Abstract
LF3872 was isolated from the milk of a healthy lactating and breastfeeding woman. Earlier, the genome of LF3872 was sequenced, and a gene encoding unique bacteriocin was discovered. We have shown here that the LF3872 strain produces a novel thermolabile class III bacteriolysin (BLF3872), exhibiting antimicrobial activity against antibiotic-resistant Staphylococcus aureus strains. Sequence analysis revealed the two-domain structural (lysozyme-like domain and peptidase M23 domain) organization of BLF3872. At least 25% residues of this protein are expected to be intrinsically disordered. Furthermore, BLF3872 is predicted to have a very high liquid-liquid phase separation. According to the electron microscopy data, the bacterial cells of LF3872 strain form co-aggregates with the S. aureus 8325-4 bacterial cells. LF3872 produced bacteriolysin BLF3872 that lyses the cells of the S. aureus 8325-4 mastitis-inducing strain. The sensitivity of the antibiotic-resistant S. aureus collection strains and freshly isolated antibiotic-resistant strains was tested using samples from women with lactation mastitis; the human nasopharynx and oral cavity; the oropharynx of pigs; and the cows with a diagnosis of clinical mastitis sensitive to the lytic action of the LF3872 strain producing BLF3872. The co-cultivation of LF3872 strain with various antibiotic-resistant S. aureus strains for 24 h reduced the level of living cells of these pathogens by six log. The LF3872 strain was found to be able to co-aggregate with all studied S. aureus strains. The cell-free culture supernatant of LF3872 (CSLF3872) induced S. aureus cell damage and ATP leakage. The effectiveness of the bacteriolytic action of LF3872 strain did not depend on the origin of the S. aureus strains. The results reported here are important for the creation of new effective drugs against antibiotic-resistant strains of S. aureus circulating in humans and animals.
1. Introduction
Staphylococci are typical representatives of the normal microbiota, colonizing the skin and mucous membranes of healthy people and farm animals, while the frequency of colonization by Staphylococcus aureus of open biocenoses is 25–30% [1,2]. At the same time, S. aureus could be widespread pathogens that cause various infections in humans and animals, ranging from minor skin lesions to life-threatening conditions, such as meningitis, endocarditis, and pneumonia [2,3,4]. Antibiotics containing the β-lactam ring (natural and synthetic penicillins, cephalosporins of I, II, III, and IV generations, carbapenems, and monobactams) are widely used for the treatment of infectious diseases caused by S. aureus [5,6,7]. Penicillins, cephalosporins, and monobactams are sensitive to the hydrolyzing action of special β-lactamase enzymes produced by various pathogens, including S. aureus strains [8]. To overcome the β-lactamase resistance of pathogens, protected penicillins (clavulanic acid, sulbactam, and tazobactam) were developed. The bactericidal effect of β-lactam antibiotics is associated with the selective inhibition of various stages of the construction of peptidoglycan of the cell wall of pathogenic microorganisms, including S. aureus. The destruction of cell wall synthesis under the action of β-lactams causes inhibition of S. aureus cell growth [9]. The growth of S. aureus resistant to antimicrobial drugs is currently one of the most acute global public health problems, leading to the failure in the treatment of infectious endocarditis, meningitis, osteomyelitis, and septic arthritis [10]. S. aureus, being considered one of the most significant etiological factors of infection, developed multiple mechanisms of antibiotic resistance, which are transferred rapidly between strains in both hospital and community settings [10,11,12].
In the case of methicillin-resistant S. aureus (MRSA), the problem is particularly evident: the MRSA was previously predominant in hospital settings as hospital-acquired MRSA (HA-MRSA), but now, it is being detected in community settings as community-acquired MRSA (CA-MRSA), showing high infectivity and virulence [13,14,15,16,17,18]. CA-MRSA strains differ from HA-MRSA strains: they have an extra genome, carry different elements of the staphylococcal cassette chromosome mec (SCCmec), affect different populations, and cause clinical symptoms that differ from those of the HA-MRSA infection [13,14,15,19,20,21,22,23,24,25,26,27,28].
The increase in the number of multi-drug resistant S. aureus strains and the shortage of new antibiotics on the market are becoming a serious public health problem. Therefore, experts of the World Health Organization (WHO) have included MRSA in the list of microorganisms that require the urgent creation of new antibiotics to combat these pathogens [29].
The ineffectiveness of previously used antibiotic therapies warrants research on alternative treatments, such as phage therapy [30,31,32] and probiotic therapy [33,34]. The sensitivity of eradicated bacteria to bacteriophages is one of the prerequisites of successful phage therapy. However, similar to antibiotic resistance, bacteria can also develop resistance to bacteriophages [32].
Strains of lactic acid bacteria isolated from the breast milk of healthy women have shown a high efficiency as oral probiotics for the treatment of such conditions [35,36]. Lactiplantibacillus plantarum ATCC 8014 strain and its antimicrobial compounds showed great potential to inhibit the growth of S. aureus isolated from bovine mastitis [37]. However, the molecular mechanisms of the antibacterial effects of lactic acid bacteria against pathogenic staphylococci have not been sufficiently studied. Earlier, we showed that co-cultivation of S. aureus 8325-4 cells with the LF3872 cells for 24 h reduced the level of living test culture by six log and that such an effect was alleviated by heat treatment of LF3872 [38]. The S. aureus 8325-4 strain is used to simulate lactation mastitis in laboratory animals [39].
In a previous study [40], analysis of the LF3872 genome discovered a gene encoding a class III bacteriocin BLF3872. Bacteriocins have antimicrobial activity against pathogenic microorganisms, which determines their biotechnological potential [41,42]. Bacteriocins inhibit the growth of target microorganisms: they affect the cell wall and influence the gene expression and protein production within cells [43]. Class III bacteriocins are antimicrobial thermos-sensitive proteins with a molecular weight of more than 30 kDa that are capable of degrading cell wall murein [44]. This group includes some colicins, zoocins, megacins (Bacillus megaterium), klebicin (Klebsiella pneumonia), helveticins I and J (Lactobacillus helveticus), and enterolysin A (Enterococcus faecalis) [45]. The new class III bacteriocin BLF3872 from LF3872 has not yet been studied.
Here, we analyze the primary structure of a new class III bacteriocin BLF3872 and model its spatial organization. Moreover, we studied the changes in the cellular morphology of S. aureus 8325-4 cells after the co-cultivation with LF3872 cells using scanning electron microscopy (SEM), and the co-aggregative activity, as well as the antibacterial properties of LF3872 against antibiotic-resistant collection strains and freshly isolated strains in samples from women with lactation mastitis; from the human nasopharynx and oral cavity; from the oropharynx of pigs; and from cows with a diagnosis of clinical mastitis.
2. Results and Discussion
2.1. A Novel Class III Bacteriocin BLF3872
The predicted size and molecular weight of the identified BLF3872 corresponds to a protein with a length of 404 residues, a molecular weight of approximately 42.5 kDa, and an isoelectric point of 5.15. The protein contains two cysteine residues and is classified by using ProtParam as stable (resistance index is 36.0). As was shown earlier [38], the 280–404 region of the BLF3872 corresponds to the peptidase M23 domain (residues 279–377; PF01551). Furthermore, the N-terminal half of this protein includes a lysozyme-like domain (residues 70–199; PF18013). Sequence comparison of the BLF3872 with the class III bacteriocins revealed the conservative motif (e.g., His280 and Asp284 in the HXXXD motif, and His361 and His363 in the HXH motif) potentially capable of coordinating the Zn2+ ion.
Figure 1A shows the 3D structure of BLF3872 modeled by using AlphaFold and shows that this protein has two spatially well-separated N- and C-terminal domains (residues 70–199 and 279–377, respectively), with a long N-terminal unordered region (residues 1–60) and a flexible linker between domains (Figure 1A). In line with the earlier expectations [38], the structures of the N- and C-domains resemble lysozyme-like enzymes and metalloendopeptidases (peptidase M23 domain), respectively.
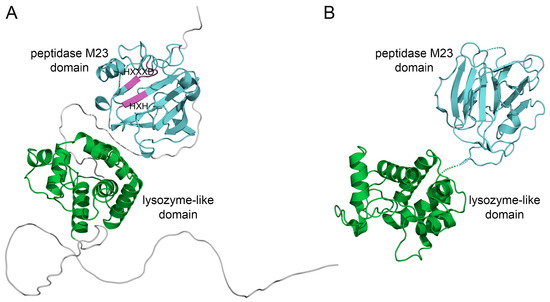
Figure 1.
(A) Model structure of BLF3872 obtained by using AlphaFold [46]. The N- and C-terminal domains are highlighted using green and cyan, respectively. Conservative motifs HXXXD and HXH in the peptidase M23 domain are marked using magenta. (B) The 3D structure of Zn2+ loaded Morphogenesis protein 1 from Bacillus phage phi29 (chain A of 3CSQ, X-RAY). The lysozyme-like domain and peptidase M23 domain are highlighted using green and cyan, respectively.
The BLAST sequence search identified the closest similarity of the BLF3872 to Morphogenesis protein 1 (365 a.a.) from Bacillus phage phi29 (UniProt ID P15132). Figure 1B shows a 3D X-ray crystal structure of the Morphogenesis protein 1 and shows that BLF3872 and Morphogenesis protein 1 are structurally rather similar despite a low level of the amino acid sequence identity (~30% according to EMBOSS Needle). Note that the amino acid sequence of the BLF3872 protein is depicted in Figure 2A. Based on these findings, one can hypothesizes that BLF3872 is capable of a cleavage of both peptide cross-links and the polysaccharide backbone of the peptidoglycan cell, thereby acting as a peptidoglycan-degrading enzyme [47].
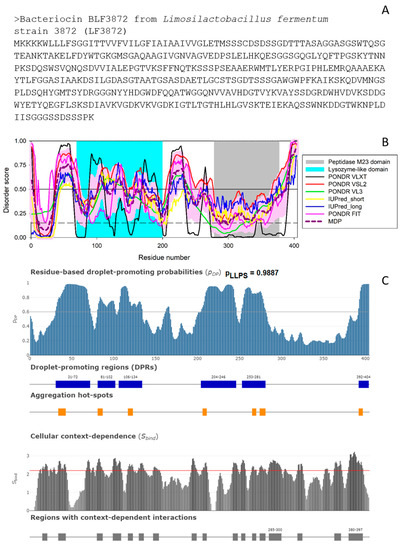
Figure 2.
(A). Amino acid sequence of the bacteriocin BLF3872. (B). Intrinsic disorder predisposition evaluated using RIDAO [48] that aggregates the results from a number of well-known disorder predictors: PONDR® VLXT (black line), PONDR® VLS2 (red line), PONDR® VL3 (green line), PONDR® FIT (pink line), IUPred2_long (blue line), and IUPred2_short (yellow line). A thick dashed dark-pink line represents the mean disorder prediction (MDP) profile calculated by averaging the outputs of these six predictors, whereas a light pink shadow shows the corresponding error distribution. The results of residual disorder propensity estimation using these tools are presented as real numbers from 0 (ideal order prediction) to 1 (ideal disorder prediction). To identify disordered residues and regions in the proteins of interest, a threshold of ≥0.5 was used. Solid and dashed horizontal lines at disorder scores 0.5 and 0.15 correspond to the disorder and flexibility thresholds. (C). Evaluation of the LLPS propensity of the bacteriocin BLF3872 using FuzDrop. The top plot represents the sequence distribution of the droplet- or LLPS-promoting probability (pLLPS) and shows that BLF3872 is characterized by a very high overall pLLPS of 0.9887. Furthermore, this protein contains six droplet-promoting regions and seven aggregation hot-spots. The bottom plot represents the cellular context dependence of BLF3872, where the SBIND values characterize the ability of residues to switch between different binding modes. Here, regions with the context-dependent interactions change protein interaction behavior and binding modes under different cellular conditions. The corresponding residues/regions can be ordered or disordered with SBIND ≥ 2.25 (red line) [49].
According to AlphaFold, as BLF3872 is expected to have long flexible regions, we evaluated the intrinsic disorder predisposition of this protein using a set of commonly utilized disorder predictors. The results of these analyses are summarized in Figure 2B, which clearly shows that at least 25% residues of this protein are expected to be disordered, with most disordered regions being located outside the lysozyme and peptidase M23 domains.
Curiously, the N-terminal lysozyme-like domain is expected to be more flexible than the C-terminal peptidase M23 domain. Since intrinsically disordered proteins are commonly involved in liquid-liquid phase separation (LLPS) and related biogenesis of various membrane-less organelles (MLOs) or biomolecular condensates [50,51,52,53,54,55,56,57], at the next stage, we checked the predisposition of BLF3872 for phase separation. Figure 2C shows that this bacteriocin is characterized by a very high LLPS propensity of 0.9887 and contains six DPRs (residues 31–72, 82–102, 106–134, 204–246, 253–281, and 392–494). Therefore, it is likely that BLF3872 can phase separate by itself. This conclusion is further supported by the results of the PSPredictor [58] analysis, according to which this protein has a very high PSP score of 0.9955 as well. This is an interesting observation suggesting that at least some of the functions of the bacteriocin BLF3872 can be associated with the formation of biomolecular condensates.
In addition to the evaluation of the LLPS predisposition of a query protein, FuzDrop is capable of finding aggregation hot-spots, which are defined as residues/regions that may promote the conversion of the liquid-like condensed state into a solid-like amyloid state [59,60]. Figure 2C shows that the bacteriocin BLF3872 contains seven such aggregation hot-spots (residues 34–43, 81–87, 117–123, 206–211, 265–270, 274–281, and 292–297). Furthermore, FuzDrop can evaluate the probability of a protein to have residues/regions with cellular context dependence that are capable of switching between different binding modes depending on the peculiarities of cellular environment [49]. Figure 2C demonstrates that BLF3872 contains 17 regions with the context-dependent interactions (residues 15–21, 34–43, 80–87, 99–109, 117–123, 130–136, 147–154, 193–199206-211, 242–248, 252–257, 265–270, 274–281, 285–300, 319–325, 363–371, and 380–397). Importantly, these regions overlap with, are included into, or are located in close proximity to the disordered or flexible regions found in BLF3872. This is in line with known notion that the heterogeneity or disorder of the bound structure can vary with cellular conditions [49].
2.2. Scanning Electron Microscopy
The results of antibacterial action of LF3872 strain producing a novel class III bacteriocin (bacteriolysin) BLF3872 on S. aureus 8325-4 strain are shown in Figure 3. The cell wall of intact S. aureus 8325-4 strain is a homogeneous electron-dense layer (Figure 3A). It is known that peptidoglycan (PG) is a key component of S. aureus cell wall that helps maintain cell shape and prevents bacteria from lysis through osmotic rupture [61]. PG is composed of polysaccharide chains cross-linked by short peptides. The polysaccharide chains contain N-acetylmuramic acid (NAM) and N-acetylglucosamide (NAG). In addition to these basic polymers, the cell walls of S. aureus contain small amounts of lipids, polysaccharides, and proteins. The initial stage of the antibacterial action of LF3872 cells is their co-aggregation with S. aureus 8325-4 cells (Figure 3B–D). The genome of LF3872 contains a gene, the product of which is responsible for the co-aggregation of the cells of this strain with pathogens [40,62]. Lysozyme (or muramidase or N-acetylmuramic acid hydrolase, EC 3.2.1.17) is known to be a protein with the enzymatic activity through the hydrolysis of β-1,4-glycosidic bonds between NAM and NAG in the PG of S. aureus bacterial cell wall [63,64].
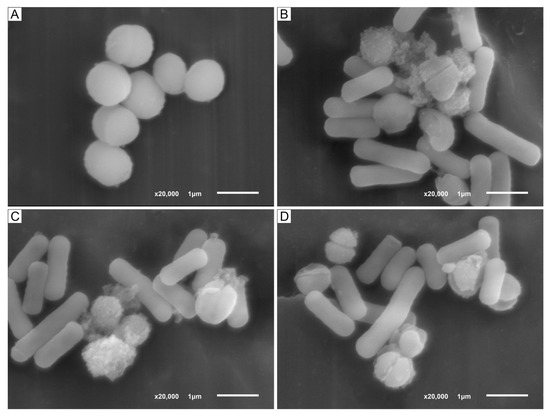
Figure 3.
(A). SEM images of intact S. aureus 8325-4 cells. (B–D). Co-aggregation of LF3872 cells with S. aureus 8325-4 cells; destruction of cell wall and death of the pathogen. Note: the ratio of S. aureus 8325-4 cells/ LF3872 cells is 1/10.
We assume that the lysozyme-like N-terminal domain of BLF3872 hydrolyzes the β-1,4-glycosidic bonds between NAM and NAG in PG of S. aureus 8325-4 bacterial cell wall and metalloendopeptidase-like C-domain of BLF3872 hydrolyzes amide bonds between amino acids of short peptides connecting polysaccharide chains of PG. The BLF3872 destroys the PG of the cell wall of S. aureus 8325-4 strain and induces the cell death (Figure 3B–D). BLF3872 has a homology with phage lysins, in which one domain has the muramidase activity and the second domain has the amidase activity. These domains exhibit synergistic antibacterial action on the pathogens and, at the same time, practically do not affect commensal bacteria [65]. Thus, BLF3872 secreted by the LF3872 strain reaches the PG layer of S. aureus 8325-4 cells, destroys it, and causes cell death.
2.3. Antibacterial Activity of LF3872 against Collection ATCC Methicillin-Resistant S. aureus (MRSA) Strains
The antagonistic activity of LF3872 against a collection of the ATCC methicillin-resistant S. aureus (MRSA) strains is shown in Table 1. In the co-culture experiments, MRSA strains ATCC BAA-1683, ATCC BAA-2313, ATCC 33592, and ATCC BAA-2094 were highly sensitive to the bacteriolytic action of LF3872. The co-cultivation of ATCC MRSA strains with the LF3872 for 24 h reduced the level of live test cultures by six log, while the heat-treated LF3872 has no effect on the level of live test cultures of ATCC MRSA strains.

Table 1.
Antagonistic activity of LF3872 against a collection of the ATCC methicillin-resistant S. aureus strains.
WHO experts have included MRSA in the list of pathogens that require the urgent creation of new effective antibiotics [29]. BLF3872 effectively destroys MRSA strains ATCC BAA-1683, ATCC BAA-2313, ATCC 33592, and ATCC BAA-2094. It is known that S. aureus strains stored in the collection may be less resistant to the antibacterial drugs [66]. To address this point, in the following experiments, we used clinical strains directly isolated from humans and farm animals.
2.4. Antibacterial Activity of LF3872 against S. aureus Clinical Isolates from the Milk of Women with Mastitis
The antagonistic activity of LF3872 against S. aureus clinical isolates from the milk of women with mastitis is shown in Table 2. In the co-culture experiments, antibiotic-resistant S. aureus IIE CI-SA Hu 1246 strain, S. aureus IIE CI-SA Hu 1247 strain, S. aureus IIE CI-SA Hu 1248 strain, and S. aureus IIE CI-SA Hu 1249 strain were highly sensitive to the bacteriolytic action of LF3872. The co-cultivation of S. aureus clinical isolates from the milk of women with mastitis with the LF3872 for 24 h reduced the level of living test cultures by six log. However, the heat-treated LF3872 did not reduce the level of live S. aureus clinical isolates from the milk of women with mastitis.

Table 2.
Antagonistic activity of LF3872 against S. aureus clinical isolates from the milk of women with mastitis.
S. aureus is the main etiological agent of acute mastitis [67,68] but more frequently causes subclinical infections that tend to become chronic and difficult to eradicate with conventional anti-microbial therapies [69]. The vertical transmission of multidrug-resistant S. aureus from mother to child has been established [70]. LF3872 is promising for the prevention of S. aureus-induced lactation mastitis in women during breast-feeding of a child and can be used to implement a strategy for preventive control of this widespread disease in the world.
2.5. Antibacterial Activity of LF3872 against S. aureus Strains Isolated from the Nasopharynx and Oral Cavity of Humans
The antagonistic activity of LF3872 against S. aureus strains isolated from the nasopharynx and oral cavity of humans is shown in Table 3. In the co-culture experiments, the antibiotic-resistant S. aureus IIE CI-SA Hu 1261 strain, S. aureus IIE CI-SA Hu 1262 strain, S. aureus IIE CI-SA Hu 1263 strain, and S. aureus IIE CI-SA Hu 1264 strain isolated from the nasopharynx and oral cavity of humans were highly sensitive to the bacteriolytic action of LF3872. The co-cultivation of S. aureus strains isolated from the human nasopharynx and oral cavity with LF3872 for 24 h reduced the level of living test cultures by six log. Heat-treated LF3872 has no effect on the level of live S. aureus strains isolated from the nasopharynx and oral cavity of humans.

Table 3.
Antagonistic activity of LF3872 against S. aureus isolated in humans from the nasopharynx and oral cavity.
Despite growing knowledge of the oral microbiome, the roles of S. aureus in oral diseases and the risk of spreading staphylococcal infection from this reservoir are still not fully understood [71]. The prevalence of S. aureus infections has increased recently. The burden of infection due to the MRSA strains is significantly more evident in children than in other age groups [72]. S. aureus colonization of the nasopharynx begins very early, in the first six months of life [73]. The use of the LF3872 strain as a probiotic may help in the prevention and eradication of antibiotic-resistant S. aureus in the nasopharynx and oral cavity in humans.
2.6. Antibacterial Activity of LF3872 against S. aureus Strains Isolated from the Oropharynx of Pigs
The antagonistic activity of LF3872 against S. aureus strains isolated from the oropharynx of pigs is shown in Table 4. In the co-culture experiments, the antibiotic-resistant S. aureus IIE CI-SA Pi 1345 strain, S. aureus IIE CI-SA Pi 1347 strain, S. aureus IIE CI-SA Pi 1356 strain, and S. aureus IIE CI-SA Pi 1357 strain isolated from the oropharynx of pigs proved to be highly sensitive to the bacteriolytic action of LF3872. The co-cultivation of S. aureus strains isolated from the oropharynx of pigs with LF3872 for 24 h reduced the level of living test cultures by six log. Heat-treated LF3872 had no effect on the level of live S. aureus strains isolated from the oropharynx of pigs.

Table 4.
Antagonistic activity of LF3872 against S. aureus isolated from the oropharynx of pigs.
Pigs are the main source of the spread of antibiotic-resistant strains of S. aureus. Numerous studies have shown that people living or working on pig farms, including farmers and their families, veterinarians and slaughterhouse workers, are at increased risk of colonization or infection with the LA-MRSA [74,75,76,77,78,79]. Direct contact is the main route of transmission of MRSA between pigs [80,81]. It has been suggested that, in the process of the transmission of these bacteria to negative animals, the MRSA-positive pigs may play a critical role (horizontal transmission) [80,81]. Only a few positive animals can lead to the spread of MRSA on or even off farms [82,83]. It is also possible that MRSA can be transmitted from sows to their offspring (vertical perinatal transmission) [80,83,84,85,86]. The use of a feed additive containing the LF3872 strain may help in the prevention and eradication of the MRSA on pig farms.
2.7. Antibacterial Activity of LF3872 against S. aureus Clinical Isolates from the Milk of Cows with Mastitis
The antagonistic activity of LF3872 against S. aureus clinical isolates from the milk of cows with mastitis is shown in Table 5. In the co-culture experiments, the antibiotic-resistant S. aureus IIE CI-SA Co 1280 strain, S. aureus IIE CI-SA Co 1281 strain, S. aureus IIE CI-SA Co 1282 strain, and S. aureus IIE CI-SA Co 1283 strain were highly sensitive to the bacteriolytic action of LF3872. The co-cultivation of clinical isolates of S. aureus from the milk of cows with mastitis, with LF3872 for 24 h, reduced the level of live test cultures by six logs. Heat-treated LF3872 had no effect on the level of live clinical isolates of S. aureus from the milk of cows with mastitis. The WHO One Health emphasizes the importance of considering human and animal health in a collaborative and interrelated manner [87].

Table 5.
Antagonistic activity of LF3872 against S. aureus clinical isolates from the milk of cows with mastitis.
The use of a feed additive containing the LF3872 strain may help in the prevention and treatment of lactation mastitis in cows.
2.8. Cells of LF3872 Co-Aggregate with Antibiotic-Resistant S. aureus Strains Isolated from Humans and Animals
We evaluated the co-aggregation capacities of LF3872 with antibiotic-resistant S. aureus strains isolated from humans and animals and ATCC MRSA strains. LF3872 is a strong co-aggregator of antibiotic-resistant S. aureus strains isolated from humans and animals and ATCC MRSA strains.
At pH 7.0 in phosphate-buffered saline (PBS) buffer, LF3872 co-aggregated with all S. aureus strains analyzed in this study. The percentage of co-aggregation ranged from 45.9 ± 6.5% to 54.5 ± 4.7%. LF3872 also co-aggregated with all tested S. aureus strains at pH 5.0 in 2-(N-morpholino)ethanesulfonic acid (MES) buffer.
At lower pH values, the percentage of co-aggregation significantly increased (p < 0.05), ranging from 80.6 ± 5.8% to 89.3 ± 7.6% (Table 6). Co-aggregation increased with decreasing pH. Some lactobacilli use co-aggregation mechanisms to realize their probiotic properties [88,89,90,91]. The ability to co-aggregate with pathogens is one of the most important manifestations of the antagonistic activity of lactobacilli [92,93]. The formation of lactobacilli co-aggregates with pathogens leads to inhibition of the adhesion of these pathogens to the host epithelial cells [94,95]. Lactobacilli use co-aggregation mechanisms to create a hostile microenvironment around the pathogen [95]. They secrete antibacterial metabolites inside the co-aggregates, such as bacteriocins [94,96], organic acids, and H2O2 [97]. In various biotopes of the macro-organism, the co-aggregation process of lactobacilli with pathogens is important for maintaining a homeostatic microbiota [98]. Due to the increase of antibiotic resistance in pathogens, it is relevant to identify lactobacilli with high co-aggregation activity [95,99]. Thus, the co-aggregation ability of LF3872 may prevent the colonization of human and animal organs and tissues by antibiotic-resistant strains of S. aureus.

Table 6.
Co-aggregative abilities of LF3872 with antibiotic-resistant S. aureus strains isolated from humans and animals.
2.9. CSLF3872 Induces Cell Damage and ATP Leakage in Antibiotic-Resistant S. aureus Strains
Cell-free culture supernatant of LF3872 (CSLF3872) induced cell damage and ATP leakage in antibiotic-resistant S. aureus strains isolated from humans, animals, and in ATCC MRSA strains. ATP leakage levels were examined based on non-selective pore formation and cell damage indexes. The research results are presented in Table 7.

Table 7.
Extracellular ATP levels in antibiotic-resistant S. aureus strains treated with CSLF3872.
The cultivation of various antibiotic-resistant S. aureus strains in the presence of CSLF3872 for 2.5 h increased the level of extracellular ATP on average from 6.5 (nm/OD) to 346.0 (nm/OD) (p < 0.001). Since the LF3872 strain produces thermolabile class III bacteriocin BLF3872, heating of the CSLF3872 leads to the loss of its ability to damage the cell wall and ATP leakage in antibiotic-resistant S. aureus strains. The boiled CSLF3872 did not induce cell damage and ATP leakage in antibiotic-resistant S. aureus strains isolated from humans, animals, and in ATCC MRSA strains. In a previous study [100], the ability of the thermolabile recombinant class III bacteriocin Helveticin-M from L. crispatus to destroy the cell wall of S. aureus was demonstrated. Helveticin-M also disrupted the inner membrane, as confirmed by leakage of intracellular ASP from S. aureus cells. As CSLF3872 may additionally contain lactic acid and hydrogen peroxide, which in high concentrations are also able, along with BLF3872, to inhibit the growth of S. aureus and damage the inner membrane [101,102], we studied lactic acid and hydrogen peroxide production by LF3872. The results of the studies are given in (2.10; 2.11).
2.10. Lactic Acid Production by LF3872 Strain in the Process of Its Cultivation in the MRS Broth
The results of the studies shown in Table 8 demonstrate that the LF3872 strain produces an insignificant amount of lactic acid during 20 h of observation (210 ± 8 µg/mL). In a previous study [103], the lytic effect of CS strain L. fermentum 1912 and strain L. plantarum 8513 on S. aureus cells was studied. The lytic effect of lactic acid and bacteriocins as separate components of CS strain L. fermentum 1912 and CS strain L. plantarum 8513 on S. aureus cells was also studied. Both strains of lactobacilli were super-producers of lactic acid. Strain L. fermentum 1912 increased the concentration of lactic acid in CS to 6.430 ± 0.063 mg/mL; strain L. plantarum 8513 increased the concentration of lactic acid in CS to 10.796 ± 0.074 mg/mL. It was found that bacteriocins from CS strain L. fermentum 1912 and strain L. plantarum 8513, but not lactic acid cause lysis of S. aureus cells [103]. According to [103], CS of LF3872 strain with such a low lactic acid level cannot cause S. aureus cell lysis. Therefore, it can be assumed that LF3872 strain lyses S. aureus cells (Figure 3A) due to the production of BLF3872.

Table 8.
Dynamics of lactic production in the process of LF3872 strain cultivation in the MRS broth.
2.11. Hydrogen Peroxide Production by LF3872 Strain in the Process of Its Cultivation in the MRS Broth
The results of the studies shown in Table 9 demonstrate that the LF3872 strain produces an insignificant amount of H2O2 during 20 h of observation (0.038 mM/107 CFU). In a previous study [104], the lytic effect of the CS of L. gasseri CRL 1421 strain, a superproducer of H2O2, and the CS of L. gasseri CRL 1412 strain, producing a small amount of H2O2, on S. aureus cells was studied. The mean amount of H2O2 generated by L. gasseri CRL 1421 was greater than that produced by LF3872 (1517.92 mM/107 CFU vs. 0.038 mM/107 CFU, p < 0.01). The CS of L. gasseri CRL 1421 strain induced intensive damage to the cell structure of S. aureus; disintegration of the cell wall observed using transmission electron microscopy [104]. At the same time, L. gasseri CRL 1412 strain producing a small amount of H2O2 (0.041 mM/107 CFU) at the level of LF3872 strain did not cause damage to S. aureus cells. Based on the research results available in the literature [104], the CS of LF3872 strain with such a low H2O2 level cannot cause S. aureus cell lysis. Therefore, it can be assumed that LF3872 strain lyses S. aureus cells (Figure 3A) due to the production of BLF3872.

Table 9.
Hydrogen peroxide in the process of LF3872 strain cultivation in the MRS broth.
3. Materials and Methods
3.1. Used Microorganisms and Their Growth Conditions
LF3872 [40,62,105,106] was deposited in the Collection of Microorganisms of the Institute of Immunological Engineering, Lyubuchany, Moscow region, Russia.
A full list of used microorganisms and their growth conditions are given in Table 10.

Table 10.
Microorganisms used in this study.
3.2. Sequence Alignment and Bioinformatic Analysis
ProtParam (https://web.expasy.org/protparam/; accessed on 2 January 2023) was used for sequence analysis [107]. A sequence similarity search was performed using the Basic Local Alignment Search Tool (BLAST) (http://www.ncbi.nlm.nih.gov/BLAST; accessed on 2 January 2023). The pairwise sequence alignment of BLF3872 and Morphogenesis protein 1 from Bacillus phage phi29 (UniProt ID: P15132) was implemented by the EMBOSS Needle (https://www.ebi.ac.uk/Tools/psa/emboss_needle/; accessed on 2 January 2023). The tertiary structure of Morphogenesis protein 1 from Bacillus phage phi29 (chain A of 3CSQ, X-RAY) was taken from the PDB bank (https://www.rcsb.org/; accessed on 2 January 2023). The tertiary structure of BLF3872 was modeled using AlphaFold (https://alphafold.ebi.ac.uk/; accessed on 2 January 2023) [46]. The tertiary structure models were drawn with molecular graphics system PyMOL v.1.8 (https://pymol.org/; accessed on 2 January 2023).
Intrinsic disorder predisposition was quantified on a per-amino acid residue basis using the Rapid Intrinsic Disorder Analysis Online (RIDAO) web server (https://ridao.app/; accessed on 12 January 2023) [48]. In these the analyses, values of 0 and 1 correspond to the most ordered and most disordered residues, respectively.
The FuzDrop web server (https://fuzdrop.bio.unipd.it/predictor; accessed on 12 January 2023), which is capable of finding droplet promoting regions (DPRs), the aggregation-promoting regions (aggregation hot spots), and residues/regions with the cellular context dependence [49] in a protein, was utilized to evaluate the propensity of a query protein for phase separation [60,108,109]. The FuzDrop output shows an interactive graph showing distribution of the per single residue droplet-promoting probabilities (pDP) and gives an estimated value of the probability of spontaneous liquid-liquid phase separation (LLPS; pLLPS), which is used for protein classification as droplet drivers (pLLPS ≥ 0.60), droplet-clients (pLLPS < 0.60 and the presence of at least one DPR), or unrelated to LLPS (pLLPS < 0.60 and no DPRs) [60,108,109]. The phase separation potential of a query protein was further validated using PSPredictor (http://www.pkumdl.cn:8000/PSPredictor/; accessed on 12 January 2023) [58].
For a query protein, PSPredictor generates a PSP score ranging from 0 to 1 in a tabulated form and shows the PSP status of a query protein in a “Yes/No” form. A query protein can be considered as a potential phase separating protein if its PSP score is ≥0.50.
3.3. Scanning Electron Microscopy
For experiments on SEM, intact S. aureus 8325-4 cells or co-aggregates of LF3872 cells with S. aureus 8325-4 cells were preliminarily fixed in a 2.5% solution of glutaraldehyde in the cacodylate buffer overnight at 4 °C. Additionally, after three washes in same buffer, the cells were fixed in 1% solution of OsO4 in cacodylate buffer at 20 °C for 3 h. After fixation and subsequent washing in buffer, the cells were dehydrated in increasing concentrations of ethanol (from 30 to 100%) for 20 min at each stage. Next, the cells were placed in tert-butanol and left at 4 °C for 12 h. The studied samples were freeze-dried in JFD-320 (JEOL, Tokyo, Japan) and were placed on a disk and sputtered with a gold in a JEE-4X (JEOL, Tokyo, Japan) vacuum evaporator. Images were taken on the JSM-6510LV (JEOL, Tokyo, Japan) microscope.
3.4. Determination of Antibacterial Activity of LF3872 against the Studied Groups of S. aureus Strains
The antibacterial activity of the LF3872 strain against the studied groups of S. aureus strains was determined by using a previously described method [38]. Briefly, in 20 mL of the TGVC medium, 1 mL of the LF3872 inoculum grown on MRC medium and the test S. aureus strains were co-cultivated. After 24 h, the amount of different S. aureus cells (grown on TGVC medium in monoculture (control) and in the presence of lactobacilli) was counted. Aliquots were taken under aseptic conditions, serially diluted, and put on TGVC agar. After incubation, S. aureus strains colonies were counted and expressed as colony-forming units per milliliter (CFU/mL). Incubation was performed under aerobic conditions (24 h at 37 °C).
3.5. Co-Aggregation Assay for the Determination of Interactions between LF3872 and S. aureus Strains
Co-aggregation between LF3872 and S. aureus strains of the studied groups was determined by measuring the optical density (OD) at 600 nm, as previously described [38]. As pH can affect the co-aggregation, the MES buffer pH 5 (Sigma-Aldrich, St. Louis, MO, USA) for cell suspension was used instead of PBS.
3.6. Preparation of Cell-Free Culture Supernatant (CSLF3872)
CSLF3872 was prepared as previously described [38]. The lyophilized sediment of CSLF3872 was used to study cytoplasmic membrane permeability in S. aureus cells of various strains. Briefly, under anaerobic conditions at 37 °C, LF3872 was grown in MRS broth. Then, the culture was diluted in MRS broth and was grown anaerobically for two days. CSLF3872 was sequentially collected via centrifugation, and then filter-sterilized and concentrated via speed-vacuum drying.
3.7. Assessment of Cytoplasmic Membrane Permeability by Measurement of Extracellular ATP in S. aureus Strains of the Studied Groups
Measurement of extracellular ATP in S. aureus strains of the studied groups for the assessment of cytoplasmic membrane permeability was performed as previously described [38]. Briefly, S. aureus strains were centrifuged and resuspended (PBS (pH 7.0), OD600 = 1.0 (5 × 108 CFU/mL)). Then, bacteria were processed in the presence of CSLF3872 (for 2.5 h at 37 °C). After CSLF3872 treatment, the ATP detection kit (Beyotime, Shanghai, China) was used for the detection of extracellular ATP levels. For luminescence detection, an Infinite 200 PRO microplate reader (Tecan, Männedorf, Switzerland) was used.
3.8. Lactic Acid Determination
The suspension of LF3872 cells grown in the MRS broth was centrifuged at 5000× g for 20 min at 4 °C. The culture supernatant was filtered using a 0.22 µm pore size filter (Millipore, Burlington, MA, USA).
The concentration of lactic acid in CSLF3872 produced by the LF3872 strain was determined according to the method [110] using a high performance liquid chromatography (HPLC) system equipped with an ultraviolet-visible detector (Shidmadzu, Kyoto, Japan) set at 220 nm and a Luna C18(2) column (150 mm × 4.6 mm, 5 µm; Phenomenex, Torrance, CA, USA). HPLC grade lactic acid (Sigma-Aldrich, St. Louis, MO, USA) was used as the standard.
3.9. Hydrogen Peroxide Determination
The suspension of LF3872 cells grown in the MRS broth was centrifuged at 5000× g for 20 min at 4 °C. The culture supernatant was filtered using a 0.22 µm pore size filter (Millipore, Burlington, MA, USA). The concentration of hydrogen peroxide was determined as described previously [101], calorimetrically through the detection of the absorbance induced with chromophore: 3,3′,5,5′-tetramethylbenzidine (TMB) in the presence of a horseradish peroxidase (Sigma-Aldrich, St. Louis, MO, USA).
3.10. Statistical Analysis
The results were analyzed using one-way analysis of variance (ANOVA) and represented as the Mean ± SD of six independent experiments, tested in triplicate. Statistical significance was evaluated by using the Student’s t-tests. Results were considered significant at p < 0.05.
4. Conclusions
The LF3872 strain has the potential of producing a novel class III bacteriocin BLF3872 with two predicted domains: an N-terminal domain with a lysozyme-like structure and a C-terminal domain with a metalloendopeptidase-like structure. BLF3872 is predicted to have noticeable levels of intrinsic disorder and is expected to serve as an LLPS driver, suggesting that at least some functions of this protein are related to the formation of biomolecular condensates. LF3872 is shown to form co-aggregates with both collection antibiotic-resistant S. aureus strains and with S. aureus strains freshly isolated from humans and animals. The co-aggregation of the LF3872 strain with various S. aureus strains is the initial stage of its antibacterial activity. According to electron microscopy data, LF3872 producing bacteriocin BLF3872 lyses the cells of S. aureus 8325-4 strain. Co-cultivation of the LF3872 strain with various antibiotic-resistant S. aureus strains for 24 h reduced the level of living cells of this pathogen by six log. The cell-free culture supernatant of LF3872 (CSLF3872) induced S. aureus cell damage and ATP leakage. Probiotic drugs and feed additives created in future on the basis of the LF3872 strain may be promising for the prevention of lactation mastitis induced by S. aureus in women during breastfeeding and in cows during the lactation period.
Author Contributions
Conceptualization, V.M.A., A.V.M., T.V.P., G.T.S., A.N.P. and A.V.K.; methodology, V.M.A.; validation, I.V.K., A.V.M., I.O.C., E.I.D., E.L.N., T.N.A., V.G.M., N.E.S., I.N.N., M.V.S., V.S.K., V.K.S., V.A.S., A.B.G., V.N.U. and A.V.K.; investigation, I.V.K., A.V.M., I.O.C., E.I.D., E.L.N., T.N.A., V.G.M., N.E.S., I.N.N., M.V.S., V.S.K., V.K.S., V.N.U. and V.A.S.; data curation, V.M.A. and I.V.K.; writing—original draft preparation, V.M.A., I.V.K. and A.V.M.; writing—review and editing, E.I.D., A.V.M., T.V.P., V.N.U. and A.V.K.; visualization, V.N.U. and A.V.M.; supervision, V.M.A., V.N.U. and A.V.K.; project administration, V.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Government of the Russian Federation (Agreement No. 075-15-2022-1124 dated 1 July 2022).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mainous, A.G.; Hueston, W.J.; Everett, C.J.; Diaz, V.A. Nasal Carriage of Staphylococcus aureus and Methicillin-Resistant S. aureus in the United States, 2001–2002. Ann. Fam. Med. 2006, 4, 132–137. [Google Scholar] [CrossRef]
- Rasigade, J.-P.; Vandenesch, F. Staphylococcus aureus: A pathogen with still unresolved issues. Infect. Genet. Evol. 2014, 21, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Mandell, G.L.; Bennett, J.E.; Dolin, R. Staphylococcus aureus (including Staphylococcal toxic shock). In Bennett’s-Principles and Practice of Infectious Diseases; Mandell, G.L., Bennett, J.E., Dolin, R., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 2000. [Google Scholar]
- Hermans, K.; Devriese, L.A.; Haesebrouck, F. Staphylococcus in Gyles CL. In Pathogenesis of Bacterial Infections in Animals; Prescott, J.F., Songer, J.G., Thoen, C.O., Eds.; Wiley-Blackwell: Oxford, UK, 2010; pp. 75–89. [Google Scholar]
- Holten, K.B.; Onusko, E.M. Appropriate prescribing of oral beta-lactam antibiotics. Am. Fam. Physician 2000, 62, 611–620. [Google Scholar]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.V.; Kuti, J.L. Pharmacodynamic Thresholds for Beta-Lactam Antibiotics: A Story of Mouse Versus Man. Front. Pharmacol. 2022, 13, 833189. [Google Scholar] [CrossRef]
- Silago, V. Beta-lactam antibiotics and extended spectrum beta-lactamases. GSC Adv. Res. Rev. 2021, 9, 015–024. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; O’Driscoll, N.H.; Lamb, A.J. Morphological and ultrastructural changes in bacterial cells as an indicator of antibacterial mechanism of action. Cell. Mol. Life. Sci. 2016, 73, 4471–4492. [Google Scholar] [CrossRef]
- Boswihi, S.S.; Udo, E.E. Methicillin-resistant Staphylococcus aureus: An update on the epidemiology, treatment options and infection control. Curr. Med. Res. Pract. 2018, 8, 18–24. [Google Scholar] [CrossRef]
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef]
- Malachowa, N.; DeLeo, F.R. Mobile genetic elements of Staphylococcus aureus. Cell Mol. Life Sci. 2010, 67, 3057–3071. [Google Scholar] [CrossRef]
- Witte, W.; Strommenger, B.; Stanek, C.; Cuny, C. Methicillin-resistant Staphylococcus aureus ST398 in Humans and Animals, Central Europe. Emerg. Infect. Dis. 2007, 13, 255–258. [Google Scholar] [CrossRef]
- Tacconelli, E.; De Angelis, G.; Cataldo, M.A.; Pozzi, E.; Cauda, R. Does antibiotic exposure increase the risk of methicillin-resistant Staphylococcus aureus (MRSA) isolation? A systematic review and meta-analysis. J. Antimicrob. Chemother. 2008, 61, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Nishiyama, A.; Takano, T.; Yabe, S.; Higuchi, W.; Razvina, O.; Shi, D. Community-acquired methicillin-resistant Staphylococcus aureus: Community transmission, pathogenesis, and drug resistance. J. Infect. Chemother. 2010, 16, 225–254. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Liu, D.; Ma, Y.; Gao, W. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus isolates at a Chinese tertiary hospital, 2012–2017. Sci. Rep. 2018, 8, 17916. [Google Scholar] [CrossRef]
- Udo, E.E. Community-Acquired Methicillin-Resistant Staphylococcus aureus: The New Face of an Old Foe? Med. Princ. Pract. 2013, 22 (Suppl. 1), 20–29. [Google Scholar] [CrossRef] [PubMed]
- Kwapisz, E.; Garbacz, K.; Kosecka-Strojek, M.; Schubert, J.; Bania, J.; Międzobrodzki, J. Presence of egc-positive major clones ST 45, 30 and 22 among methicillin-resistant and methicillin-susceptible oral Staphylococcus aureus strains. Sci. Rep. 2020, 10, 18889. [Google Scholar] [CrossRef] [PubMed]
- Grundmann, H.; Hori, S.; Winter, B.; Tami, A.; Austin, D.J. Risk Factors for the Transmission of Methicillin-Resistant Staphylococcus aureus in an Adult Intensive Care Unit: Fitting a Model to the Data. J. Infect. Dis. 2002, 185, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Naimi, T.S.; LeDell, K.H.; Como-Sabetti, K.; Borchardt, S.M.; Boxrud, D.J.; Etienne, J.; Johnson, S.K.; Vandenesch, F.; Fridkin, S.; O’Boyle, C.; et al. Comparison of Community- and Health Care–Associated Methicillin-Resistant Staphylococcus aureus Infection. JAMA 2003, 290, 2976–2984. [Google Scholar] [CrossRef]
- Tenover, F.C.; McDougal, L.K.; Goering, R.V.; Killgore, G.; Projan, S.J.; Patel, J.B.; Dunman, P.M. Characterization of a Strain of Community-Associated Methicillin-Resistant Staphylococcus aureus Widely Disseminated in the United States. J. Clin. Microbiol. 2006, 44, 108–118. [Google Scholar] [CrossRef]
- Enright, M.C.; Robinson, D.A.; Randle, G.; Feil, E.J.; Grundmann, H.; Spratt, B.G. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 2002, 99, 7687–7692. [Google Scholar] [CrossRef]
- Graffunder, E.M.; Venezia, R.A. Risk factors associated with nosocomial methicillin-resistant Staphylococcus aureus (MRSA) infection including previous use of antimicrobials. J. Antimicrob. Chemother. 2002, 49, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Okuma, K.; Iwakawa, K.; Turnidge, J.D.; Grubb, W.B.; Bell, J.M.; O’Brien, F.G.; Coombs, G.W.; Pearman, J.W.; Tenover, F.C.; Kapi, M.; et al. Dissemination of New Methicillin-Resistant Staphylococcus aureus Clones in the Community. J. Clin. Microbiol. 2002, 40, 4289–4294. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.A.; Enright, M.C. Evolutionary Models of the Emergence ofMethicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 3926–3934. [Google Scholar] [CrossRef] [PubMed]
- Vandenesch, F.; Naimi, T.; Enright, M.C.; Lina, G.; Nimmo, G.R.; Heffernan, H.; Liassine, N.; Bes, M.; Greenland, T.; Reverdy, M.-E.; et al. Community-Acquired Methicillin-Resistant Staphylococcus aureus Carrying Panton-Valentine Leukocidin Genes: Worldwide Emergence. Emerg. Infect. Dis. 2003, 9, 978–984. [Google Scholar] [CrossRef]
- Ito, T.; Ma, X.X.; Takeuchi, F.; Okuma, K.; Yuzawa, H.; Hiramatsu, K. Novel Type V Staphylococcal Cassette Chromosome mec Driven by a Novel Cassette Chromosome Recombinase, ccrC. Antimicrob. Agents Chemother. 2004, 48, 2637–2651. [Google Scholar] [CrossRef] [PubMed]
- Wijaya, L.; Hsu, L.-Y.; Kurup, A. Community-associated methicillin-resistant Staphylococcus aureus: Overview and local situation. Ann. Acad. Med. Singap. 2006, 35, 479–486. [Google Scholar] [PubMed]
- WHO. Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 27 November 2022).
- Garvey, M. Bacteriophages and the One Health Approach to Combat Multidrug Resistance: Is This the Way? Antibiotics 2020, 9, 414. [Google Scholar] [CrossRef]
- Zalewska-Piątek, B.; Piątek, R. Bacteriophages as Potential Tools for Use in Antimicrobial Therapy and Vaccine Development. Pharmaceuticals 2021, 14, 331. [Google Scholar] [CrossRef]
- Broncano-Lavado, A.; Santamaría-Corral, G.; Esteban, J.; García-Quintanilla, M. Advances in Bacteriophage Therapy against Relevant MultiDrug-Resistant Pathogens. Antibiotics 2021, 10, 672. [Google Scholar] [CrossRef] [PubMed]
- Busarcevic, M.; Kojic, M.; Dalgalarrondo, M.; Chobert, J.-M.; Haertlé, T.; Topisirovic, L. Purification of bacteriocin LS1 produced by human oral isolate Lactobacillus salivarius BGHO1. Oral Microbiol. Immunol. 2008, 23, 254–258. [Google Scholar] [CrossRef]
- Chen, L.-J.; Tsai, H.-T.; Chen, W.-J.; Hsieh, C.-Y.; Wang, P.-C.; Chen, C.-S.; Wang, L.; Yang, C.-C. In vitro antagonistic growth effects of Lactobacillus fermentum and Lactobacillus salivarius and their fermentative broth on periodontal pathogens. Braz. J. Microbiol. 2012, 43, 1376–1384. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, R.; Martín, V.; Maldonado, A.; Jiménez, E.; Fernández, L.; Rodríguez, J.M. Treatment of Infectious Mastitis during Lactation: Antibiotics versus Oral Administration of Lactobacilli Isolated from Breast Milk. Clin. Infect. Dis. 2010, 50, 1551–1558. [Google Scholar] [CrossRef]
- Fernández, L.; Arroyo, R.; Espinosa, I.; Marín, M.; Jiménez, E.; Rodríguez, J. Probiotics for human lactational mastitis. Benef. Microbes 2014, 5, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, N.A.; Kermanshahi, R.K.; Yakhchali, B.; Sattari, T.N. Antagonistic activity of probiotic lactobacilli against Staphylococcus aureus isolated from bovine mastitis. African J. Microbiol. Res. 2010, 4, 2169–2173. [Google Scholar] [CrossRef]
- Abramov, V.M.; Kosarev, I.V.; Machulin, A.V.; Priputnevich, T.V.; Chikileva, I.O.; Deryusheva, E.I.; Abashina, T.N.; Donetskova, A.D.; Panin, A.N.; Melnikov, V.G.; et al. Limosilactobacillus fermentum Strain 3872: Antibacterial and Immunoregulatory Properties and Synergy with Prebiotics against Socially Significant Antibiotic-Resistant Infections of Animals and Humans. Antibiotics 2022, 11, 1437. [Google Scholar] [CrossRef] [PubMed]
- Bramley, A.J.; Patel, A.H.; O’Reilly, M.; Foster, R.; Foster, T.J. Roles of alpha-toxin and beta-toxin in virulence of Staphylococcus aureus for the mouse mammary gland. Infect. Immun. 1989, 57, 2489–2494. [Google Scholar] [CrossRef]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Potential probiotic-associated traits revealed from completed high quality genome sequence of Lactobacillus fermentum 3872. Stand. Genom. Sci. 2017, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Güllüce, M.; Karadayı, M.; Barış, Ö. Bacteriocins: Promising Natural Antimicrobials. In Microbial Pathogens and Strategies for Combating Them: Science, Technology and Education; Formatex Research Center: Badajoz, Spain, 2013; pp. 1016–1027. [Google Scholar]
- Deegan, L.H.; Cotter, P.D.; Hill, C.; Ross, P. Bacteriocins: Biological tools for bio-preservation and shelf-life extension. Int. Dairy J. 2006, 16, 1058–1071. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Bateman, A. MEROPS: The peptidase database. Nucleic Acids Res. 2010, 38, D227–D233. [Google Scholar] [CrossRef]
- Kaur, S.; Kaur, S. Bacteriocins as Potential Anticancer Agents. Front. Pharmacol. 2015, 6, 272. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Xiang, Y.; Morais, M.C.; Cohen, D.N.; Bowman, V.D.; Anderson, D.L.; Rossmann, M.G. Crystal and cryoEM structural studies of a cell wall degrading enzyme in the bacteriophage φ29 tail. Proc. Natl. Acad. Sci. USA 2008, 105, 9552–9557. [Google Scholar] [CrossRef]
- Dayhoff, G.W.; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein Sci. 2022, 31, e4496. [Google Scholar] [CrossRef]
- Horvath, A.; Miskei, M.; Ambrus, V.; Vendruscolo, M.; Fuxreiter, M. Sequence-based prediction of protein binding mode landscapes. PLOS Comput. Biol. 2020, 16, e1007864. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Antifeeva, I.A.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B.Y.; Kulkarni, P.; Uversky, V.N. Biological soft matter: Intrinsically disordered proteins in liquid–liquid phase separation and biomolecular condensates. Essays Biochem. 2022, 66, 831–847. [Google Scholar] [CrossRef]
- Antifeeva, I.A.; Fonin, A.V.; Fefilova, A.S.; Stepanenko, O.V.; Povarova, O.I.; Silonov, S.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Liquid–liquid phase separation as an organizing principle of intracellular space: Overview of the evolution of the cell compartmentalization concept. Cell. Mol. Life Sci. 2022, 79, 251. [Google Scholar] [CrossRef] [PubMed]
- Bliven, S.E.; Lafita, A.; Rose, P.W.; Capitani, G.; Prlić, A.; Bourne, P.E. Analyzing the symmetrical arrangement of structural repeats in proteins with CE-Symm. PLOS Comput. Biol. 2019, 15, e1006842. [Google Scholar] [CrossRef] [PubMed]
- Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.; Darling, A.L.; Zaslavsky, B.; Uversky, V.N. Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation. Trends Biochem. Sci. 2019, 44, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2018, 18, e1700193. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017, 44, 18–30. [Google Scholar] [CrossRef]
- Uversky, V.N. Protein intrinsic disorder-based liquid–liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv. Colloid Interface Sci. 2017, 239, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015, 589, 15–22. [Google Scholar] [CrossRef]
- Chu, X.; Sun, T.; Li, Q.; Xu, Y.; Zhang, Z.; Lai, L.; Pei, J. Prediction of liquid–liquid phase separating proteins using machine learning. BMC Bioinform. 2022, 23, 72. [Google Scholar] [CrossRef] [PubMed]
- Horvath, A.; Vendruscolo, M.; Fuxreiter, M. Sequence-based Prediction of the Cellular Toxicity Associated with Amyloid Aggregation within Protein Condensates. Biochemistry 2022, 61, 2461–2469. [Google Scholar] [CrossRef]
- Vendruscolo, M.; Fuxreiter, M. Sequence Determinants of the Aggregation of Proteins Within Condensates Generated by Liquid-liquid Phase Separation. J. Mol. Biol. 2022, 434, 167201. [Google Scholar] [CrossRef] [PubMed]
- Höltje, J.-V. Growth of the Stress-Bearing and Shape-Maintaining Murein Sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998, 62, 181–203. [Google Scholar] [CrossRef]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Lactobacillus fermentum 3872 as a potential tool for combatting Campylobacter jejuni infections. Virulence 2017, 8, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Ferraboschi, P.; Ciceri, S.; Grisenti, P. Applications of Lysozyme, an Innate Immune Defense Factor, as an Alternative Antibiotic. Antibiotics 2021, 10, 1534. [Google Scholar] [CrossRef]
- Callewaert, L.; Michiels, C.W. Lysozymes in the animal kingdom. J. Biosci. 2010, 35, 127–160. [Google Scholar] [CrossRef]
- Fischetti, V.A. Development of Phage Lysins as Novel Therapeutics: A Historical Perspective. Viruses 2018, 10, 310. [Google Scholar] [CrossRef]
- Sutton, J.A.F.; Carnell, O.T.; Lafage, L.; Gray, J.; Biboy, J.; Gibson, J.F.; Pollitt, E.J.G.; Tazoll, S.C.; Turnbull, W.; Hajdamowicz, N.H.; et al. Staphylococcus aureus cell wall structure and dynamics during host-pathogen interaction. PLOS Pathog. 2021, 17, e1009468. [Google Scholar] [CrossRef]
- Delgado, S.; García, P.; Fernández, L.; Jiménez, E.; Rodríguez-Baños, M.; del Campo, R.; Rodríguez, J.M. Characterization ofStaphylococcus aureus strains involved in human and bovine mastitis. FEMS Immunol. Med Microbiol. 2011, 62, 225–235. [Google Scholar] [CrossRef]
- Reddy, P.; Qi, C.; Zembower, T.; Noskin, G.A.; Bolon, M. Postpartum Mastitis and Community-acquired Methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2007, 13, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Sears, P.M.; McCarthy, K.K. Management and treatment of staphylococcal mastitis. Vet. Clin. North Am. Food Anim. Pract. 2003, 19, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yao, Z. Maternal-Infant Correlation of Multidrug-Resistant Staphylococcus aureus Carriage: A Prospective Cohort Study. Front. Pediatr. 2018, 6, 384. [Google Scholar] [CrossRef] [PubMed]
- Garbacz, K.; Kwapisz, E.; Wierzbowska, M. Denture stomatitis associated with small-colony variants of Staphylococcus aureus: A case report. BMC Oral Health 2019, 19, 219. [Google Scholar] [CrossRef]
- Gorwitz, R.J. A Review of Community-Associated Methicillin-Resistant Staphylococcus aureus Skin and Soft Tissue Infections. Pediatr. Infect. Dis. J. 2008, 27, 1–7. [Google Scholar] [CrossRef]
- Patel, J.B.; Kilbride, H.; Paulson, L. Neonatal Presentation of an Air-Filled Neck Mass that Enlarges with Valsalva: A Case Report. Am. J. Perinatol. Rep. 2015, 5, e207–e211. [Google Scholar] [CrossRef]
- Voss, A.; Loeffen, F.; Bakker, J.; Klaassen, C.; Wulf, M. Methicillin-resistant Staphylococcus aureus in Pig Farming. Emerg. Infect. Dis. 2005, 11, 1965–1966. [Google Scholar] [CrossRef]
- Lewis, H.C.; Mølbak, K.; Reese, C.; Aarestrup, F.; Selchau, M.; Sørum, M.; Skov, R.L. Pigs as Source of Methicillin-Resistant Staphylococcus aureus CC398 Infections in Humans, Denmark. Emerg. Infect. Dis. 2008, 14, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Denis, O.; Suetens, C.; Hallin, M.; Catry, B.; Ramboer, I.; Dispas, M.; Willems, G.; Gordts, B.; Butaye, P.; Struelens, M.J. Methicillin-Resistant Staphylococcus aureus ST398 in Swine Farm Personnel, Belgium. Emerg. Infect. Dis. 2009, 15, 1098–1101. [Google Scholar] [CrossRef]
- Huber, H.; Koller, S.; Giezendanner, N.; Stephan, R.; Zweifel, C. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill. 2010, 15, 19542. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20430001 (accessed on 23 February 2023). [CrossRef]
- Mulders, M.N.; Haenen, A.P.J.; Geenen, P.L.; Vesseur, P.C.; Poldervaart, E.S.; Bosch, T.; Huijsdens, X.W.; Hengeveld, P.D.; Dam-Deisz, W.D.C.; Graat, E.A.M.; et al. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol. Infect. 2010, 138, 743–755. [Google Scholar] [CrossRef]
- Bisdorff, B.; Scholhölter, J.L.; Claußen, K.; Pulz, M.; Nowak, D.; Radon, K. MRSA-ST398 in livestock farmers and neighbouring residents in a rural area in Germany. Epidemiol. Infect. 2012, 140, 1800–1808. [Google Scholar] [CrossRef]
- Broens, E.M.; Graat, E.A.; van de Giessen, A.W.; Broekhuizen-Stins, M.J.; de Jong, M.C. Quantification of transmission of livestock-associated methicillin resistant Staphylococcus aureus in pigs. Vet. Microbiol. 2012, 155, 381–388. [Google Scholar] [CrossRef]
- Broens, E.M.; Espinosa-Gongora, C.; Graat, E.A.; Vendrig, N.; Van Der Wolf, P.J.; Guardabassi, L.; Butaye, P.; Nielsen, J.P.; De Jong, M.C.; Van De Giessen, A.W. Longitudinal study on transmission of MRSA CC398 within pig herds. BMC Vet. Res. 2012, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- van Duijkeren, E.; Ikawaty, R.; Broekhuizen-Stins, M.; Jansen, M.; Spalburg, E.; de Neeling, A.; Allaart, J.; van Nes, A.; Wagenaar, J.; Fluit, A. Transmission of methicillin-resistant Staphylococcus aureus strains between different kinds of pig farms. Vet. Microbiol. 2008, 126, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Gongora, C.; Larsen, J.; Moodley, A.; Nielsen, J.P.; Skov, R.L.; Andreasen, M.; Guardabassi, L. Farm-specific lineages of methicillin-resistant Staphylococcus aureus clonal complex 398 in Danish pig farms. Epidemiol. Infect. 2012, 140, 1794–1799. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Zwambag, A.; Rosendal, T.; Reid-Smith, R.; Friendship, R. Longitudinal Investigation of Methicillin-Resistant Staphylococcus aureus in Piglets. Zoonoses Public Health 2011, 58, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Verhegghe, M.; Pletinckx, L.J.; Crombé, F.; Van Weyenberg, S.; Haesebrouck, F.; Butaye, P.; Heyndrickx, M.; Rasschaert, G. Cohort study for the presence of livestock-associated MRSA in piglets: Effect of sow status at farrowing and determination of the piglet colonization age. Vet. Microbiol. 2013, 162, 679–686. [Google Scholar] [CrossRef]
- Moodley, A.; Latronico, F.; Guardabassi, L. Experimental colonization of pigs with methicillin-resistant Staphylococcus aureus (MRSA): Insights into the colonization and transmission of livestock-associated MRSA. Epidemiol. Infect. 2011, 139, 1594–1600. [Google Scholar] [CrossRef]
- One Health. Available online: https://www.who.int/health-topics/one-health#tab=tab_1 (accessed on 27 December 2022).
- Kos, B.; Šušković, J.; Vuković, S.; Šimpraga, M.; Frece, J.; Matošić, S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003, 94, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Surono, I.; Meriluoto, J.; Salminen, S. Indigenous Dadih Lactic Acid Bacteria: Cell-Surface Properties and Interactions with Pathogens. J. Food Sci. 2007, 72, M89–M93. [Google Scholar] [CrossRef]
- Hojjati, M.; Behabahani, B.A.; Falah, F. Aggregation, adherence, anti-adhesion and antagonistic activity properties relating to surface charge of probiotic Lactobacillus brevis gp104 against Staphylococcus aureus. Microb. Pathog. 2020, 147, 104420. [Google Scholar] [CrossRef]
- Fonseca, H.C.; de Sousa Melo, D.; Ramos, C.L.; Dias, D.R.; Schwan, R.F. Probiotic Properties of Lactobacilli and Their Ability to Inhibit the Adhesion of Enteropathogenic Bacteria to Caco-2 and HT-29 Cells. Probiotics Antimicrob. Proteins 2021, 13, 102–112. [Google Scholar] [CrossRef]
- Boris, S.; Suárez, J.E.; Vázquez, F.; Barbés, C. Adherence of Human Vaginal Lactobacilli to Vaginal Epithelial Cells and Interaction with Uropathogens. Infect. Immun. 1998, 66, 1985–1989. [Google Scholar] [CrossRef] [PubMed]
- Osset, J.; Bartolomé, R.M.; García, E.; Andreu, A. Assessment of the Capacity of Lactobacillus to Inhibit the Growth of Uropathogens and Block Their Adhesion to Vaginal Epithelial Cells. J. Infect. Dis. 2001, 183, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Mastromarino, P.; Brigidi, P.; Macchia, S.; Maggi, L.; Pirovano, F.; Trinchieri, V.; Conte, U.; Matteuzzi, D. Characterization and selection of vaginal Lactobacillus strains for the preparation of vaginal tablets. J. Appl. Microbiol. 2002, 93, 884–893. [Google Scholar] [CrossRef]
- Younes, J.A.; Van Der Mei, H.C.; van den Heuvel, E.; Busscher, H.J.; Reid, G. Adhesion Forces and Coaggregation between Vaginal Staphylococci and Lactobacilli. PLoS ONE 2012, 7, e36917. [Google Scholar] [CrossRef]
- Todorov, S.D.; Furtado, D.N.; Saad, S.M.I.; de Melo Franco, B.D.G. Bacteriocin production and resistance to drugs are advantageous features for Lactobacillus acidophilus La-14, a potential probiotic strain. New Microbiol. 2011, 34, 357–370. [Google Scholar]
- Verdenelli, M.; Coman, M.; Cecchini, C.; Silvi, S.; Orpianesi, C.; Cresci, A. Evaluation of antipathogenic activity and adherence properties of human Lactobacillus strains for vaginal formulations. J. Appl. Microbiol. 2014, 116, 1297–1307. [Google Scholar] [CrossRef]
- Rickard, A.H.; Gilbert, P.; High, N.J.; Kolenbrander, P.E.; Handley, P.S. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003, 11, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Reid, G.; McGroarty, J.A.; Gil Domingue, P.A.; Chow, A.W.; Bruce, A.W.; Eisen, A.; Costerton, J.W. Coaggregation of urogenital bacteria in vitro and in vivo. Curr. Microbiol. 1990, 20, 47–52. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, X.; Zhang, X.; Wu, H.; Zou, Y.; Li, P.; Sun, C.; Xu, W.; Liu, F.; Wang, D. Class III bacteriocin Helveticin-M causes sublethal damage on target cells through impairment of cell wall and membrane. J. Ind. Microbiol. Biotechnol. 2018, 45, 213–227. [Google Scholar] [CrossRef]
- Martiín, R.; Suárez, J.E. Biosynthesis and Degradation of H2O2 by Vaginal Lactobacilli. Appl. Environ. Microbiol. 2010, 76, 400–405. [Google Scholar] [CrossRef]
- Tachedjian, G.; Aldunate, M.; Bradshaw, C.S.; Cone, R.A. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res. Microbiol. 2017, 168, 782–792. [Google Scholar] [CrossRef]
- Wong, C.-B.; Khoo, B.-Y.; Sasidharan, S.; Piyawattanametha, W.; Kim, S.; Khemthongcharoen, N.; Ang, M.-Y.; Chuah, L.-O.; Liong, M.-T. Inhibition of Staphylococcus aureus by crude and fractionated extract from lactic acid bacteria. Benef. Microbes 2015, 6, 129–139. [Google Scholar] [CrossRef]
- Otero, M.C.; Nader-Macias, M.E.F. Inhibition of Staphylococcusaureus by H2O2-producing Lactobacillus gasseri isolated from the vaginal tract of cattle. Anim. Reprod. Sci. 2006, 96, 35–46. [Google Scholar] [CrossRef]
- Karlyshev, A.V.; Raju, K.; Abramov, V.M. Draft Genome Sequence of Lactobacillus fermentum Strain 3872. Genome Announc. 2013, 1, 01006-13. [Google Scholar] [CrossRef] [PubMed]
- Lehri, B.; Seddon, A.M.; Karlyshev, A.V. Lactobacillus fermentum 3872 genome sequencing reveals plasmid and chromosomal genes potentially involved in a probiotic activity. FEMS Microbiol. Lett. 2015, 362, fnv068. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. In 2-D Proteome Analysis Protocols. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1999; Volume 112, pp. 531–552. [Google Scholar] [CrossRef]
- Hardenberg, M.; Horvath, A.; Ambrus, V.; Fuxreiter, M.; Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl. Acad. Sci. USA 2020, 117, 33254–33262. [Google Scholar] [CrossRef] [PubMed]
- Hatos, A.; Tosatto, S.C.E.; Vendruscolo, M.; Fuxreiter, M. FuzDrop on AlphaFold: Visualizing the sequence-dependent propensity of liquid–liquid phase separation and aggregation of proteins. Nucleic Acids Res. 2022, 50, W337–W344. [Google Scholar] [CrossRef] [PubMed]
- Dubey, U.; Mistry, V. Growth Characteristics of Bifidobacteria in Infant Formulas. J. Dairy Sci. 1996, 79, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).