Bioinformatics Approaches Applied to the Discovery of Antifungal Peptides
Abstract
1. Introduction
2. Results and Discussions
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The changing epidemiology of invasive fungal infections. Methods Mol. Biol. 2017, 1508, 17–65. [Google Scholar] [CrossRef] [PubMed]
- Stewart, A.G.; Paterson, D.L. How urgent is the need for new antifungals? Expert Opin. Pharmacother. 2021, 22, 1857–1870. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Guarner, J. Emerging and reemerging fungal infections. Semin. Diagn. Pathol. 2019, 36, 177–181. [Google Scholar] [CrossRef]
- Li, T.; Li, L.; Du, F.; Sun, L.; Shi, J.; Long, M.; Chen, Z. Activity and mechanism of action of AFPs from microorganisms: A Review. Molecules 2021, 26, 3438. [Google Scholar] [CrossRef]
- Fernández de Ullivarri, M.; Arbulu, S.; Garcia-Gutierrez, E.; Cotter, P.D. Antifungal peptides as therapeutic agents. Front. Cell. Infect. Microbiol. 2020, 10, 105. [Google Scholar] [CrossRef]
- Tyagi, A.; Roy, S.; Singh, S.; Semwal, M.; Shasany, A.K.; Sharma, A.; Provazník, I. Phytoafp: In Silico approaches for designing plant-derived antifungal peptides. Antibiotics 2021, 10, 815. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Rodríguez-Cerdeira, C.; Gregorio, M.C.; Molares-Vila, A.; López-Barcenas, A.; Fabbrocini, G.; Bardhi, B.; Sinani, A.; Sánchez-Blanco, E.; Arenas-Guzmán, R.; Hernandez-Castro, R. Biofilms and vulvovaginal candidiasis. Colloids Surf. B Biointerfaces 2019, 174, 110–125. [Google Scholar] [CrossRef]
- Mousavizadegan, M.; Mohabatkar, H. An evaluation on different machine learning algorithms for classification and prediction of antifungal peptides. Med. Chem. 2016, 12, 795–800. [Google Scholar] [CrossRef]
- Singh, V.; Shrivastava, S.; Kumar Singh, S.; Kumar, A.; Saxena, S. Accelerating the discovery of antifungal peptides using deep temporal convolutional networks. Brief. Bioinform. 2022, 23, bbac008. [Google Scholar] [CrossRef]
- Wang, G. improved methods for classification, prediction and design of antimicrobial peptides. Methods Mol. Biol. 2015, 1268, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Menousek, J.; Mishra, B.; Hanke, M.L.; Heim, C.E.; Kielian, T.; Wang, G. Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300. Int. J. Antimicrob. Agents 2012, 39, 402–406. [Google Scholar] [CrossRef]
- Wang, G.; Watson, K.M.; Peterkofsky, A.; Buckheit, R.W. Identification of novel human immunodeficiency virus type 1-inhibitory peptides based on the antimicrobial peptide database. Antimicrob. Agents Chemother. 2010, 54, 1343–1346. [Google Scholar] [CrossRef]
- Wang, G. Database-guided discovery of potent peptides to combat HIV-1 or superbugs. Pharmaceuticals 2013, 6, 728–758. [Google Scholar] [CrossRef]
- Mishra, B.; Wang, G. Ab Initio Design of potent anti-MRSA peptides based on database filtering technology. J. Am. Chem. Soc. 2012, 134, 12426–12429. [Google Scholar] [CrossRef] [PubMed]
- Hadley, E.B.; Hancock, R.E.W. Strategies for the Discovery and advancement of novel cationic antimicrobial peptides. Curr. Top. Med. Chem. 2010, 10, 1872–1881. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Pankaj, V.; Singh, S.; Roy, S.; Semwal, M.; Shasany, A.K.; Sharma, A. PlantAFP: A curated database of plant-origin antifungal peptides. Amino Acids 2019, 51, 1561–1568. [Google Scholar] [CrossRef] [PubMed]
- Doytchinova, I.A.; Walshe, V.A.; Jones, N.A.; Gloster, S.E.; Borrow, P.; Flower, D.R. Coupling in Silico And In Vitro Analysis of Peptide-Mhc Binding: A Bioinformatic Approach Enabling Prediction of Superbinding Peptides And Anchorless Epitopes. J. Immunol. 2004, 172, 7495–7502. [Google Scholar] [CrossRef]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Nizet, V.; Dorrestein, P.C. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 2011, 157, 2485–2492. [Google Scholar] [CrossRef]
- Polanco, C.; Samaniego, J.L.; Buhse, T.; Mosqueira, F.G.; Negron-Mendoza, A.; Ramos-Bernal, S.; Castanon-Gonzalez, J.A. Characterization of selective antibacterial peptides by polarity index. Int. J. Pept. 2012, 2012, 1–11. [Google Scholar] [CrossRef]
- Polanco, C.; Samaniego-Mendoza, J.L.; Buhse, T.; Castañón-González, J.A.; Leopold-Sordo, M. Polar characterization of antifungal peptides from apd2 database. Cell Biochem. Biophys. 2014, 70, 1479–1488. [Google Scholar] [CrossRef] [PubMed]
- Mousavizadegan, M.; Mohabatkar, H. Computational prediction of antifungal peptides via Chou’s PseAAC and SVM. J. Bioinform. Comput. Biol. 2018, 16, 1850016. [Google Scholar] [CrossRef]
- Shen, H.-B.; Chou, K.-C. PseAAC: A flexible web server for generating various kinds of protein pseudo amino acid composition. Anal. Biochem. 2008, 373, 386–388. [Google Scholar] [CrossRef] [PubMed]
- Schölkopf, B.; Smola, A.J.; Bach, F. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambridge, MA, USA, 2002; ISBN 9780262536578/9780262194754. [Google Scholar]
- Brown, M.P.S.; Grundy, W.N.; Lin, D.; Cristianini, N.; Sugnet, C.W.; Furey, T.S.; Ares, M.; Haussler, D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. USA 2000, 97, 262–267. [Google Scholar] [CrossRef]
- Thomas, S.; Karnik, S.; Barai, R.S.; Jayaraman, V.K.; Idicula-Thomas, S. CAMP: A useful resource for research on antimicrobial peptides. Nucleic Acids Res. 2009, 38, D774–D780. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Ben Hamida, J.; Vergoten, G.; Fliss, I. PhytAMP: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2008, 37, D963–D968. [Google Scholar] [CrossRef]
- Ojeda, F.; Suykens, J.A.K.; De Moor, B. Low rank updated LS-SVM classifiers for fast variable selection. Neural Netw. 2008, 21, 437–449. [Google Scholar] [CrossRef]
- Mierswa, I.; Wurst, M.; Klinkenberg, R.; Scholz, M.; Euler, T. yale: Rapid prototyping for complex data mining tasks. In Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Philadelphia, PA, USA, 20–23 August 2006; Association for Computing Machinery: New York, NY, USA, 2006; pp. 935–940. [Google Scholar]
- Amaral, A.C.; Silva, O.N.; Mundim, N.C.C.R.; De Carvalho, M.J.A.; Migliolo, L.; Leite, J.R.S.A.; Prates, M.V.; Bocca, A.L.; Franco, O.L.; Felipe, M.S.S. Predicting antimicrobial peptides from eukaryotic genomes: In silico strategies to develop antibiotics. Peptides 2012, 37, 301–308. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmann, J.A.C.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and Procheck-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Sumathi, K.; Ananthalakshmi, P.; Roshan, M.N.A.M.; Sekar, K. 3dSS: 3D structural superposition. Nucleic Acids Res. 2006, 34, W128–W132. [Google Scholar] [CrossRef] [PubMed]
- Proaño-Bolaños, C.; Zhou, M.; Wang, L.; Coloma, L.A.; Chen, T.; Shaw, C. Peptidomic approach identifies cruzioseptins, a new family of potent antimicrobial peptides in the splendid leaf frog, Cruziohyla calcarifer. J. Proteom. 2016, 146, 1–13. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Peptide Mass Calculator V3.2. Available online: http://rna.rega.kuleuven.be/masspec/pepcalc.htm (accessed on 16 May 2022).
- Garnier, J.; Gibrat, J.-F.; Robson, B. The GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996, 266, 540–553. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Peptide Calculator. Available online: https://www.bachem.com/knowledge-center/peptide-calculator/ (accessed on 16 May 2022).
- Neelabh; Singh, K.; Rani, J. Sequential and structural aspects of antifungal peptides from animals, bacteria and fungi based on bioinformatics tools. Probiotics Antimicrob. Proteins 2016, 8, 85–101. [Google Scholar] [CrossRef]
- Poirot, O.; O’Toole, E.; Notredame, C. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003, 31, 3503–3506. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhalla, S.; Chaudhary, K.; Kumar, R.; Sharma, M.; Raghava, G.P.S. In silico approach for prediction of antifungal peptides. Front. Microbiol. 2018, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An Enhanced Comprehensive Data Repository of Antimicrobial Peptides. Nucleic Acids Res. 2021, 50, D488–D496. [Google Scholar] [CrossRef] [PubMed]
- Beaufays, J.; Lins, L.; Thomas, A.; Brasseur, R. In silico predictions of 3D structures of linear and cyclic peptides with natural and non-proteinogenic residues. J. Pept. Sci. 2011, 18, 17–24. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical pka predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- BIOVIA Discovery Studio. 2016. Available online: https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/ (accessed on 7 November 2022).
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD—Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Amaral, J.L.; Souza, P.F.N.; Oliveira, J.T.A.; Freire, V.N.; Sousa, D.O.B. Computational approach, scanning electron and fluorescence microscopies revealed insights into the action mechanisms of anticandidal peptide Mo-CBP3-PepIII. Life Sci. 2021, 281, 119775. [Google Scholar] [CrossRef]
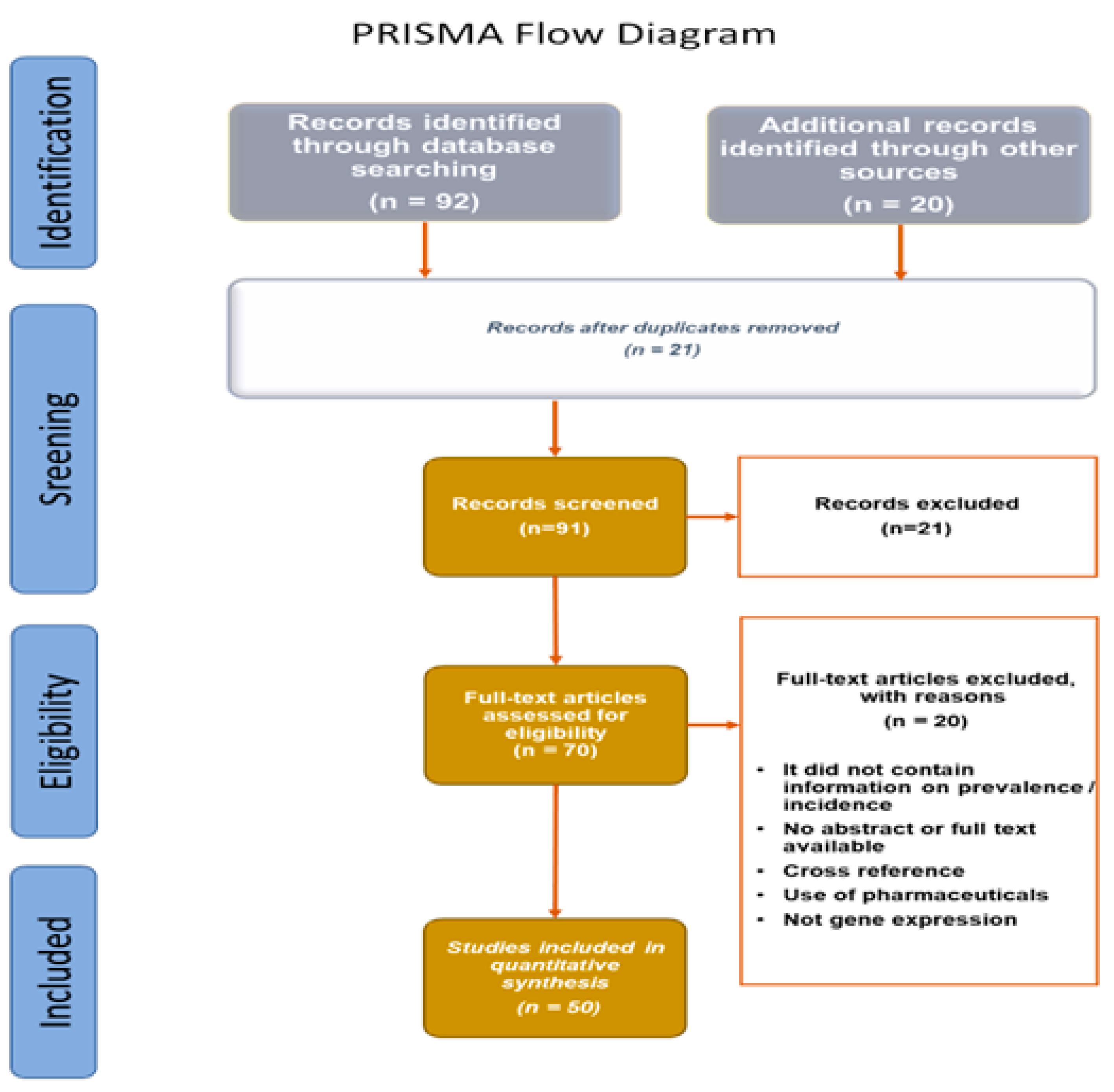
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]

| Computational Resources | Utility | Discussion | References |
|---|---|---|---|
| APD | Reference site for AFPs | Complete database | [11] |
| PlantAFP | Repository for plant-derived AFPs | Complete database | [17] |
| Polarity Index | Identification of AFPs | More suitable for bacteria than fungi; efficiency > 90% | [20] |
| In-house method | AFP classification and prediction | Positive validation assays performed in vitro for peptides with high antifungal prediction score (>0.95) | [22] |
| In-house method | Identification of AFP sequences from P. brasiliensis and H. sapiens | Four highest-scoring peptides were selected in silico and checked in vitro; two peptides had weak antifungal activity against Candida albicans | [30] |
| In-house method | Identification of AFP sequences from C. calcarifer | Antimicrobial activity against C. albicans found in three synthetic peptides | [33] |
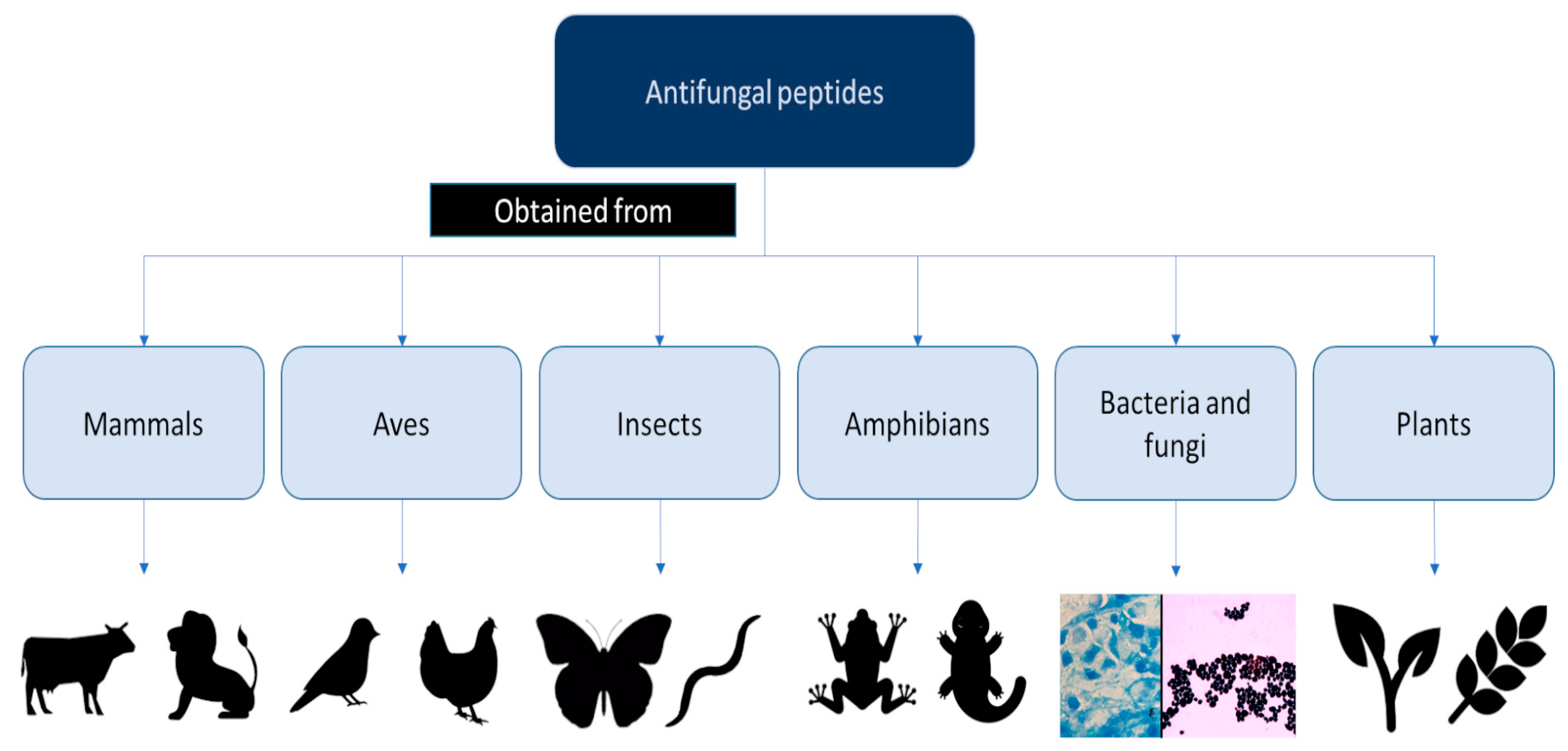
| In-house method | Discovery of AFPs produced naturally by prokaryotes and eukaryotes | Review of some AFPs produced in mammals, birds, insects, amphibians, and microbes based on their structural characterization | [42] |
| Antifp | Class-specific prediction web server for AFPs | Differentiates with good accuracy between sequences that are very similar in identity but possess different activities | [45] |
| PhytoAFP | Web prediction server | Prediction and design of plant-derived antifungal peptides | [6] |
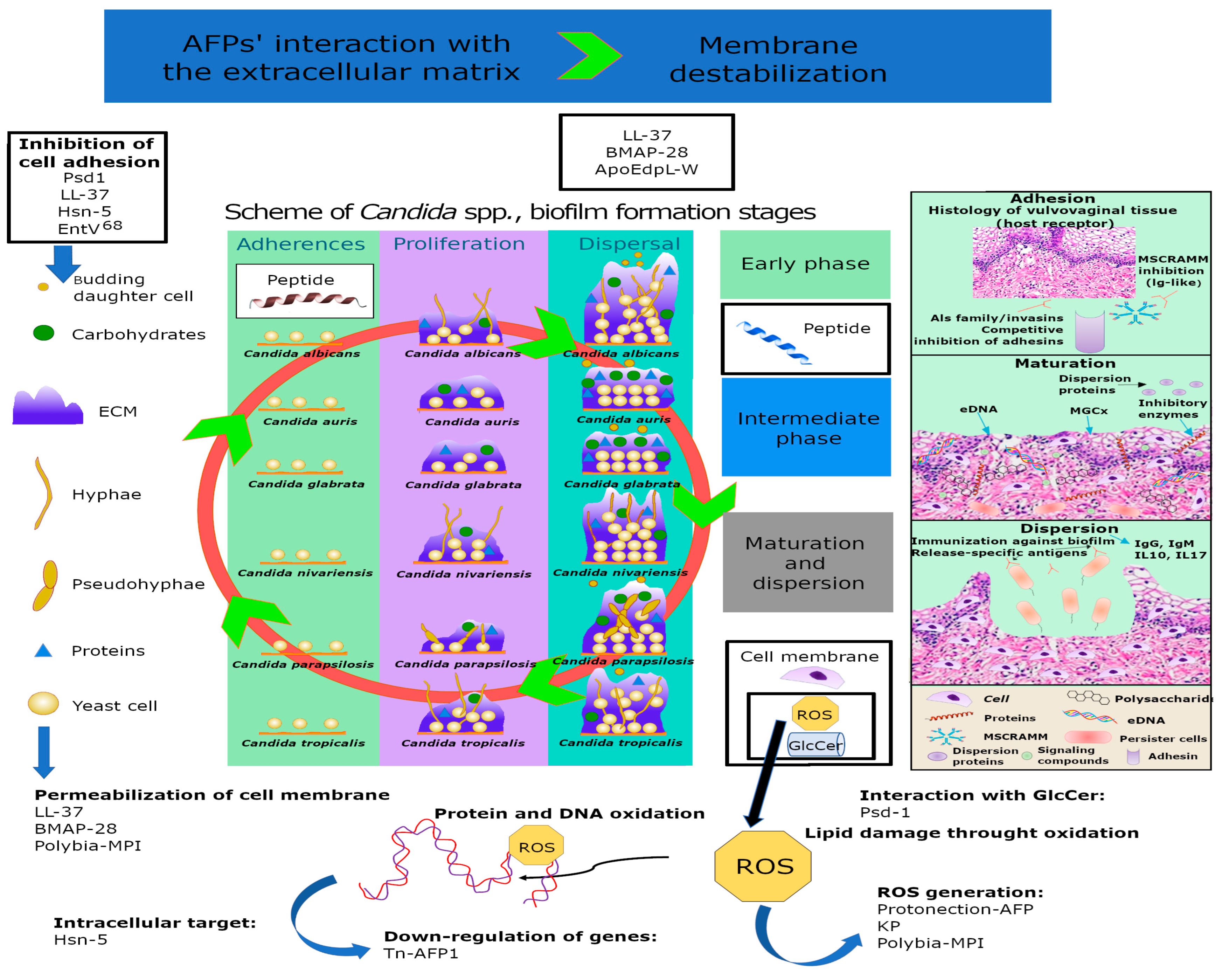
| In-house method | Mechanism-of-action analysis of antifungal agents | In silico assays (molecular docking and dynamics simulations) indicated that cell wall and membrane of C. albicans are targeted by Mo-CBP3-PepIII | [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Cerdeira, C.; Molares-Vila, A.; Sánchez-Cárdenas, C.D.; Velásquez-Bámaca, J.S.; Martínez-Herrera, E. Bioinformatics Approaches Applied to the Discovery of Antifungal Peptides. Antibiotics 2023, 12, 566. https://doi.org/10.3390/antibiotics12030566
Rodríguez-Cerdeira C, Molares-Vila A, Sánchez-Cárdenas CD, Velásquez-Bámaca JS, Martínez-Herrera E. Bioinformatics Approaches Applied to the Discovery of Antifungal Peptides. Antibiotics. 2023; 12(3):566. https://doi.org/10.3390/antibiotics12030566
Chicago/Turabian StyleRodríguez-Cerdeira, Carmen, Alberto Molares-Vila, Carlos Daniel Sánchez-Cárdenas, Jimmy Steven Velásquez-Bámaca, and Erick Martínez-Herrera. 2023. "Bioinformatics Approaches Applied to the Discovery of Antifungal Peptides" Antibiotics 12, no. 3: 566. https://doi.org/10.3390/antibiotics12030566
APA StyleRodríguez-Cerdeira, C., Molares-Vila, A., Sánchez-Cárdenas, C. D., Velásquez-Bámaca, J. S., & Martínez-Herrera, E. (2023). Bioinformatics Approaches Applied to the Discovery of Antifungal Peptides. Antibiotics, 12(3), 566. https://doi.org/10.3390/antibiotics12030566









