A Six-Year Retrospective Study of Microbiological Characteristics and Antimicrobial Resistance in Specimens from a Tertiary Hospital’s Surgical Ward
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Type and Ethics Approval
2.2. Sample Collection, Transport, and Processing
2.3. Statistics
3. Results
3.1. Types of Cultures and Microbiological Characteristics
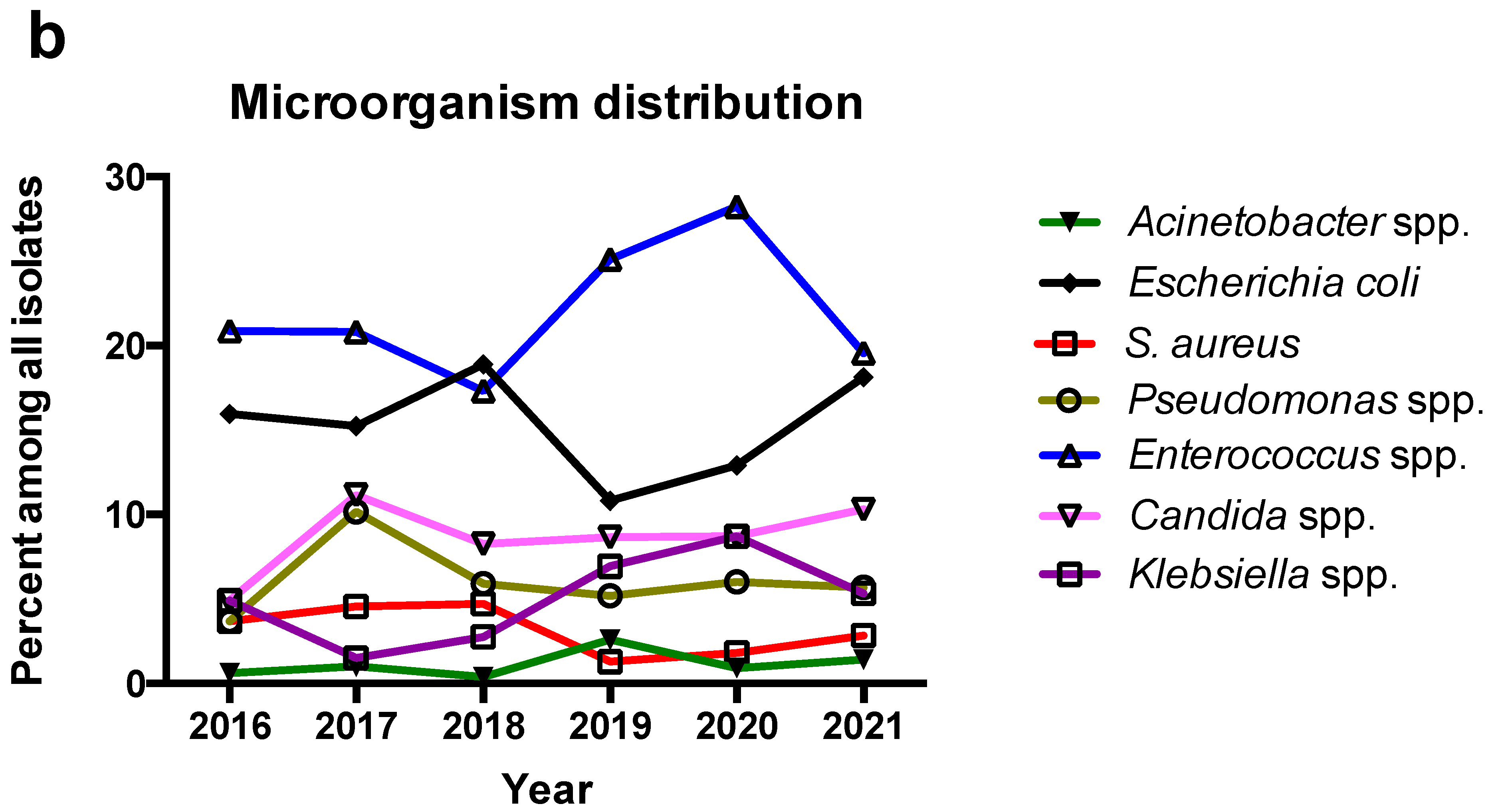
3.2. Trends of Microbiological Characteristics during the Period of the Study
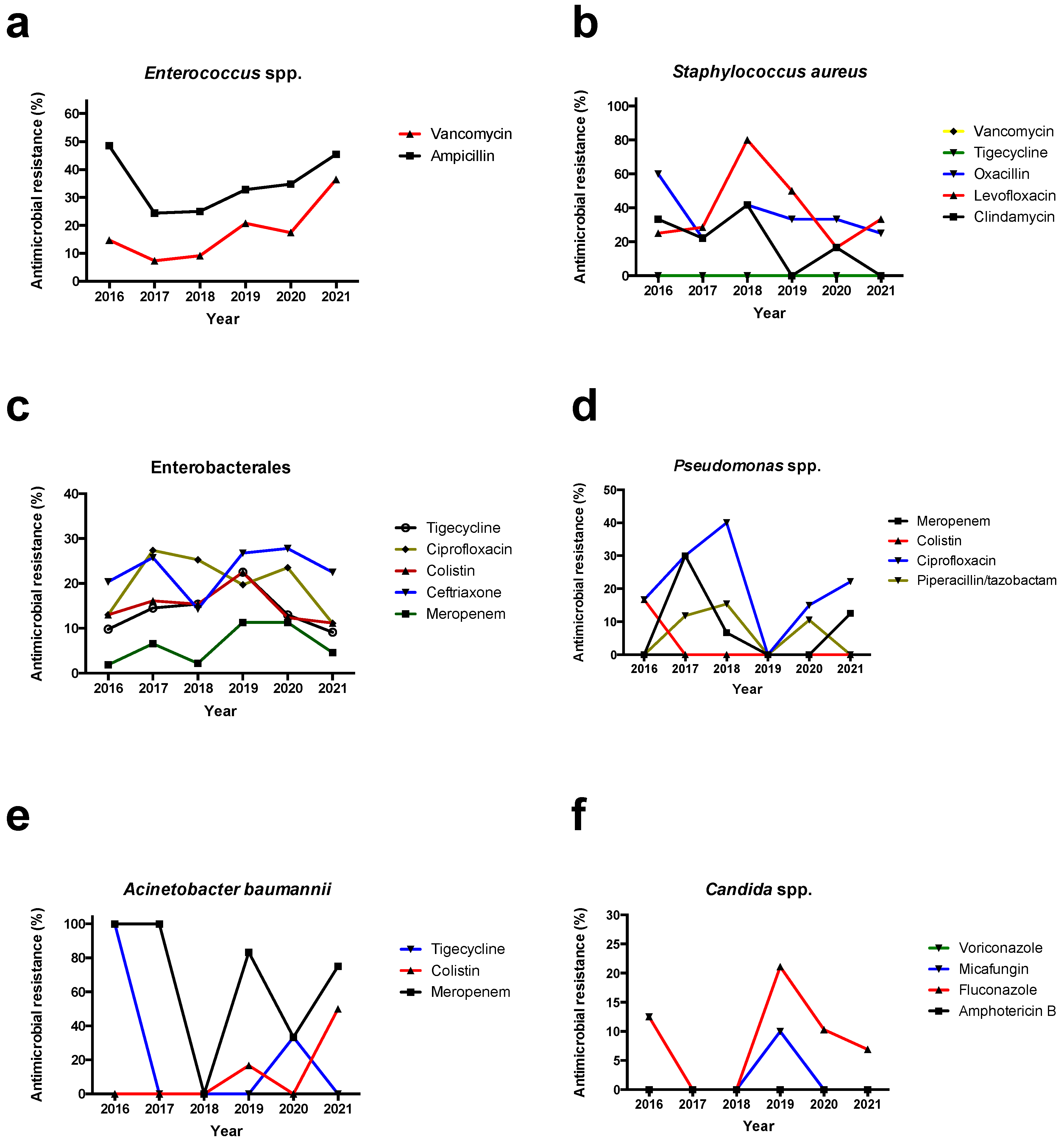
3.3. Antimicrobial Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gawande, A. Two Hundred Years of Surgery. N. Engl. J. Med. 2012, 366, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, W. The Changing Face of Surgical Oncology. Ann. Surg. Oncol. 2017, 24, 546–548. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, S.I.; Vadmal, M.; Tang, P. A Note from History: Landmarks in History of Cancer, Part 7. Cancer 2015, 121, 2480–2513. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, S.I. A Note from History: Landmarks in History of Cancer, Part 1. Cancer 2011, 117, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, S.I.; Darvishian, F. A Note from History: Landmarks in History of Cancer, Part 5. Cancer 2013, 119, 1450–1466. [Google Scholar] [CrossRef] [PubMed]
- Strobel, R.M.; Leonhardt, M.; Förster, F.; Neumann, K.; Lobbes, L.A.; Seifarth, C.; Lee, L.D.; Schineis, C.H.W.; Kamphues, C.; Weixler, B.; et al. The Impact of Surgical Site Infection-a Cost Analysis. Langenbecks Arch. Surg. 2022, 407, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, S.; Kelly, E.; Rogers, S.O.; Gates, J. Urinary Tract Infection in Surgical Patients. Am. J. Surg. 2003, 186, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Viel-Thériault, I.; Bettolli, M.; Toye, B.; Harrison, M.-A.; Le Saux, N. Contemporary Microbiology and Antimicrobial Treatment of Complicated Appendicitis: The Value of a Short-Term Study. Pediatr. Infect. Dis. J. 2019, 38, e290–e294. [Google Scholar] [CrossRef]
- Obinwa, O.; Casidy, M.; Flynn, J. The Microbiology of Bacterial Peritonitis Due to Appendicitis in Children. Ir. J. Med. Sci. 2014, 183, 585–591. [Google Scholar] [CrossRef]
- Suh, S.-W.; Choi, Y.S.; Choi, S.-H.; Do, J.H.; Oh, H.-C.; Kim, H.J.; Lee, S.E. Antibiotic Selection Based on Microbiology and Resistance Profiles of Bile from Gallbladder of Patients with Acute Cholecystitis. Sci. Rep. 2021, 11, 2969. [Google Scholar] [CrossRef]
- Shafagh, S.; Rohani, S.H.; Hajian, A. Biliary Infection; Distribution of Species and Antibiogram Study. Ann. Med. Surg. 2021, 70, 102822. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Ashley, E.A.; Hamers, R.L.; Turner, P.; Kesteman, T.; Akech, S.; Corso, A.; Mayxay, M.; Okeke, I.N.; Limmathurotsakul, D.; et al. Surveillance Strategies Using Routine Microbiology for Antimicrobial Resistance in Low- and Middle-Income Countries. Clin. Microbiol. Infect. 2021, 27, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- López-Hernández, I.; López-Cerero, L.; Fernández-Cuenca, F.; Pascual, Á. The Role of the Microbiology Laboratory in the Diagnosis of Multidrug-Resistant Gram-Negative Bacilli Infections. The Importance of the Determination of Resistance Mechanisms. Med. Intensiv. (Engl. Ed.) 2022, 46, 455–464. [Google Scholar] [CrossRef]
- Mangram, A.J.; Horan, T.C.; Pearson, M.L.; Silver, L.C.; Jarvis, W.R. Guideline for Prevention of Surgical Site Infection, 1999. Hospital Infection Control Practices Advisory Committee. Infect. Control Hosp. Epidemiol. 1999, 20, 250–278, quiz 279–280. [Google Scholar] [CrossRef] [PubMed]
- Owens, C.D.; Stoessel, K. Surgical Site Infections: Epidemiology, Microbiology and Prevention. J. Hosp. Infect. 2008, 70 (Suppl. S2), 3–10. [Google Scholar] [CrossRef]
- Foschi, D.; Yakushkina, A.; Cammarata, F.; Lamperti, G.; Colombo, F.; Rimoldi, S.; Antinori, S.; Sampietro, G.M. Surgical Site Infections Caused by Multi-Drug Resistant Organisms: A Case–Control Study in General Surgery. Updates Surg. 2022, 74, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Călina, D.; Docea, A.O.; Rosu, L.; Zlatian, O.; Rosu, A.F.; Anghelina, F.; Rogoveanu, O.; Arsene, A.L.; Nicolae, A.C.; Drăgoi, C.M.; et al. Antimicrobial Resistance Development Following Surgical Site Infections. Mol. Med. Rep. 2017, 15, 681–688. [Google Scholar] [CrossRef]
- Temkin, E.; Margalit, I.; Nutman, A.; Carmeli, Y. Surgical Antibiotic Prophylaxis in Patients Colonized with Multidrug-Resistant Gram-Negative Bacteria: Practical and Conceptual Aspects. J. Antimicrob. Chemother. 2021, 76, i40–i46. [Google Scholar] [CrossRef]
- Righi, E.; Mutters, N.T.; Guirao, X.; Del Toro, M.D.; Eckmann, C.; Friedrich, A.W.; Giannella, M.; Kluytmans, J.; Presterl, E.; Christaki, E.; et al. ESCMID/EUCIC Clinical Practice Guidelines on Perioperative Antibiotic Prophylaxis in Patients Colonized by Multidrug-Resistant Gram-Negative Bacteria before Surgery. Clin. Microbiol. Infect. 2022. [Google Scholar] [CrossRef]
- Falagas, M.E.; Alexiou, V.G.; Peppas, G.; Makris, G.C. Do Changes in Antimicrobial Resistance Necessitate Reconsideration of Surgical Antimicrobial Prophylaxis Strategies? Surg. Infect. 2009, 10, 557–562. [Google Scholar] [CrossRef]
- Polemis, M.; Mandilara, G.; Pappa, O.; Argyropoulou, A.; Perivolioti, E.; Koudoumnakis, N.; Pournaras, S.; Vasilakopoulou, A.; Vourli, S.; Katsifa, H.; et al. COVID-19 and Antimicrobial Resistance: Data from the Greek Electronic System for the Surveillance of Antimicrobial Resistance-WHONET-Greece (January 2018–March 2021). Life 2021, 11, 996. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, P.; Astrinaki, E.; Vitsaxaki, E.; Bolikas, E.; Christofaki, D.; Salvaraki, A.; Lagoudaki, E.; Ioannidou, E.; Karakonstantis, S.; Saplamidou, S.; et al. A Point Prevalence Survey of Healthcare-Associated Infections and Antimicrobial Use in Public Acute Care Hospitals in Crete, Greece. Antibiotics 2022, 11, 1258. [Google Scholar] [CrossRef] [PubMed]
- M100-S31; Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021.
- Badia, J.M.; Casey, A.L.; Petrosillo, N.; Hudson, P.M.; Mitchell, S.A.; Crosby, C. Impact of Surgical Site Infection on Healthcare Costs and Patient Outcomes: A Systematic Review in Six European Countries. J. Hosp. Infect. 2017, 96, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nitzan, O.; Brodsky, Y.; Edelstein, H.; Hershko, D.; Saliba, W.; Keness, Y.; Peretz, A.; Chazan, B. Microbiologic Data in Acute Cholecystitis: Ten Years’ Experience from Bile Cultures Obtained during Percutaneous Cholecystostomy. Surg. Infect. 2017, 18, 345–349. [Google Scholar] [CrossRef]
- Sula, S.; Han, T.; Marttila, H.; Haijanen, J.; Löyttyniemi, E.; Sippola, S.; Grönroos, J.; Hakanen, A.J.; Salminen, P. Blood Culture Positivity in Patients with Acute Appendicitis: A Propensity Score-Matched Prospective Cohort Study. Scand. J. Surg. 2022, 111, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Ballus, J.; Lopez-Delgado, J.C.; Sabater-Riera, J.; Perez-Fernandez, X.L.; Betbese, A.J.; Roncal, J.A. Surgical Site Infection in Critically Ill Patients with Secondary and Tertiary Peritonitis: Epidemiology, Microbiology and Influence in Outcomes. BMC Infect. Dis. 2015, 15, 304. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.; Beggs, J.J.; Walsh, T.R.; Gandra, S.; Laxminarayan, R. Anthropological and Socioeconomic Factors Contributing to Global Antimicrobial Resistance: A Univariate and Multivariable Analysis. Lancet Planet. Health 2018, 2, e398–e405. [Google Scholar] [CrossRef] [PubMed]
- Castro-Sánchez, E.; Moore, L.S.P.; Husson, F.; Holmes, A.H. What Are the Factors Driving Antimicrobial Resistance? Perspectives from a Public Event in London, England. BMC Infect. Dis. 2016, 16, 465. [Google Scholar] [CrossRef]
- Woolhouse, M.; Ward, M.; van Bunnik, B.; Farrar, J. Antimicrobial Resistance in Humans, Livestock and the Wider Environment. Phil. Trans. R. Soc. B 2015, 370, 20140083. [Google Scholar] [CrossRef]
- Murray, B.E. The Life and Times of the Enterococcus. Clin. Microbiol. Rev. 1990, 3, 46–65. [Google Scholar] [CrossRef]
- Vesić, D.; Kristich, C.J. MurAA Is Required for Intrinsic Cephalosporin Resistance of Enterococcus Faecalis. Antimicrob. Agents Chemother. 2012, 56, 2443–2451. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Giard, J.-C. Antibiotic Resistance in Enterococcus Faecium Clinical Isolates. Expert Rev. Anti-Infect Ther. 2014, 12, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Solomkin, J.S.; Mazuski, J.E.; Bradley, J.S.; Rodvold, K.A.; Goldstein, E.J.C.; Baron, E.J.; O’Neill, P.J.; Chow, A.W.; Dellinger, E.P.; Eachempati, S.R.; et al. Diagnosis and Management of Complicated Intra-Abdominal Infection in Adults and Children: Guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clin. Infect. Dis. 2010, 50, 133–164. [Google Scholar] [CrossRef] [PubMed]
- Sartelli, M.; Cristini, F.; Coccolini, F.; Labricciosa, F.M.; Siquini, W.; Catena, F. A Proposal for a Classification Guiding the Selection of Appropriate Antibiotic Therapy for Intra-Abdominal Infections. Antibiotics 2022, 11, 1394. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.M.; Tessier, J.M.; Sawyer, R.; Dellinger, E.P.; Miller, P.R.; Namias, N.; West, M.A.; Cook, C.H.; O’Neill, P.J.; Napolitano, L.; et al. Does Isolation of Enterococcus Affect Outcomes in Intra-Abdominal Infections? Surg. Infect. 2017, 18, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.K.; Kim, J.; Moon, C.; Lee, M.S.; Hur, J.; Lee, H.; Kim, S.W. Antimicrobial Susceptibility of Microorganisms Isolated from Patients with Intraabdominal Infection in Korea: A Multicenter Study. J. Korean Med. Sci. 2019, 34, e309. [Google Scholar] [CrossRef] [PubMed]
- Cantón, R.; Loza, E.; Aznar, J.; Castillo, F.J.; Cercenado, E.; Fraile-Ribot, P.A.; González-Romo, F.; López-Hontangas, J.L.; Rodríguez-Lozano, J.; Suárez-Barrenechea, A.I.; et al. Monitoring the Antimicrobial Susceptibility of Gram-Negative Organisms Involved in Intraabdominal and Urinary Tract Infections Recovered during the SMART Study (Spain, 2016 and 2017). Rev. Esp. Quimioter. 2019, 32, 145–155. [Google Scholar]
- Zhang, H.; Yang, Q.; Liao, K.; Ni, Y.; Yu, Y.; Hu, B.; Sun, Z.; Huang, W.; Wang, Y.; Wu, A.; et al. Update of Incidence and Antimicrobial Susceptibility Trends of Escherichia Coli and Klebsiella Pneumoniae Isolates from Chinese Intra-Abdominal Infection Patients. BMC Infect. Dis. 2017, 17, 776. [Google Scholar] [CrossRef]
- Dafopoulou, K.; Tsakris, A.; Pournaras, S. Changes in Antimicrobial Resistance of Clinical Isolates of Acinetobacter Baumannii Group Isolated in Greece, 2010–2015. J. Med. Microbiol. 2018, 67, 496–498. [Google Scholar] [CrossRef]
- Moschou, A.; Maraki, S.; Giormezis, N.; Moraitaki, H.; Stafylaki, D.; Militsopoulou, M.; Spiliopoulou, I.; Papadakis, J.A.; Samonis, G.; Kofteridis, D.P. Prevalence and Molecular Epidemiology of Staphylococcus Aureus Nasal Colonization in Four Nursing Home Residents in Crete, Greece. J. Infect. Chemother. 2020, 26, 199–204. [Google Scholar] [CrossRef]
- Ioannou, P.; Tsagkaraki, E.; Athanasaki, A.; Tsioutis, C.; Gikas, A. Gram-Negative Bacteria as Emerging Pathogens Affecting Mortality in Skin and Soft Tissue Infections. Hippokratia 2018, 22, 23–28. [Google Scholar] [PubMed]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/ECDC-WHO-AMR-report.pdf (accessed on 21 January 2023).
- de Graaff, M.R.; Hogenbirk, R.N.M.; Janssen, Y.F.; E Elfrink, A.K.; Liem, R.S.L.; Nienhuijs, S.W.; de Vries, J.-P.P.M.; Elshof, J.-W.; Verdaasdonk, E.; Melenhorst, J.; et al. Impact of the COVID-19 pandemic on surgical care in the Netherlands. Br. J. Surg. 2022, 109, 1282–1292. [Google Scholar] [CrossRef] [PubMed]
- Tham, N.; Fazio, T.; Johnson, D.; Skandarajah, A.; Hayes, I.P. Hospital Acquired Infections in Surgical Patients: Impact of COVID-19-Related Infection Prevention Measures. World J. Surg. 2022, 46, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.B.; Bosch, W.; O’Horo, J.C.; Girardo, M.E.; Bolton, P.B.; Murray, A.W.; Hirte, I.L.; Singbartl, K.; Martin, D.P. Surgical Site Infections during the COVID-19 Era: A Retrospective, Multicenter Analysis. Am. J. Infect. Control 2022. [Google Scholar] [CrossRef] [PubMed]
- Fleenor-Ford, A.; Hayden, M.K.; Weinstein, R.A. Vancomycin-Resistant Enterococci: Implications for Surgeons. Surgery 1999, 125, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The Many Faces of Enterococcus Spp.—Commensal, Probiotic and Opportunistic Pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Vasilakopoulou, A.; Karakosta, P.; Vourli, S.; Tarpatzi, A.; Varda, P.; Kostoula, M.; Antoniadou, A.; Pournaras, S. Gastrointestinal Carriage of Vancomycin-Resistant Enterococci and Carbapenem-Resistant Gram-Negative Bacteria in an Endemic Setting: Prevalence, Risk Factors, and Outcomes. Front. Public Health 2020, 8, 55. [Google Scholar] [CrossRef]
- Joshi, S.; Shallal, A.; Zervos, M. Vancomycin-Resistant Enterococci: Epidemiology, Infection Prevention, and Control. Infect. Dis. Clin. N. Am. 2021, 35, 953–968. [Google Scholar] [CrossRef]
- O’Riordan, F.; Shiely, F.; Byrne, S.; O’Brien, D.; Ronayne, A.; Fleming, A. Antimicrobial Use and Antimicrobial Resistance in Enterobacterales and Enterococcus Faecium: A Time Series Analysis. J. Hosp. Infect. 2022, 120, 57–64. [Google Scholar] [CrossRef]
- Webb, B.J.; Majers, J.; Healy, R.; Jones, P.B.; Butler, A.M.; Snow, G.; Forsyth, S.; Lopansri, B.K.; Ford, C.D.; Hoda, D. Antimicrobial Stewardship in a Hematological Malignancy Unit: Carbapenem Reduction and Decreased Vancomycin-Resistant Enterococcus Infection. Clin. Infect. Dis. 2020, 71, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Darwish, R.M.; Matar, S.G.; Snaineh, A.A.A.; Alsharif, M.R.; Yahia, A.B.; Mustafa, H.N.; Hasabo, E.A. Impact of Antimicrobial Stewardship on Antibiogram, Consumption and Incidence of Multi Drug Resistance. BMC Infect. Dis. 2022, 22, 916. [Google Scholar] [CrossRef] [PubMed]

| Type of Sample | Number of Isolates (%) |
|---|---|
| Pus from surgical trauma | 384 (26.3) |
| Pus from abscess | 205 (14.1) |
| Pus (not specified) | 203 (13.9) |
| Peritoneal fluid | 174 (11.9) |
| Urine | 104 (7.1) |
| Blood | 96 (6.6) |
| Fluid (not specified) | 82 (5.6) |
| Bile | 81 (5.6) |
| Tissue | 57 (3.9) |
| Vascular catheter | 36 (2.5) |
| Unknown | 8 (0.5) |
| Bronchial secretions | 5 (0.3) |
| Graft | 4 (0.3) |
| Prosthetic materials | 4 (0.3) |
| Catheter (not specified) | 3 (0.2) |
| Drain catheter | 3 (0.2) |
| Vaginal | 2 (0.1) |
| Stool | 2 (0.1) |
| Bone | 2 (0.1) |
| Sputum | 2 (0.1) |
| Duodenal fluid | 1 (0.1) |
| Pleural fluid | 1 (0.1) |
| All samples | 1459 (100) |
| Pathogen | 2016–2021 (%) | Pre-COVID-19 (%) | Post-COVID-19 (%) | p-Value |
|---|---|---|---|---|
| Gram-positives | 675 (46.26) | 391 (46.27) | 284 (45.88) | 0.9155 |
| Enterococcus spp. | 326 (22.34) | 177 (21.02) | 149 (23.90) | 0.1622 |
| Coagulase-negative Staphylococcus | 152 (10.42) | 94 (11.12) | 58 (6.86) | 0.2985 |
| Streptococcus spp. | 105 (7.20) | 65 (7.69) | 40 (4.73) | 0.4124 |
| Staphylococcus aureus | 44 (2.95) | 30 (3.57) | 14 (2.32) | 0.1660 |
| Corynebacterium spp. | 10 (0.69) | 1 (0.12) | 9 (1.07) | 0.0026 |
| Clostridium spp. | 9 (0.62) | 6 (0.71) | 3 (0.36) | 0.3168 |
| Gram-negatives | 655 (44.89) | 383 (45.33) | 277 (44.75) | 0.8318 |
| Enterobacterales * | 482 (32.95) | 278 (32.79) | 204 (33.10) | 0.9103 |
| Escherichia coli | 223 (15.38) | 129 (15.22) | 94 (15.53) | 1 |
| Enterobacter spp. | 90 (6.17) | 54 (6.39) | 36 (4.26) | 0.7413 |
| Pseudomonas spp. | 89 (6.04) | 53 (6.23) | 36 (5.85) | 0.8248 |
| Klebsiella spp. | 78 (5.53) | 34 (4.03) | 44 (7.02) | 0.0094 |
| Proteus spp. | 40 (2.74) | 32 (3.79) | 8 (0.95) | 0.0033 |
| Citrobacter spp. | 28 (1.92) | 19 (2.25) | 9 (1.07) | 0.3359 |
| Morganella morganii | 16 (1.10) | 8 (0.95) | 8 (1.29) | 0.6138 |
| Acinetobacter spp. | 17 (1.16) | 10 (1.15) | 7 (1.16) | 1 |
| Fungi ** | 129 (8.84) | 71 (8.40) | 58 (9.37) | 0.5155 |
| Candida albicans | 72 (4.93) | 36 (4.26) | 36 (5.82) | 0.1802 |
| Candida glabrata | 20 (1.37) | 10 (1.18) | 10 (1.62) | 0.5017 |
| Candida tropicalis | 18 (1.23) | 14 (1.66) | 4 (0.65) | 0.0960 |
| Candida parapsilosis | 11 (0.75) | 6 (0.71) | 5 (0.81) | 1 |
| All microorganisms | 1459 (100) | 845 (100) | 619 (100) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ioannou, P.; Maraki, S.; Koumaki, D.; Manios, G.A.; Koumaki, V.; Kassotakis, D.; Zacharopoulos, G.V.; Kofteridis, D.P.; Manios, A.; de Bree, E. A Six-Year Retrospective Study of Microbiological Characteristics and Antimicrobial Resistance in Specimens from a Tertiary Hospital’s Surgical Ward. Antibiotics 2023, 12, 490. https://doi.org/10.3390/antibiotics12030490
Ioannou P, Maraki S, Koumaki D, Manios GA, Koumaki V, Kassotakis D, Zacharopoulos GV, Kofteridis DP, Manios A, de Bree E. A Six-Year Retrospective Study of Microbiological Characteristics and Antimicrobial Resistance in Specimens from a Tertiary Hospital’s Surgical Ward. Antibiotics. 2023; 12(3):490. https://doi.org/10.3390/antibiotics12030490
Chicago/Turabian StyleIoannou, Petros, Sofia Maraki, Dimitra Koumaki, Georgios A. Manios, Vasiliki Koumaki, Dimitrios Kassotakis, Georgios V. Zacharopoulos, Diamantis P. Kofteridis, Andreas Manios, and Eelco de Bree. 2023. "A Six-Year Retrospective Study of Microbiological Characteristics and Antimicrobial Resistance in Specimens from a Tertiary Hospital’s Surgical Ward" Antibiotics 12, no. 3: 490. https://doi.org/10.3390/antibiotics12030490
APA StyleIoannou, P., Maraki, S., Koumaki, D., Manios, G. A., Koumaki, V., Kassotakis, D., Zacharopoulos, G. V., Kofteridis, D. P., Manios, A., & de Bree, E. (2023). A Six-Year Retrospective Study of Microbiological Characteristics and Antimicrobial Resistance in Specimens from a Tertiary Hospital’s Surgical Ward. Antibiotics, 12(3), 490. https://doi.org/10.3390/antibiotics12030490










