Abstract
The cfr genes encode for a 23S rRNA methyltransferase, conferring a multiresistance phenotype to phenicol, lincosamide, oxazolidinone, pleuromutilin, and streptogramin A antibiotics. These genes have been described in staphylococci, including methicillin-resistant Staphylococcus aureus (MRSA). In this study, we retrospectively performed an in-depth genomic characterisation of three cfr-positive, multidrug-resistant (MDR) livestock-associated (LA) MRSA clonal complexes (CCs) 1 and 398 detected in different Italian pig holdings (2008–2011) during population studies on Italian livestock (2008–2014). We used a combined Illumina and Oxford Nanopore Technologies (ONT) whole genome sequencing (WGS) approach on two isolates (the 2008 CC1 and the 2010 CC398 isolates, but not the 2011 CC1 isolate). Interestingly, the three isolates presented different cfr variants, with only one displaying a linezolid-resistant phenotype. In isolate 2008 CC1, the cfr gene was identified within a Tn558 composite transposon-like structure flanked by IS elements located on a novel 44,826 bp plasmid. This represents the first report of CC1 LA-MRSA harbouring the cfr gene in its functional variant. Differently, cfr was chromosomally located in isolate 2010 CC398. Our findings have significant public health implications, confirm the need for the continuous genomic surveillance of cfr-positive zoonotic LA-MRSA, and backdate cfr presence in LA-MRSA from Italian pigs to at least 2008.
Keywords:
Staphylococcus aureus; LA-MRSA; linezolid; cfr; WGS; long reads; short reads; bioinformatics analysis; CC1; CC398 1. Introduction
Linezolid was the first oxazolidinone antimicrobial available exclusively for human use from April 2000, and it represents one of the few therapeutic options to treat methicillin-resistant Staphylococcus aureus (MRSA) skin and soft-tissue infections, osteomyelitis, and pneumonia [1]. Seven mobilisable oxazolidinone resistance genes, including cfr, cfr(B), cfr(C), cfr(D), cfr(E), optrA, and poxtA, have been described so far [2]. In particular, after being identified for the first time about 20 years ago in Staphylococcus sciuri of bovine origin in Germany [3], cfr genes have been found in 19 staphylococcal species from both human and animal sources as well as in other bacterial species [2]. These genes encode for a 23S rRNA methyltransferase, which confers a multiresistance phenotype (phenotype PhLOPSA), including resistance to phenicol, lincosamide, oxazolidinone, pleuromutilin, and streptogramin A antibiotic classes [4]. The cfr genes have been detected in both methicillin-susceptible S. aureus (MSSA) and MRSA isolates from human clinical samples worldwide [2], including Italy [5]. The cfr(B) gene has recently been described for the first time in MRSA isolates from French clinical human samples [6]. In livestock, cfr was first described in S. aureus of porcine origin in Germany in 2006 [7]. Since then, it has sporadically been detected in pig samples from Belgium [8], Spain [9], Portugal [10], the Netherlands [11], China [12], and South Korea [13], and in bovine samples from Egypt [14]. These genes are most frequently located on mobile genetic elements (MGEs), especially plasmids, which were proven to harbour cfr-carrying transposons and translocatable units (TUs), playing a crucial role in their dissemination [15]. Additionally, TUs and integrative and conjugative elements (ICEs) have also been detected at a chromosomal level [2].
In this study, we retrospectively report the in-depth genomic characterisation of three cfr-positive, multidrug-resistant (MDR) livestock-associated (LA) MRSA isolates belonging to clonal complexes (CCs) 1 (two isolates) and 398 (one isolate), isolated between 2008 and 2011 from Italian pig holdings, combining Illumina short-read and Oxford Nanopore Technologies (ONT) long-read sequencing approaches.
2. Results
2.1. Antimicrobial Susceptibility Testing (AST)
The three LA-MRSA isolates were phenotypically MDR, displaying a common resistance pattern to beta-lactams, amphenicols, fluoroquinolones, and tetracyclines (Table 1). Both CC1 isolates (IDs 21554/1 and 55864/23) were also macrolide (erythromycin)- and lincosamide (clindamycin)-resistant. IDs 76669/3 (CC398) and 21554/1 (CC1) were pleuromutilin (tiamulin)-resistant. Based on the epidemiological cut-off value and clinical breakpoint of linezolid (MIC value > 4 mg/L), only one CC1 isolate (ID 21554/1) was phenotypically resistant to linezolid (MIC value = 8 mg/L); the other two were susceptible (MIC value = 1 mg/L).

Table 1.
Genomic and phenotypic characteristics of the three cfr-positive LA-MRSA isolates analysed by WGS (Illumina short reads).
2.2. WGS and Bioinformatics Analysis (Illumina Short Reads)
The previous results of multilocus sequence typing (MLST), spa typing, and staphylococcal cassette chromosome mec (SCCmec) typing were confirmed by the results obtained by in silico typing for all the three de novo assembled genomes. Isolates IDs 21554/1 and 55864/23 belonged to spa type t127, sequence type (ST) 1, CC1, and isolate ID 76669/3 belonged to spa type t034, ST398.
Except for linezolid resistance, all MDR phenotypes were consistent with the genetic analysis. All three isolates tested cfr-positive, but only one CC1 isolate (ID 21554/1) was phenotypically linezolid-resistant (Table 1).
The variant calling analysis indicated that both linezolid-susceptible isolates, ID 55864/23 (CC1) and 76669/3 (CC398), presented a cfr gene with a single nucleotide deletion corresponding with position 10,692 bp in the Tn558 transposon of plasmid pSCFS6. S. warneri (AM408573) was used as a reference sequence. This single base-pair deletion caused a reading frameshift and, consequently, a truncated protein. The linezolid-resistant CC1 LA-MRSA isolate (ID 21554/1) harboured a cfr sequence identical to AM408573 from S. warneri (Figure 1).

Figure 1.
Visualisation of the variant calling analysis obtained for cfr gene of the three LA-MRSA isolates using as reference the Tn558 transposon of plasmid pSCFS6 (S. warneri (AM408573) (CGE database)).
2.3. WGS and Bioinformatics Analysis (Hybrid Assembly)
The complete genomes of isolates ID 76669/3 (CC398) and 21554/1 (CC1) subjected to a hybrid (Illumina-ONT) assembly were obtained and analysed. The results of the Bakta annotation and MobileElementFinder analyses are reported in detail in Tables S1 and S2. In particular, the annotation of the assembly of isolate ID 76669/3 (CC398) identified 2649 coding sequences (CDSs), and cfr was located on contig 9 (160,836 bp; 30227..31275 nt) together with the fexA gene. These AMR genes were surrounded by IS elements within a Tn558 composite transposon-like structure, including truncated tnpA (ΔtnpA), tnp-IS21, and istB (IS21-558 element). A BLAST analysis revealed that contig 9 of isolate ID 76669/3 had the BLAST best match (according to Max Score) with the chromosome sequence of LA-MRSA ST398 (strain 17Gst354; CP073065.1) isolated from the nasal swab of a healthy Swiss farmer in 2017, showing a 96.00% coverage and a 99.78% identity. However, this strain did not harbour the cfr gene. The BLAST best match (98.00% coverage and 99.72% identity) with cfr-positive MRSA was with the chromosome sequence of the LA-MRSA ST398 strain (RIVM_M047065; CP096539) isolated from a human nasal swab in the Netherlands in 2019 [11].
The results of the average nucleotide identity (ANI) for the alignment coverage and identity of the complete sequence of isolate ID 76669/3 (2,843,032 bp) compared with the complete chromosomes of the LA-MRSA ST398 strain 17Gst354 (2,783,931 bp) and LA-MRSA ST398 strain RIVM_M047065 (2,806,671 bp) showed that they shared an identity of 99.76% and 99.86% with a 93.24% and 96.00% coverage, respectively.
From the NCBI database interrogation, we also observed that the same single base-pair deletion (c.381delA) found in the cfr gene of the CC398 LA-MRSA isolate ID 76669/3 was recently described in a cfr gene located in a 18,240 bp chromosomal region (cfr gene cluster; MW298531) of a CC398 LA-MRSA sample isolated in 2019 from a single pig nasal swab in Central Italy [18]. Multiple alignment confirmed this deletion by comparing our sequence with the cfr reference sequences of S. sciuri (NG_047631) and S. warneri (AM408573) (Figure 2).
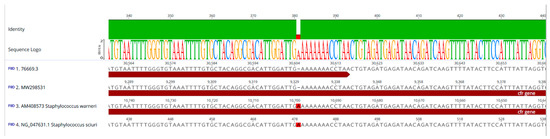
Figure 2.
Multiple alignment of the contig-harbouring cfr of the CC398 isolate (ID 76669/3) (1) with (2) a 18,240 bp chromosomal region containing cfr (cfr gene cluster) from S. aureus G322 (MW29853), (3) a 22,010 bp reference sequence of the Tn558 variant of pSCFS6 plasmid from S. warneri (AM408573), and (4) a 1250 bp cfr sequence from pSCFS1 plasmid of S. sciuri (NG_047631).
An almost (99.99%) identical cfr gene cluster of MRSA G322 (MW298531), described by Fioriti and colleagues in 2021 [18], was identified within the transposon harboured on contig 9 of our isolate ID 76669/3 (21276..39515 nt). Both cfr gene clusters differed by only one nucleotide in the fexA gene encoding a phenicol resistance. In our isolate, this gene was identical to the fexA1 reference sequence in the ResFinder database (AJ549214) whereas the fexA gene within the cfr gene cluster of isolate MRSA G322 presented a single-point mutation (G to C) at position 1192 (G1192C). MRSA G322 remained phenotypically resistant to chloramphenicol and florfenicol [18].
Resolved plasmids were obtained only for isolate ID 21554/1 (CC1) (circular contigs 3, 4, 5, and 6; Tables S1 and S2). The cfr gene was located on a novel 44,826 bp plasmid (contig 3; 23694..24743 nt) named pMOL21554 (Figure 3). Similar to contig 9 of isolate ID 76669/3, cfr was flanked by IS elements in this plasmid within a Tn558 composite transposon-like structure (~13,774 bp). This structure presented an identity of 99.70% with a coverage of 100% with the reference sequence of IS21-558 (AM086211), composed by truncated tnpA (ΔtnpA), tnp-IS21, istB (IS21-558 element), cfr, and fexA. The major difference with the reference sequence was an insertion in our IS21-558-like structure, which included additional mobile elements (25432..30683 nt) represented by two transposases, an ATP-binding protein, and two recombinases (Figure 3).
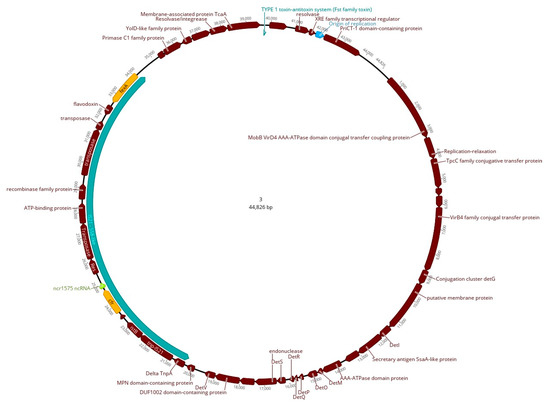
Figure 3.
Graphical representation of the cfr-carrying plasmid (pMOL21554) of isolate ID 21554/1. Coding regions are indicated in dark red, fexA and cfr genes are marked in yellow, and the 1575 non-coding RNA (ncRNA) sequence is in light green. The Tn558 composite transposon-like structure (~13,774 bp) is indicated in greenish-blue, and the origin of replication is in light blue.
As reported for contig 9 of isolate ID 76669/3, this plasmid only harboured fexA and cfr as AMR resistance genes. Isolate ID 21554/1 harboured other AMR genes such as blaZ, bleO, aadD located in a rep20 plasmid and the tet(K) gene in a rep7a plasmid.
The BLAST analysis revealed that pMOL21554 of isolate ID 21554/1 had the BLAST best match with: (i) a 39,287 bp plasmid (88% coverage and 100% identity) named pSA737 (KC206006) from ST239 MSSA (strain 004-737X) isolated in 2007 from a human clinical sample in the United States [19]; and (ii) a 39,212 bp plasmid (88% coverage and 99.99% identity) named pY96A (CP065516.1) from ST1 MSSA (strain GDY8P96A) isolated in 2019 from a pig nasal swab in China. The main differences between the 3 plasmid sequences were related to the presence in our plasmid of a 5249 bp (25430..30679 nt) region harbouring several transposable elements represented by a transposase, a recombinase, and an ATP-binding protein encoding gene. This region was absent in the two previously deposited plasmid sequences (Figure 4).
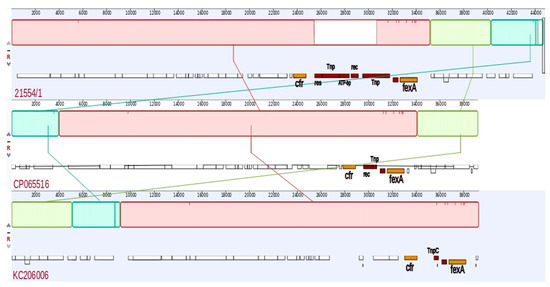
Figure 4.
Graphical representation of the alignment of the cfr-carrying plasmid (pMOL21554) of isolate ID 21554/1 compared with selected already-deposited plasmid sequences publicly available: plasmid pY96A (CP065516.1) from an ST1 MSSA strain (pig; 2019; China) and plasmid pSA737 (KC206006) from an ST239 MSSA strain (human; 2017; Unites States).
The complete sequences of the cfr-positive isolates and the resolved cfr-carrying plasmid were deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under project accession number PRJEB59270.
3. Discussion
In this study, we report the in-depth molecular characterisation of CC1 (two isolates) and CC398 (one isolate) cfr-positive MDR LA-MRSA isolates of porcine origin detected for the first time in Italian animal production in 2008–2011 [16,17] by using the combined approach of short- and long-read sequencing. Based on previous population studies (2008–2014) conducted on Italian animal production and according to other European-wide studies on cfr-positive MRSA isolated from pig holdings, the occurrence of cfr-positive LA-MSSA and -MRSA in animal production is emerging, but seems to be sporadic [8,9,10,11]. Accordingly, in a recent study (2019–2020) conducted on dairy cattle farms located in Italian regions representing around 80% of Italian milk production, 22 LA-MRSA samples isolated from 316 bulk tank milk samples and belonging to different CCs tested negative for the presence of cfr [20].
The isolates characterised in this study presented different cfr variants/locations and linezolid-resistant phenotypes. The cfr gene was demonstrated to be located on a novel 44,826 bp plasmid in the CC1 isolate (ID 21554/1) and, to the best of our knowledge, this represents the first report of CC1 LA-MRSA harbouring the cfr gene in its functional variant; i.e., displaying both a microbiological and a clinical resistance. The presence of the cfr gene was previously detected only in a 39,212 bp plasmid (CP065516.1) from one CC1 MSSA sample isolated in 2019 from a pig nasal swab in China. Differently, in our CC398 LA-MRSA isolate (ID 76669/3), cfr seemed to be chromosomally located. This result was supported by a comparison with a previously deposited complete chromosome sequence of cfr-positive CC398 MRSA of human origin (strain RIVM_M047065; CP096539), considering the obtained coverage (96.00%) and identity (99.86%). This isolate, together with the CC1 isolate ID 55864/23, presented a frameshift mutation caused by a single base-pair deletion in the cfr gene and, consequently, a truncated protein, corroborating their phenotypic susceptibility to linezolid (MIC value = 1 mg/L). Prior to our findings, linezolid-susceptible MSSA and MRSA associated with cfr variants with a single base-pair deletion or mutation were isolated from two human clinical cases (one in Brazil [21] and one in the Netherlands [11]), and also from two nasal swabs of porcine origin (one in Italy [18] and the other in Australia (CP029172)). All of them belonged to CC398.
As for the genetic environment of cfr insertion sequences and mobile elements found close to cfr, those of the IS21 family (e.g., IS21-558) in particular have been reported to have an important role in its mobilisation [15]. The importance of the IS21 family appears to be consistent, irrespective of whether the cfr gene is found in plasmids or in chromosomal structures. IS21-558, also called ISSau9, was first identified on plasmid pSCFS3 from S. aureus of porcine origin in Germany [7], and a cfr-harbouring segment bound by two directly oriented copies of IS21-558 was first identified in plasmid pSCFS6 from both S. warneri and S. simulans of porcine origin isolated in Denmark [22], suggesting that this structure could be transferred and spread among different staphylococcal species. In this regard, we identified in both isolates subjected to the hybrid assembly approach (IDs 76669/3-CC398 and 21554/1-CC1) a IS21-558-like structure near to the fexA and cfr genes. In plasmid pMOL21554 from isolate ID 21554/1, this structure was very similar (99.7% identity with 100% coverage) to the reference sequence of IS21-558 (AM086211), suggesting that the cfr gene may have been transmitted through the Tn558 transposition. Considering the potential transmissibility of cfr and its emergence in animal production in Europe, these results have significant public health implications and backdate the cfr presence in LA-MRSA from Italian pigs at least to 2008. However, we could speculate that LA-MRSA of swine origin has had, so far, a mild attitude to acquire and maintain such resistance genes along the production cycles. This may reflect the low frequency of detection of cfr-positive MRSA observed so far in humans [11], even in case they may represent an important reservoir of exposure for animal production.
Moreover, it has to be considered that even if oxazolidinones and streptogramin A antibiotic classes are not used in food-producing animals, the selective pressure exerted by the non-prudent use of PhLOPSA antibiotic classes in both human and veterinary sectors (e.g., in humans, lincosamides and oxazolidinones; in veterinary medicine, phenicols, lincosamides, and pleuromutilins) may lead to the further dissemination of the cfr gene.
Although our identified plasmid harboured only fexA and cfr, the potential coexistence of cfr and other acquired AMR genes in the same plasmids is worrying as co-selection mechanisms and the persistence of the cfr gene may occur under selective pressure induced by the use of non-PhLOPSA antibiotic classes [15].
Notably, after the first European survey conducted in 2008 [23], a new European Union-wide baseline survey is to be carried out in the next years in order to provide updated and representative information on the hazard of LA-MRSA from pigs [24]. This type of harmonised study will help provide data at both European and national levels on linezolid-resistant LA-MRSA.
4. Conclusions
In conclusion, our findings confirm the need for the continuous genomic surveillance of cfr-positive zoonotic LA-MRSA and other oxazolidinone-resistant bacteria in livestock. This approach will help the understanding of the genetic environment and the transmission patterns of MGEs involved in the dissemination of powerful genetic determinants, mediating multiresistance to last-resort drugs for human medicine.
5. Materials and Methods
5.1. LA-MRSA Isolates
Three cfr-positive LA-MRSA samples (isolate IDs 21554/1, 76669/3, and 55864/23) were retrospectively characterised in depth by a hybrid WGS approach and a bioinformatics analysis. They had been isolated from two dust samples (IDs 21554/1 and 55864/23) and one nasal swab (ID 76669/3) collected from not epidemiologically related breeding, farrow-to-finish, and fattening (finishing) Italian commercial pig holdings (range: 2500–6500 heads) in 2008, 2011, and 2010, respectively. The three isolates, stored at the National Reference Laboratory for Antimicrobial Resistance (NRL-AR), Istituto Zooprofilattico Sperimentale del Lazio e della Toscana “M. Aleandri”, Rome (Italy), had been previously characterised by a DNA microarray analysis and molecular typing through spa typing, multilocus sequence typing, and SCCmec typing, as previously described [16,17].
These isolates were the only cfr-positive ones from a collection of LA-MRSA isolates (n = 433) available and screened at the NRL-AR, and were obtained from different animal production methods such as fattening pigs (n = 172), dairy cows (n = 118), veal calves (n = 142), and broiler chicken (n = 1) in the context of national population studies conducted in Italy between 2008 and 2014 [16,17,25,26,27,28,29].
5.2. Antimicrobial Susceptibility Testing (AST)
The three cfr-positive LA-MRSA isolates were previously tested for their antimicrobial susceptibility by broth microdilution (Trek Diagnostic Systems, Westlake, OH, USA). The MICs were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST; http://www.eucast.org (accessed on 20 June 2022) using epidemiological cut-offs. The following panel of drugs was tested: penicillin, cefoxitin, ciprofloxacin, chloramphenicol, clindamycin, erythromycin, gentamicin, kanamycin, streptomycin, linezolid, quinopristin/dalfopristin, fusidic acid, mupirocin, rifampicin, tetracycline, tiamulin, sulfamethoxazole, trimethoprim, and vancomycin [16].
5.3. Library Preparation and Whole Genome Sequencing (WGS)
DNA extraction and library preparation were performed as previously reported [30]. Briefly, genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The libraries for short-read pair-end sequencing were prepared for the three isolates using a NexteraXT DNA library preparation kit (Illumina, Inc., San Diego, CA, USA) and sequenced on an Illumina platform (MiSeq sequencer, Illumina, Inc., San Diego, CA, USA)). In parallel, the libraries of two isolates (IDs 21554/1 and 76669/3) were prepared with a ligation sequencing kit (SQK-LSK109) and sequenced using a nanopore-based MinION device (ONT) [31].
5.4. Bioinformatics Analysis (Illumina Short Reads)
Illumina raw reads were analysed using an internal pipeline for the assembly, which included the following tools: FastQC v0.11.9, Trimmomatic v0.39 [32], Spades v3.13.0 [33], and Quast v5.0.2 [34]. In silico molecular characterisation was performed on all the assembled genomes for the MLST analysis [35,36] to confirm the ST. Center for Genomic Epidemiology (CGE) online tools with default parameters (http://www.genomicepidemiology.org/services/ (accessed on 19 July 2022)) were used to confirm the SCCmec type [37,38], the spa type [39], and the genetic basis of AMR [40,41,42]. The presence of cfr mutations was investigated by mapping the trimmed reads to the cfr reference sequence of the CGE ResFinder database (updated on 24 May 2022) AM408573 (S. warneri), and variant calling was performed using BWA mem v0.7.12 [43] and SAMtools v1.7 [44], and then visualised by IGV 2.0.1 [45].
5.5. Bioinformatics Analysis (Hybrid Assembly)
For the two isolates subjected to both short- and long-read sequencing, high accuracy base-calling was performed on the long reads obtained from the MinION device according to the ONT workflow. A hybrid (Illumina-ONT) assembly was implemented using the Unicycler pipeline [46] with default parameters [47]. The obtained assemblies were also annotated using the online version of the Bakta tool (https://bakta.computational.bio/ (accessed on 19 July 2022) [48]. Additionally, a manual curation for the obtained annotation was performed. The identification of the mobile genetic elements and their relation to the AMR genes and virulence factors was performed with the online version of the CGE tool, MobileElementFinder v1.0.3 (last accessed 19 July 2022) [49] using default parameters. From the hybrid assemblies, the obtained contigs containing cfr were also compared with the BLAST algorithm using the online tool against the nr/nt database with default parameters. The complete sequence of isolate ID 76669/3 was also compared with selected complete chromosome sequences of cfr-positive S. aureus obtained from publicly available databases showing the BLAST best match and calculating the ANI for the alignment coverage and identity using the BLAST algorithm [50].
The presence of mutations of the cfr genes was confirmed, and their genetic environment was investigated by comparing the multiple alignment of the contig-harbouring cfr of isolate ID 76669/3 with publicly available selected sequences harbouring cfr from: (i) S. warneri (AM408573) [22]; (ii) S. sciuri (NG_047631) [51]; and (iii) S. aureus G322 (MW29853) [18]. This was achieved using Geneious Prime 2023.0.1 software.
The graphical representation of the general structures and of the genetic regions of the cfr-carrying plasmid of isolate ID 21554/1 as well as the comparison with selected already-deposited plasmid sequences were performed using Geneious Prime 2023.0.1 and the Mauve algorithm for the alignment [52].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12030530/s1, Table S1: Bakta annotations; Table S2: MobileElementFinder results.
Author Contributions
Conceptualisation, M.I., E.L.D., V.C., A.F. and A.B; methodology, M.I., E.L.D., P.A. and V.C.; validation, M.I., E.L.D. and V.C.; formal analysis, M.I., E.L.D. and P.A.; investigation, F.F., C.B., R.O., A.G., M.I., E.L.D. and P.A.; resources, A.F. and A.B.; data curation, M.I. and E.L.D.; writing—original draft preparation, M.I., E.L.D., V.C. and A.C.; writing—review and editing, all authors; visualisation, M.I. and E.L.D.; supervision, V.C., A.F. and A.B.; project administration, A.F. and A.B.; funding acquisition, A.F. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded partly by grants from the EMIDA ERA-NET (FP7-KBBE) first transnational research call, project title: “Methicillin-resistant Staphylococcus aureus lineages in primary productions: multi-host pathogen, spill-over and spill-back between animals and humans?”; “LA-MRSA” (acronym title) ID 68. The genomic work on the use of the combined approach of short- and long-read sequencing to resolve the cfr-carrying plasmid was conducted in the framework of the Full Force project, supported by the European Union’s Horizon 2020 Research and Innovation programme under grant agreement No 773830: One Health European Joint Programme.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The whole genome sequence data presented in this study were deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena (accessed on 27 February 2023)) under the study accession number PRJEB59270 with the following individual accession numbers: ERS14687029 (pMOL21554); ERS14687032 (55864/23); ERS14687030 (21554/1); and ERS14687031 (76669/3).
Acknowledgments
The authors wish to thank Daniele Smedile, Ilaria Congiu, and Flaminia Riggio from the Istituto Zooprofilattico Sperimentale del Lazio e Toscana “M. Aleandri” for their technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gu, B.; Kelesidis, T.; Tsiodras, S.; Hindler, J.; Humphries, R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2013, 68, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Zhang, W.; Du, X.D.; Krüger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile Oxazolidinone Resistance Genes in Gram-Positive and Gram-Negative Bacteria. Clin. Microbiol. Rev. 2021, 34, e0018820. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Werckenthin, C.; Kehrenberg, C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 2000, 44, 2530–2533. [Google Scholar] [CrossRef]
- Long, K.S.; Poehlsgaard, J.; Kehrenberg, C.; Schwarz, S.; Vester, B. The cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006, 50, 2500–2505. [Google Scholar] [CrossRef]
- Antonelli, A.; D’Andrea, M.M.; Galano, A.; Borchi, B.; Brenciani, A.; Vaggelli, G.; Cavallo, A.; Bartoloni, A.; Giovanetti, E.; Rossolini, G.M. Linezolid-resistant cfr-positive MRSA, Italy. J. Antimicrob. Chemother. 2016, 71, 2349–2351. [Google Scholar] [CrossRef]
- Youenou, B.; Martins Simoes, P.; Tristan, A.; Farfour, E.; Beauruelle, C.; Kolenda, C.; Ranc, A.G.; Vandenesch, F.; Laurent, F.; Dupieux, C. Linezolid resistance: Detection of the cfr(B) gene in French clinical MRSA strains. J. Antimicrob. Chemother. 2022, 13, dkac411. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Schwarz, S. Distribution of Florfenicol Resistance Genes fexA and cfr among Chloramphenicol-Resistant Staphylococcus Isolates. Antimicrob. Agents Chemother. 2006, 50, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Peeters, L.E.J.; Argudín, M.A.; Azadikhah, S.; Butaye, P. Antimicrobial resistance and population structure of Staphylococcus aureus recovered from pigs farms. Vet. Microbiol. 2015, 180, 151–156. [Google Scholar] [CrossRef]
- Ruiz-Ripa, L.; Bellés-Bellés, A.; Fernández-Fernández, R.; García, M.; Vilaró, A.; Zarazaga, M.; Torres, C. Linezolid-resistant MRSA-CC398 carrying the cfr gene, and MRSA-CC9 isolates from pigs with signs of infection in Spain. J. Appl. Microbiol. 2021, 131, 615–622. [Google Scholar] [CrossRef]
- Leão, C.; Clemente, L.; Cara d’Anjo, M.; Albuquerque, T.; Amaro, A. Emergence of Cfr-Mediated Linezolid Resistance among Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) from Healthy Pigs in Portugal. Antibiotics 2022, 11, 1439. [Google Scholar] [CrossRef]
- Schouls, L.M.; Veldman, K.; Brouwer, M.S.M.; Dierikx, C.; Witteveen, S.; van Santen-Verheuvel, M.; Hendrickx, A.P.A.; Landman, F.; Hengeveld, P.; Wullings, B.; et al. Dutch MRSA surveillance study group. cfr and fexA genes in methicillin-resistant Staphylococcus aureus from humans and livestock in the Netherlands. Commun. Med. 2022, 2, 135. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-M.; Zhou, Y.-F.; Li, L.; Fang, L.-X.; Duan, J.-H.; Liu, F.-R.; Liang, H.-Q.; Wu, Y.-T.; Gu, W.-Q.; Liao, X.-P.; et al. Characterization of the multi-drug resistance gene cfr in methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from animals and humans in China. Front. Microbiol. 2018, 9, 2925. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.Y.; Moon, D.C.; Mechesso, A.F.; Choi, J.-H.; Kim, S.-J.; Song, H.-J.; Kim, M.H.; Yoon, S.-S.; Lim, S.-K. Emergence of cfr-mediated linezolid resistance in Staphylococcus aureus isolated from pig carcasses. Antibiotics 2020, 9, 769. [Google Scholar] [CrossRef] [PubMed]
- Osman, K.M.; Amer, A.M.; Badr, J.M.; Helmy, N.M.; Elhelw, R.A.; Orabi, A.; Bakry, M.; Saad, A.S.A. Antimicrobial resistance, biofilm formation and mecA characterization of methicillin-susceptible S. aureus and non-S. aureus of beef meat origin in Egypt. Front. Microbiol. 2016, 7, 222. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, Y.; Schwarz, S. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2013, 68, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Feltrin, F.; Cordaro, G.; Porrero, M.C.; Kraushaar, B.; Argudín, M.A.; Nykäsenoja, S.; Monaco, M.; Stegger, M.; Aarestrup, F.M.; et al. Livestock-Associated Methicillin Resistant and Methicillin Susceptible Staphylococcus aureus Sequence Type (CC)1 in European Farmed Animals: High Genetic Relatedness of Isolates from Italian Cattle Herds and Humans. PLoS ONE 2015, 10, e0137143. [Google Scholar] [CrossRef]
- Feltrin, F.; Alba, P.; Amoruso, R.; Argudin, M.; Lauzat, B.; Iurescia, M.; Sorbara, L.; Dottarelli, S.; Carfora, V.; Menichini, E.; et al. Molecular characterization of the Staphylococcus aureus CC398 population in Italy. In Proceedings of the 11th International Meeting on Microbial Epidemiological Markers (ESCMID-ESGEM Conference), Estoril, Portugal, 9–12 March 2016. [Google Scholar]
- Fioriti, S.; Coccitto, S.N.; Morroni, G.; Simoni, S.; Cinthi, M.; Magi, G.; Sante, L.D.; Alvarez-Suarez, J.M.; Mingoia, M.; Giovanetti, E.; et al. Detection of a chromosomal truncated cfr gene in a linezolid-susceptible LA-MRSA ST398 isolate of porcine origin, Italy. J. Glob. Antimicrob. Resist. 2021, 26, 199–201. [Google Scholar] [CrossRef]
- Mendes, R.E.; Deshpande, L.M.; Bonilla, H.F.; Schwarz, S.; Huband, M.D.; Jones, R.N.; Quinn, J.P. Dissemination of a pSCFS3-like cfr-carrying plasmid in Staphylococcus aureus and Staphylococcus epidermidis clinical isolates recovered from hospitals in Ohio. Antimicrob. Agents Chemother. 2013, 57, 2923–2928. [Google Scholar] [CrossRef]
- Battisti, A.; (National Reference Laboratory for Antimicrobial Resistance, Department of General Diagnostics, Istituto Zooprofilattico Sperimentale del Lazio e della Toscana “M. Aleandri”, Rome, Italy). Personal Communication, 2021.
- Gales, A.C.; Deshpande, L.M.; de Souza, A.G.; Pignatari, A.C.; Mendes, R.E. MSSA ST398/t034 carrying a plasmid-mediated Cfr and Erm(B) in Brazil. J. Antimicrob. Chemother. 2015, 70, 303–305. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Aarestrup, F.M.; Schwarz, S. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr. Antimicrob. Agents Chemother. 2007, 51, 483–487. [Google Scholar] [CrossRef]
- EFSA. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008. Part A: MRSA prevalence estimates; on request from the European Commission. EFSA J. 2009, 7, 1376. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Aerts, M.; Battisti, A.; Hendriksen, R.; Larsen, J.; Nilsson, O.; Cortinas Abrahantes, J.; Guerra, B.; Papanikolaou, A.; Beloeil, P.-A. Technical specifications for a baseline survey on the prevalence of Methicillin Resistant Staphylococcus aureus (MRSA) in pigs. EFSA J. 2022, 20, e07620. [Google Scholar] [CrossRef] [PubMed]
- Battisti, A.; Franco, A.; Merialdi, G.; Hasman, H.; Iurescia, M.; Lorenzetti, R.; Feltrin, F.; Zini, M.; Aarestrup, F.M. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet. Microbiol. 2010, 142, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Cortimiglia, C.; Luini, M.; Bianchini, V.; Marzagalli, L.; Vezzoli, F.; Avisani, D.; Bertoletti, M.; Ianzano, A.; Franco, A.; Battisti, A. Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy Region (Northern Italy). Epidemiol. Infect. 2016, 144, 3046–3051. [Google Scholar] [CrossRef]
- Feltrin, F.; Alba, P.; Kraushaar, B.; Ianzano, A.; Argudín, M.A.; Di Matteo, P.; Porrero, M.C.; Aarestrup, F.M.; Butaye, P.; Franco, A.; et al. A Livestock-Associated, Multidrug-Resistant, Methicillin-Resistant Staphylococcus aureus Clonal Complex 97 Lineage Spreading in Dairy Cattle and Pigs in Italy. Appl. Environ. Microbiol. 2015, 82, 816–821. [Google Scholar] [CrossRef]
- Tomao, P.; Pirolo, M.; Agnoletti, F.; Pantosti, A.; Battisti, A.; Di Martino, G.; Visaggio, D.; Monaco, M.; Franco, A.; Pimentel de Araujo, F.; et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus from dairy farms in North-eastern Italy. Int. J. Food Microbiol. 2020, 332, 108817. [Google Scholar] [CrossRef]
- Zoppi, S.; Dondo, A.; Di Blasio, A.; Vitale, N.; Carfora, V.; Goria, M.; Chiavacci, L.; Giorgi, I.; D’Errico, V.; Irico, L.; et al. Livestock-Associated Methicillin-Resistant Staphylococcus aureus and Related Risk Factors in Holdings of Veal Calves in Northwest Italy. Microb. Drug Resist. 2021, 27, 1136–1143. [Google Scholar] [CrossRef]
- Alba, P.; Taddei, R.; Cordaro, G.; Fontana, M.C.; Toschi, E.; Gaibani, P.; Marani, I.; Giacomi, A.; Diaconu, E.L.; Iurescia, M.; et al. Carbapenemase IncF-borne blaNDM-5 gene in the E. coli ST167 high-risk clone from canine clinical infection, Italy. Vet. Microbiol. 2021, 256, 109045. [Google Scholar] [CrossRef]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- International Working Group on The Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 2009, 56, 4961–4967. [Google Scholar]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr and major difference in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef]
- Bartels, M.D.; Petersen, A.; Worning, P.; Nielsen, J.B.; Larner-Svensson, H.; Johansen, H.K.; Andersen, L.P.; Jarløv, J.O.; Boye, K.; Larsen, A.R.; et al. Comparing whole-genome sequencing with Sanger sequencing for spa typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2014, 52, 4305–4308. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2020, 72, 2764–2768. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Diaconu, E.L.; Carfora, V.; Alba, P.; Di Matteo, P.; Stravino, F.; Buccella, C.; Dell’Aira, E.; Onorati, R.; Sorbara, L.; Battisti, A.; et al. Novel IncFII plasmid harbouring blaNDM-4 in a carbapenem-resistant Escherichia coli of pig origin, Italy. J. Antimicrob. Chemother. 2020, 75, 3475–3479. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Kehrenberg, C.; Ojo, K.K.; Schwarz, S. Nucleotide sequence and organization of the multiresistance plasmid pSCFS1 from Staphylococcus sciuri. J. Antimicrob. Chemother. 2004, 54, 936–939. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).