistant | ||||
| Chloramphenicol | 5–250 mg/L | F = 1.57, p = 0.216 | F = 49.27, p = 0.000 | F = 0.33, p = 0.950 |
| Iron(III)/Phage > Phage | ||||
| Iron (III) | 100–1750 mg/L | F = 21.25, p = <0.001 | F = 60.46, p = <0.001 | F = 1.55, p = 0.154 |
| Iron (II) | 100–5000 mg/L | F = 19.44, p = 0.000 | F = 5.31, p = 0.000 | F = 5.29, p = 0.000 |
| Gallium (III) | 100–1750 mg/L | F = 743.54, p = <0.001 | F = 29.70, p = <0.001 | F = 14.25, p = <0.001 |
| Ampicillin | 20–500 mg/L | F = 120.12, p = <0.001 | F = 11.48, p = <0.001 | F = 3.36, p = 0.003 |
| Tetracycline | 5–100 mg/L | F = 24.43, p = <0.001 | F = 13.34, p = <0.001 | F = 1.87, p = 0.084 |
| Chloramphenicol | 5–250 mg/L | F = 25.65, p = 0.000 | F = 32.29, p = 0.000 | F = 4.17, p = 0.001 |
| Silver (I) | 5–100 mg/L | F = 74.49, p = <0.001 | F = 14.11, p = <0.001 | F = 10.83, p = <0.001 |
| Sulfonamide | 5–250 mg/L | F = 345.86, p = 0.000 | F = 20.55, p = 0.000 | F = 4.51, p = 0.000 |
| Iron (III)/Phage = Phage | ||||
| Rifampicin | 5–250 mg/L | F = 3.70, p = 0.060 | F = 71.98, p = 0.000 | F = 3.88, p = 0.000 |
| Sulfonamide | 10–500 mg/L | F = 0.41, p = 0.523 | F = 8.56, p = 0.000 | F = 0.48, p = 0.868 |
| Silver (I) | 5–100 mg/L | F = 0.73, p = 0.398 | F = 43.62, p = <0.001 | F = 0.37, p = 0.932 |
| Iron (III) > Iron (III)/Phage | ||||
| Rifampicin | 5–250 mg/L | F = 4.19, p = 0.046 | F = 67.85, p = 0.000 | F = 0.96, p = 0.479 |
| (b) | ||||
| Substance | Range Tested | Population | Concentration | Interaction |
| Iron (III) > Phage | ||||
| Iron (II) | 100–5000 mg/L | F = 38.18, p = <0.001 | F = 2.74, p = 0.013 | F = 2.99, p = 0.007 |
| Gallium (III) | 100–1750 mg/L | F = 696.66, p = <0.001 | F = 30.41, p = <0.001 | F = 17.44, p = <0.001 |
| Silver (I) | 5–100 mg/L | F = 207.07, p = <0.001 | F = 59.90, p = <0.001 | F = 47.65, p = <0.001 |
| Ampicillin | 20–500 mg/L | F = 12.67, p = <0.001 | F = 16.48, p = <0.001 | F = 2.82, p = 0.011 |
| Sulfonamide | 5–250 mg/L | F = 79.40, p = 0.000 | F = 3.30, p = 0.00 | F = 0.368, p = 0.933 |
| Chloramphenicol | 5–250 mg/L | F = 21.67, p = 0.000 | F = 38.99, p = 0.000 | F = 4.22, p = 0.001 |
| Phage > Iron (III) | ||||
| Iron (III) | 100–1750 mg/L | F = 9.53, p = 0.003 | F = 146.84, p = <0.001 | F = 1.95, p = 0.065 |
| Iron (III) = Phage | ||||
| Tetracycline | 5–100 mg/L | F = 0.20, p = 0.661 | F = 25.51, p = <0.001 | F = 3.78, p = <0.001 |
| Rifampicin | 5–250 mg/L | F = 0.01, p = 0.908 | F = 50.28, p = 0.000 | F = 3.13, p = 0.006 |
| Iron (III) > Control | ||||
| Iron (III) | 100–1750 mg/L | F = 10.88, p = 0.002 | F = 133.21, p = <0.001 | F = 4.67, p = <0.001 |
| Iron (II) | 100–5000 mg/L | F = 40.58, p = <0.001 | F = 1.84, p = 0.090 | F = 3.34, p = 0.004 |
| Gallium (III) | 100–1750 mg/L | F = 653.99, p = <0.001 | F = 27.88, p = <0.001 | F = 15.72, p = <0.001 |
| Tetracycline | 5–100 mg/L | F = 60.13, p = <0.001 | F = 33.82, p = <0.001 | F = 21.11, p = <0.001 |
| Ampicillin | 20–500 mg/L | F = 82.44, p = <0.001 | F = 9.62, p = <0.001 | F = 1.31, p = 0.258 |
| Chloramphenicol | 5–250 mg/L | F = 43.62, p = 0.000 | F = 65.78, p = 0.000 | F = 7.53, p = 0.000 |
| Sulfonamide | 5–250 mg/L | F = 50.85, p = 0.000 | F = 3.33, p = 0.004 | F = 0.33, p = 0.950 |
| Rifampicin | 5–250 mg/L | F = 19.17, p = 0.000 | F = 53.98, p = 0.000 | F = 4.14, p = 0.001 |
| Iron (III)>Ancestor | ||||
| Iron (III) | 100–1750 mg/L | F = 131.19, p = <0.001 | F = 65.68, p = <0.001 | F = 11.74, p = <0.001 |
| Silver (I) | 5–100 mg/L | F = 155.21, p = <0.001 | F = 42.20, p = <0.001 | F = 53.63, p = <0.001 |
| Gallium (III) | 100–1750 mg/L | F = 756.28, p = <0.001 | F = 60.10, p = <0.001 | F = 19.12, p = <0.001 |
| Ampicillin | 20–500 mg/L | F = 151.14, p = <0.001 | F = 12.71, p = <0.001 | F = 0.76, p = 0.637 |
| Tetracycline | 5–100 mg/L | F = 123.49, p = <0.001 | F = 59.99, p = <0.001 | F = 40.17, p = <0.001 |
| Chloramphenicol | 5–250 mg/L | F = 58.84, p = 0.000 | F = 32.16, p = 0.000 | F = 25.73, p = 0.000 |
| Sulfonamide | 5–250 mg/L | F = 103.22, p = 0.000 | F = 3.49, p = 0.003 | F = 0.368, p = 0.933 |
| Rifampicin | 5–250 mg/L | F = 2206.29, p = 0.000 | F = 25.69, p = 0.000 | F = 26.32, p = 0.000 |
| (c) | ||||
| Substance | Range Tested | Population | Concentration | Interaction |
| Phage > Control | ||||
| Iron (III) | 100–1750 mg/L | F = 27.05, p = <0.001 | F = 59.26, p = <0.001 | F = 1.33, p = 0.242 |
| Ampicillin | 20–500 mg/L | F = 64.16, p = <0.001 | F = 36.35, p = <0.001 | F = 1.20, p = 0.317 |
| Tetracycline | 5–100 mg/L | F = 19.44, p = <0.001 | F = 5.66, p = <0.001 | F = 3.48, p = 0.003 |
| Rifampicin | 5–250 mg/L | F = 18.29, p = 0.000 | F = 61.51, p = 0.000 | F = 1.53, p = 0.169 |
| Sulfonamide | 5–250 mg/L | F = 20.90, p = 0.000 | F = 6.99, p = 0.000 | F = 0.59, p = 0.779 |
| Phage = Control | ||||
| Iron (II) | 100–5000 mg/L | F = 3.59, p = 0.064 | F = 1.05, p = 0.414 | F = 1.40, p = 0.220 |
| Gallium (III) | 100–1750 mg/L | F = 0.015, p = 0.903 | F = 6.86, p = <0.001 | F = 1.06, p = 0.407 |
| Silver (I) | 5–100 mg/L | F = 0.536, p = 0.467 | F = 1.66, p = 0.129 | F = 2.14, p = 0.047 |
| Chloramphenicol | 5–250 mg/L | F = 0.049, p = 0.826 | F = 35.88, p = 0.000 | F = 1.27, p = 0.279 |
| Phage > Ancestor | ||||
| Iron (III) | 100–1750 mg/L | F = 149.61, p = 0.000 | F = 43.78, p = 0.000 | F = 5.66, p = 0.000 |
| Tetracycline | 5–100 mg/L | F = 26.74, p = 0.000 | F = 6.84, p = <0.000 | F = 4.09, p = 0.001 |
| Ampicillin | 20–500 mg/L | F = 168.61, p = <0.000 | F = 47.16, p = <0.000 | F = 3.05, p = 0.007 |
| Rifampicin | 5–250 mg/L | F = 2198.97, p = 0.000 | F = 36.08, p = 0.000 | F = 27.82, p = 0.000 |
| Phage = Ancestor | ||||
| Chloramphenicol | 5–250 mg/L | F = 1.68, p = 0.200 | F = 12.78, p = 0.000 | F = 9.65, p = 0.000 |
| Sulfonamide | 5–250 mg/L | F = 0.31, p = 0.580 | F = 7.36, p = 0.000 | F = 1.46, p = 0.195 |
| Ancestor > Phage | ||||
| Iron (II) | 100–5000 mg/L | F = 11.76, p = 0.001 | F = 1.32, p = 0.253 | F = 1.46, p = 0.194 |
| Gallium (III) | 100–1750 mg/L | F = 11.53, p = 0.001 | F = 31.11, p = 0.000 | F = 11.35, p = 0.000 |
| Silver (I) | 5–100 mg/L | F = 4.91, p = 0.031 | F = 2.52, p = 0.021 | F = 9.51, p = 0.000 |
| (d) | ||||
| Substance | Range tested | Population | Concentration | Interaction |
| Control > Ancestor | ||||
| Iron (III) | 100–1750 mg/L | F = 42.01, p = <0.001 | F = 30.36, p = <0.001 | F = 2.26, p = 0.033 |
| Gallium (III) | 100–1750 mg/L | F = 9.40, p = 0.003 | F = 27.20, p = <0.001 | F = 7.53, p = <0.001 |
| Ampicillin | 20–500 mg/L | F = 12.16, p = <0.001 | F = 25.87, p = <0.001 | F = 1.39, p = 0.221 |
| Chloramphenicol | 5–250 mg/L | F = 7.55, p = 0.008 | F = 58.18, p = 0.000 | F = 38.75, p = 0.000 |
| Rifampicin | 5–250 mg/L | F = 1123.30, p = 0.000 | F = 38.25, p = 0.000 | F = 30.36, p = 0.000 |
| Sulfonamide | 5–250 mg/L | F = 83.30, p = 0.000 | F = 17.64, p = 0.000 | F = 1.26, p = 0.286 |
| Control = Ancestor | ||||
| Silver (I) | 5–100 mg/L | F = 0.94, p = 0.337 | F = 2.39, p = 0.028 | F = 3.24, p = 0.004 |
| Tetracycline | 5–100 mg/L | F = 0.39, p = 0.536 | F = 3.68, p = 0.002 | F = 0.28, p = 0.970 |
| Ancestor > Control | ||||
| Iron (II) | 100–5000 mg/L | F = 15.02, p = 0.000 | F = 1.56, p = 0.160 | F = 1.28, p = 0.273 |
| Populations | Iron (III) | Phage T7 |
|---|---|---|
| Iron (III)/Phage-resistant | 1 | 1 |
| Phage-resistant | 3 | 2 |
| Iron (III)-resistant | 2 | 3 |
| Control | 4 | 4 |
| Ancestor | 5 | 5 |
| (a) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Position | Mutation | Fe1 | Fe2 | Fe3 | Fe4 | Fe5 | Fe6 | Fe7 | F8 | F9/F10 |
| rpoC → | 4,185,540 | C→T | 0.078 | 0.000 | 0.000 | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| rpoB → | 4,183,399 | Δ6 bp | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.935 | 0.000 | 1.000 |
| rpoB → | 4,183,204 | G→T | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.902 | 0.000 | 0.063 | 0.000 |
| rpoB → | 4,181,278 | C→T | 0.000 | 0.000 | 0.000 | 0.000 | 0.801 | 0.000 | 0.000 | 0.000 | 0.000 |
| rpoC → | 4,187,633 | A→C | 0.000 | 0.713 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| yeaG → | 1,868,570 | G→T | 0.000 | 0.647 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| rpoB → | 4,184,809 | G→A | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.527 | 0.000 | 0.000 |
| rpoB → | 4,184,795 | C→G | 0.000 | 0.000 | 0.526 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| ycgB ← | 1,236,863 | G→A | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.637 | 0.000 |
| (b) | |||||||||||
| Gene | Annotation | ||||||||||
| rpoC → | P64L (CCG→CTG) | ||||||||||
| rpoB → | D654Y (GAC→TAC) | ||||||||||
| rpoB → | R12C (CGT→TGT) | ||||||||||
| rpoC → | N762H (AAC→CAC) | ||||||||||
| yeaG → | E555 * (GAG→TAG) | ||||||||||
| rpoB → | G1189S (GGT→AGT) | ||||||||||
| rpoB → | T1184R (ACG→AGG) | ||||||||||
| ychE →/→ oppA | intergenic (+254/-485) | ||||||||||
| ycgB ← | H127Y (CAT→TAT) | ||||||||||
| rpoB → | coding (2155-2160/4029 nt) | ||||||||||
| (a) | ||||||
|---|---|---|---|---|---|---|
| Gene | Position | Mutation | Ph2 | Ph3 | Ph4 | Ph5 |
| clpX →/→ lon | 458,790 | IS186 (-) + 6 bp:: Δ1 bp | 1.000 | 0.000 | 1.000 | 1.000 |
| clpX →/→ lon | 458,790 | IS186 (+) +6 bp:: Δ1 bp | 1.000 | 1.000 | 0.000 | 0.000 |
| uxaB←/←yneF | 1,610,807 | T→C | 0.000 | 0.000 | 0.000 | 1.000 |
| yeaG→ | 1,868,147 | G→A | 0.000 | 1.000 | 0.000 | 0.000 |
| proQ← | 1,915,478 | Δ1 bp | 0.000 | 0.771 | 0.000 | 0.000 |
| fliR →/→ rcsA | 2,023,824 | Δ1 bp | 1.000 | 0.000 | 0.000 | 0.000 |
| [gatR]–[fbaB] | 2,171,429 | Δ6547 bp | 0.000 | 0.000 | 0.000 | 1.000 |
| yejM→ | 2,285,441 | C→T | 0.000 | 0.787 | 0.000 | 0.000 |
| ypjF→/←ypjA | 2,777,982 | Δ1 bp | 0.000 | 0.000 | 0.000 | 1.000 |
| rpoS← | 2,867,175 | C→A | 0.536 | 0.000 | 0.000 | 0.000 |
| rpoS← | 2,867,178 | A→T | 0.000 | 0.000 | 1.000 | 0.000 |
| rpoS← | 2,867,428 | C→A | 0.183 | 0.000 | 0.000 | 1.000 |
| rpoS← | 2,867,322 | +CTT | 0.000 | 0.868 | 0.000 | 0.000 |
| waaC→ | 3,796,019 | Δ1 bp | 0.000 | 1.000 | 0.000 | 0.000 |
| waaC→ | 3,796,167 | IS3 (+) +3 bp | 0.000 | 0.000 | 1.000 | 0.000 |
| rpoC→ | 4,186,532 | A→G | 0.000 | 0.000 | 1.000 | 0.000 |
| rpoC→ | 4,187,522 | A→C | 0.827 | 0.000 | 0.000 | 0.000 |
| pgi →/→ yjbE | 4,235,682 | T→C | 0.000 | 0.000 | 0.000 | 1.000 |
| yjbH→ | 4,237,938 | A→C | 0.000 | 0.000 | 0.000 | 1.000 |
| yjbH→ | 4,238,073 | G→A | 0.000 | 0.000 | 1.000 | 0.000 |
| yjbH→ | 4,239,443 | Δ5 bp | 0.704 | 0.000 | 0.000 | 0.000 |
| (b) | ||||||
| Gene | Annotation | |||||
| clpX →/→ lon | intergenic (+90/−93) | |||||
| clpX →/→ lon | intergenic (+90/−93) | |||||
| uxaB←/←yneF | intergenic (−127/+100) | |||||
| yeaG→ | E414K (GAA→AAA) | |||||
| proQ← | coding (57/699 nt) | |||||
| fliR →/→ rcsA | intergenic (+146/−144) | |||||
| [gatR]–[fbaB] | IS3-mediated | |||||
| yejM→ | Q356 * (CAG→TAG) | |||||
| ypjF→/←ypjA | intergenic (+200/+164) | |||||
| rpoS← | G126V (GGG→GTG) | |||||
| rpoS← | L125Q (CTG→CAG) | |||||
| rpoS← | E42 * (GAA→TAA) | |||||
| rpoS← | coding (230/993 nt) | |||||
| waaC→ | coding (41/960 nt) | |||||
| waaC→ | coding (189–191/960 nt) | |||||
| rpoC→ | K395E (AAA→GAA) | |||||
| rpoC→ | M725L (ATG→CTG) | |||||
| pgi →/→ yjbE | intergenic (+275/−224) | |||||
| yjbH→ | Y102S (TAT→TCT) | |||||
| (a) | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Position | Mutation | FPh1 | FPh2 | FPh4 | FPh5 | FPh6 |
| rcsA → | 2,024,505 | A→T | 0.867 | 1.000 | 0.842 | 1.000 | 0.721 |
| hldE ← | 3,195,969 | G→T | 0.000 | 0.000 | 0.000 | 1.000 | 0.000 |
| waaC → | 3,796,783 | IS1 (+) +9 bp | 1.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| rpoB → | 4,183,399 | Δ6 bp | 0.000 | 0.000 | 0.474 | 1.000 | 0.521 |
| nudE ←/→ yrfF | 3,526,449 | IS5 (+) +4 bp | 0.000 | 0.000 | 0.000 | 0.628 | 0.000 |
| hldD → | 3,794,149 | +GA:: IS3 (+) +4 bp | 0.000 | 0.895 | 0.159 | 0.000 | 0.238 |
| rpoC → | 4,187,507 | A→C | 0.726 | 0.000 | 0.000 | 0.000 | 0.000 |
| (b) | |||||||
| Gene | Annotation | ||||||
| rcsA→ | I180F (ATC→TTC) | ||||||
| hldE← | A262E (GCG→GAG) | ||||||
| waaC→ | coding (805–813/960 nt) | ||||||
| rpoB→ | coding (2155–2160/4029 nt) | ||||||
| nudE ←/→ yrfF | intergenic (−300/−17) | ||||||
| hldD→ | coding (163–166/933 nt) | ||||||
| rpoC→ | N720H (AAC→CAC) | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeje, O.; Ewunkem, A.J.; Jeffers-Francis, L.K.; Graves, J.L., Jr. Serving Two Masters: Effect of Escherichia coli Dual Resistance on Antibiotic Susceptibility. Antibiotics 2023, 12, 603. https://doi.org/10.3390/antibiotics12030603
Jeje O, Ewunkem AJ, Jeffers-Francis LK, Graves JL Jr. Serving Two Masters: Effect of Escherichia coli Dual Resistance on Antibiotic Susceptibility. Antibiotics. 2023; 12(3):603. https://doi.org/10.3390/antibiotics12030603
Chicago/Turabian StyleJeje, Olusola, Akamu J. Ewunkem, Liesl K. Jeffers-Francis, and Joseph L. Graves, Jr. 2023. "Serving Two Masters: Effect of Escherichia coli Dual Resistance on Antibiotic Susceptibility" Antibiotics 12, no. 3: 603. https://doi.org/10.3390/antibiotics12030603
APA StyleJeje, O., Ewunkem, A. J., Jeffers-Francis, L. K., & Graves, J. L., Jr. (2023). Serving Two Masters: Effect of Escherichia coli Dual Resistance on Antibiotic Susceptibility. Antibiotics, 12(3), 603. https://doi.org/10.3390/antibiotics12030603






