Surveillance on ESBL-Escherichia coli and Indicator ARG in Wastewater and Reclaimed Water of Four Regions of Spain: Impact of Different Disinfection Treatments
Abstract
1. Introduction
2. Materials and Methods
2.1. Urban Wastewater Treatment Plants (WWTPs)
2.2. Wastewater Sample Collection
2.3. Enumeration and Isolation of Extended-Spectrum β-Lactamase-Producing E. coli (ESBL E. coli)
2.4. Wastewater DNA Extraction
2.5. Antimicrobial Resistance Genes (ARGs) Detection
2.6. Antibiotic Susceptibility Testing (AST)
2.7. Statistical Analysis
3. Results and Discussion
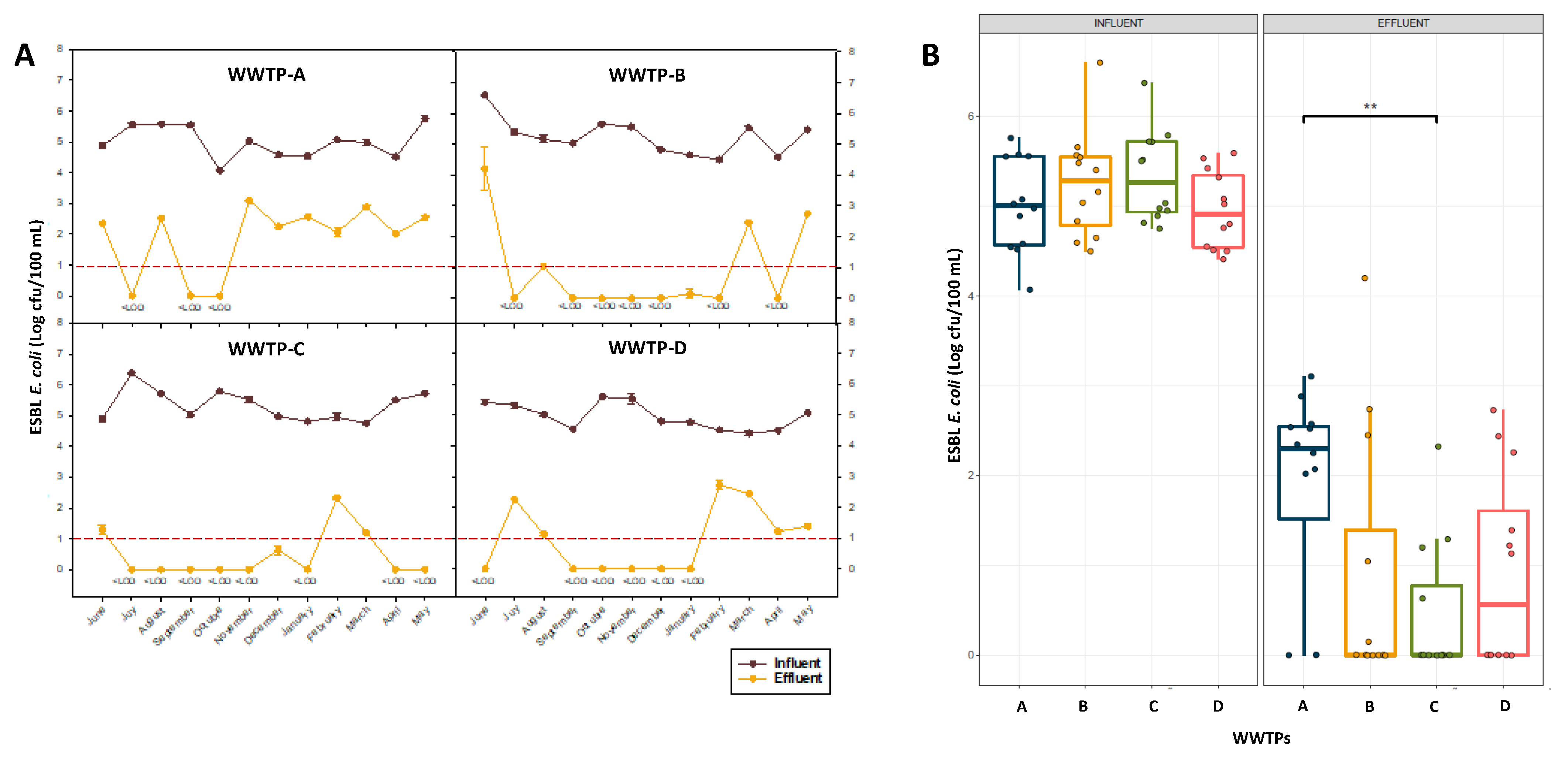
3.1. Occurrence of ESBL Producing E. coli in Wastewater Samples
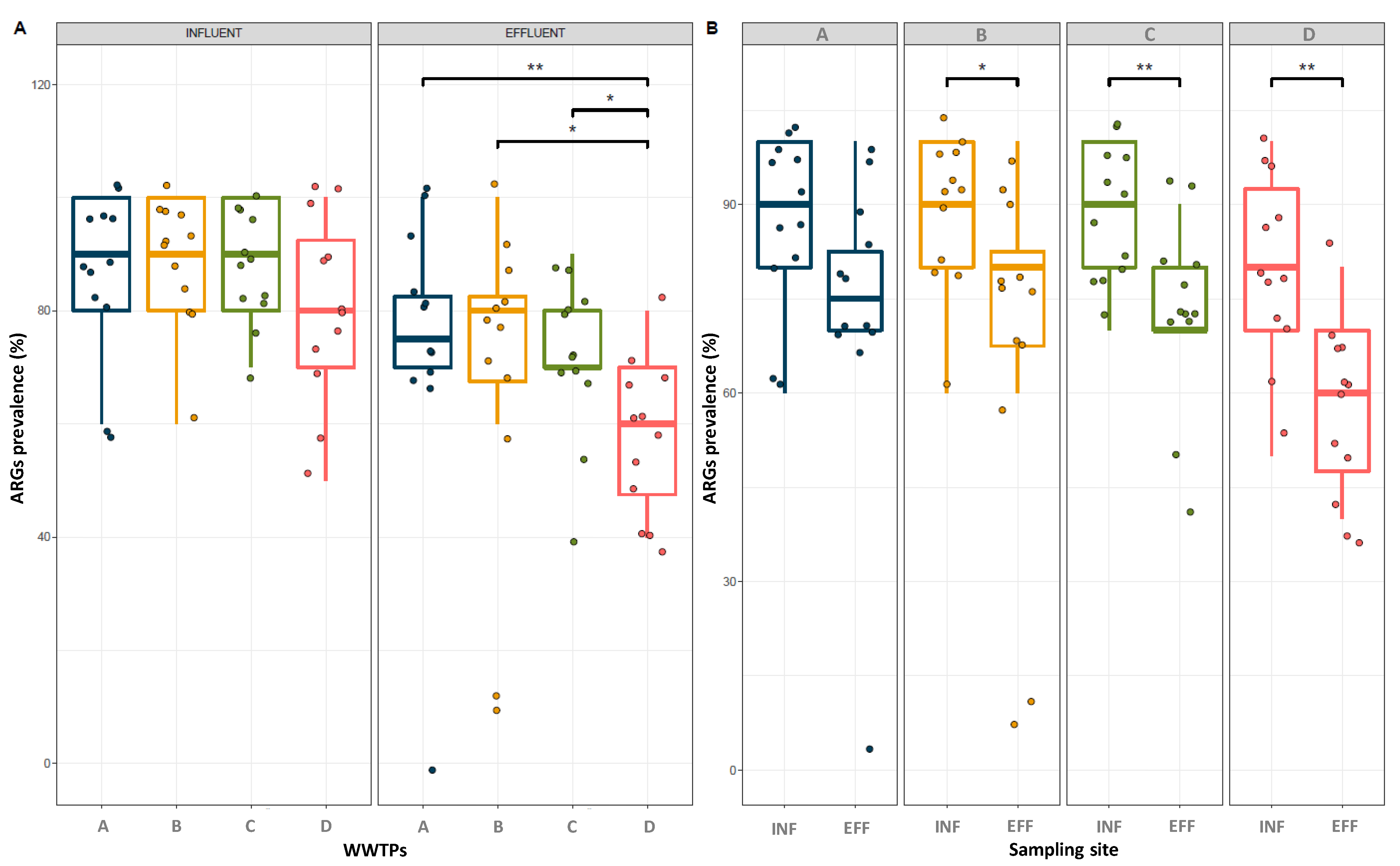
3.2. Prevalence of Indicator ARGs in the Wastewater Collected from WWTPs
3.3. Antibiotic Resistance Profile of ESBL E. coli Isolates
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Report of the 6th Meeting of the WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance with AGISAR 5-Year Strategic Framework to Support Implementation of the Global Action Plan on Antimicrobial Resistance (2015–2019). 2015. Available online: https://apps.who.int/iris/bitstream/handle/10665/190954/9789241509534_eng.pdf (accessed on 26 July 2022).
- Michael, I.; Rizzo, L.; McArdell, C.S.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef] [PubMed]
- Pärnänen, K.M.M.; Narciso-Da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Wong, D.M.L.F.; Dalsgaard, A. The effects of tertiary wastewater treatment on the prevalence of antimicrobial resistant bacteria. Water Res. 2002, 36, 1955–1964. [Google Scholar] [CrossRef]
- Jury, K.L.; Khan, S.J.; Vancov, T.; Stuetz, R.M.; Ashbolt, N.J. Are sewage treatment plants promoting antibiotic resistance? Crit. Rev. Environ. Sci. Technol. 2011, 41, 243–270. [Google Scholar] [CrossRef]
- Moura, A.; Henriques, I.; Smalla, K.; Correia, A. Wastewater bacterial communities bring together broad-host range plasmids, integrons and a wide diversity of uncharacterized gene cassettes. Res. Microbiol. 2010, 161, 58–66. [Google Scholar] [CrossRef]
- Rodríguez, E.A.; Ramirez, D.; Balcázar, J.L.; Jiménez, J.N. Metagenomic analysis of urban wastewater resistome and mobilome: A support for antimicrobial resistance surveillance in an endemic country. Environ. Pollut. 2021, 276, 116736. [Google Scholar] [CrossRef] [PubMed]
- Zaatout, N.; Bouras, S.; Slimani, N. Prevalence of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in wastewater: A systematic review and meta-analysis. J. Water Health 2021, 19, 705–723. [Google Scholar] [CrossRef]
- Truchado, P.; Gil, M.I.; López, C.; Garre, A.; López-Aragón, R.F.; Böhme, K.; Allende, A. New standards at European Union level on water reuse for agricultural irrigation: Are the Spanish wastewater treatment plants ready to produce and distribute reclaimed water within the minimum quality requirements? Int. J. Food Microbiol. 2021, 16, 109352. [Google Scholar] [CrossRef]
- EU (European Union). Regulation (EU) 2020/741 of the European Parliament and of the Council of 25 May 2020 on minimum requirements for water reuse. Off. J. Eur. Union L 2020, 177, 32–55. [Google Scholar]
- Cai, Y.; Sun, T.; Li, G.; An, T. Traditional and Emerging Water Disinfection Technologies Challenging the Control of Antibiotic-Resistant Bacteria and Antibiotic Resistance Genes. ACS ES&T Eng. 2021, 1, 1046–1064. [Google Scholar] [CrossRef]
- Pei, M.; Zhang, B.; He, Y.; Su, J.; Gin, K.; Lev, O.; Shen, G.; Hu, S. State of the art of tertiary treatment technologies for controlling antibiotic resistance in wastewater treatment plants. Environ. Int. 2019, 131, 105026. [Google Scholar] [CrossRef]
- Sharma, V.K.; Johnson, N.; Cizmas, L.; McDonald, T.J.; Kim, H. A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 2016, 150, 702–714. [Google Scholar] [CrossRef]
- Liu, Y.; Fan, Q.; Wang, J. Zn-Fe-CNTs catalytic in situ generation of H2O2 for Fenton-like degradation of sulfamethoxazole. J. Hazard. Mater. 2018, 342, 166–176. [Google Scholar] [CrossRef]
- Pariente, M.; Segura, Y.; Álvarez-Torrellas, S.; Casas, J.; de Pedro, Z.; Diaz, E.; García, J.; López-Muñoz, M.; Marugán, J.; Mohedano, A.; et al. Critical review of technologies for the on-site treatment of hospital wastewater: From conventional to combined advanced processes. J. Environ. Manag. 2022, 320, 115769. [Google Scholar] [CrossRef]
- Tang, J.T.; Wang, J.L. MOF-derived three-dimensional flower-like FeCu@C composite as an efficient Fenton-like catalyst for sulfamethazine degradation. Chem. Eng. J. 2019, 375, 122007. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H. Catalytic ozonation for water and wastewater treatment: Recent advances and perspective. Sci. Total Environ. 2020, 704, 135249. [Google Scholar] [CrossRef]
- Wang, J.; Chu, L.; Wojnárovits, L.; Takács, E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview. Sci. Total Environ. 2020, 744, 140997. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, S. Removal of various pollutants from water and wastewater by modified chitosan adsorbents. Crit. Rev. Environ. Sci. Technol. 2017, 47, 2331–2386. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, S. Covalent organic frameworks (COFs) for environmental applications. Coord. Chem. Rev. 2019, 400, 213046. [Google Scholar] [CrossRef]
- Zhuan, R.; Wang, J. Degradation of sulfamethoxazole by ionizing radiation: Kinetics and implications of additives. Sci. Total Environ. 2019, 668, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, S.T.; Liu, Y.; Wang, J.L. Covalent organic frameworks as efficient adsorbent for sulfamethazine removal from aqueous solution. J. Hazard. Mater. 2020, 383, 121126. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Książek, S.; Olańczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 2010, 44, 5089–5097. [Google Scholar] [CrossRef]
- Amador, P.P.; Fernandes, R.M.; Prudencio, M.C.; Barreto, M.P.; Duarte, I.M. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of class A and class C β-lactamases. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2015, 50, 26–39. [Google Scholar]
- Alexander, J.; Bollmann, A.; Seitz, W.; Schwartz, T. Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci. Total Environ. 2015, 512, 316–325. [Google Scholar] [CrossRef]
- Biswal, B.K.; Mazza, A.; Masson, L.; Gehr, R.; Frigon, D. Impact of wastewater treatment processes on antimicrobial resistance genes and their co-occurrence with virulence genes in Escherichia coli. Water Res. 2014, 50, 245–253. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zou, S.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef]
- Hultman, J.; Tamminen, M.; Pärnänen, K.; Cairns, J.; Karkman, A.; Virta, M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 2018, 94, fiy038. [Google Scholar] [CrossRef]
- Laht, M.; Karkman, A.; Voolaid, V.; Ritz, C.; Tenson, T.; Virta, M.; Kisand, V. Abundances of Tetracycline, Sulphonamide and Beta-Lactam Antibiotic Resistance Genes in Conventional Wastewater Treatment Plants (WWTPs) with Different Waste Load. PLoS ONE 2014, 9, e103705. [Google Scholar] [CrossRef]
- Nimonkar, Y.S.; Yadav, B.; Talreja, P.; Sharma, A.; Patil, S.; Saware, S.S.; Ranade, D.R.; Prakash, O. Assessment of the Role of Wastewater Treatment Plant in Spread of Antibiotic Resistance and Bacterial Pathogens. Indian J. Microbiol. 2019, 59, 261–265. [Google Scholar] [CrossRef]
- Ferreira da Silva, M.; Vaz-Moreira, I.; Gonzalez-Pajuelo, M.; Nunes, O.; Manaia, C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2007, 60, 166–176. [Google Scholar] [CrossRef]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Franz, E.; Veenman, C.; van Hoek, A.H.A.M.; Husman, A.D.R.; Blaak, H. Pathogenic Escherichia coli producing Extended-Spectrum β-Lactamases isolated from surface water and wastewater. Sci. Rep. 2015, 5, 14372. [Google Scholar] [CrossRef]
- Usui, M.; Ozeki, K.; Komatsu, T.; Fukuda, A.; Tamura, Y. Prevalence of Extended-Spectrum β-Lactamase–Producing Bacteria on Fresh Vegetables in Japan. J. Food Prot. 2019, 82, 1663–1666. [Google Scholar] [CrossRef]
- EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards); Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; et al. Scientific Opinion on the role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 2021, 19, e06651. [Google Scholar]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Ng, K.H.; Samuel, L.; Kathleen, M.M.; Leong, S.S.; Felecia, C. Distribution and prevalence of chloramphenicol-resistance gene in Escherichia coli isolated from aquaculture and other environment. Int. Food Res. J. 2014, 21, 1321–1325. [Google Scholar]
- Liu, X.; Wang, H.; Zhao, H. Propagation of antibiotic resistance genes in an industrial recirculating aquaculture system located at northern China. Environ. Pollut. 2020, 261, 114155. [Google Scholar] [CrossRef] [PubMed]
- Cummings, D.E.; Archer, K.F.; Arriola, D.J.; Baker, P.A.; Faucett, K.G.; Laroya, J.B.; Pfeil, K.L.; Ryan, C.R.; Ryan, K.R.U.; Zuill, D.E. Broad Dissemination of Plasmid-Mediated Quinolone Resistance Genes in Sediments of Two Urban Coastal Wetlands. Environ. Sci. Technol. 2011, 45, 447–454. [Google Scholar] [CrossRef]
- Gonçalves, D.M. Escherichia coli e Klebsiella pneumoniae, das ESBL´s às Carbapenemases, Colonização Fecal e Infeção. Influência da População Idosa Numa Região do Norte de Portugal. Ph.D. Thesis, University of Porto, Academic repository of the University of Porto, Porto, Portugal, 2013. [Google Scholar]
- Aarestrup, F.M.; Lertworapreecha, M.; Evans, M.C.; Bangtrakulnonth, A.; Chalermchaikit, T.; Hendriksen, R.S.; Wegener, H.C. Antimicrobial susceptibility and occurence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J. Antimicrob. Chemother. 2003, 52, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Sung, K.; Nawaz, M.S. Detection of aacA-aphD, qacEδ1, marA, floR, and tetA genes from multidrug-resistant bacteria: Comparative analysis of real-time multiplex PCR assays using EvaGreen® and SYBR® Green I dyes. Mol. Cell. Probes 2011, 25, 78–86. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Zhao, H. Prevalence of antibiotic resistance genes in wastewater collected from ornamental fish market in northern China. Environ. Pollut. 2021, 271, 116316. [Google Scholar] [CrossRef]
- Haberecht, H.B.; Nealon, N.J.; Gilliland, J.R.; Holder, A.V.; Runyan, C.; Oppel, R.C.; Ibrahim, H.M.; Mueller, L.; Schrupp, F.; Vilchez, S.; et al. Antimicrobial-resistant Escherichia coli from environmental waters in northern Colorado. J. Environ. Public Health 2019, 2019, 3862949. [Google Scholar] [CrossRef] [PubMed]
- Schmiege, D.; Zacharias, N.; Sib, E.; Falkenberg, T.; Moebus, S.; Evers, M.; Kistemann, T. Prevalence of multidrug-resistant and extended-spectrum beta-lactamase-producing Escherichia coli in urban community wastewater. Sci. Total Environ. 2021, 785, 147269. [Google Scholar] [CrossRef]
- Raven, K.E.; Ludden, C.; Gouliouris, T.; Blane, B.; Naydenova, P.; Brown, N.M.; Parkhill, J.; Peacock, S.J. Genomic surveillance of Escherichia coli in municipal wastewater treatment plants as an indicator of clinically relevant pathogens and their resistance genes. Microb. Genom. 2019, 5, e000267. [Google Scholar] [CrossRef]
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater Treatment Plants Release Large Amounts of Extended-Spectrum β-Lactamase–Producing Escherichia coli Into the Environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef]
- Nzima, B.; Adegoke, A.; Ofon, U.; Al-Dahmoshi, H.; Saki, M.; Ndubuisi-Nnaji, U.; Inyang, C. Resistotyping and extended-spectrum beta-lactamase genes among Escherichia coli from wastewater treatment plants and recipient surface water for reuse in South Africa. New Microbes New Infect. 2020, 38, 100803. [Google Scholar] [CrossRef]
- Solaiman, S.; Handy, E.; Brinks, T.; Goon, K.; Bollinger, C.; Sapkota, A.R.; Sharma, M.; Micallef, S.A. Extended spectrum β-lactamase activity and cephalosporin resistance in Escherichia coli from U.S. Mid-Atlantic surface and reclaimed water. Appl. Environ. Microbiol. 2022, 88, e0083722. [Google Scholar] [CrossRef]
- Caucci, S.; Karkman, A.; Cacace, D.; Rybicki, M.; Timpel, P.; Voolaid, V.; Gurke, R.; Virta, M.; Berendonk, T.U. Seasonality of antibiotic prescriptions for outpatients and resistance genes in sewers and wastewater treatment plant outflow. FEMS Microbiol. Ecol. 2016, 92, fiw060. [Google Scholar] [CrossRef]
- Lépesová, K.; Olejníková, P.; Mackuľak, T.; Tichý, J.; Birošová, L. Annual changes in the occurrence of antibiotic-resistant coliform bacteria and enterococci in municipal wastewater. Environ. Sci. Pollut. Res. 2019, 26, 18470–18483. [Google Scholar] [CrossRef] [PubMed]
- Ben, W.; Wang, J.; Cao, R.; Yang, M.; Zhang, Y.; Qiang, Z. Distribution of antibiotic resistance in the effluents of ten municipal wastewater treatment plants in China and the effect of treatment processes. Chemosphere 2017, 172, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Berglund, B.; Fick, J.; Lindgren, P.-E. Urban wastewater effluent increases antibiotic resistance gene concentrations in a receiving northern European river. Environ. Toxicol. Chem. 2015, 34, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Wong, K.; Xagoraraki, I. Release of antibiotic resistant bacteria and genes in the effluent and biosolids of five wastewater utilities in Michigan. Water Res. 2011, 45, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Ferro, G.; Guarino, F.; Castiglione, S.; Rizzo, L. Antibiotic resistance spread potential in urban wastewater effluents disinfected by UV/H2O2 process. Sci. Total Environ. 2016, 560, 29–35. [Google Scholar] [CrossRef]
- McConnell, M.M.; Hansen, L.T.; Jamieson, R.C.; Neudorf, K.D.; Yost, C.K.; Tong, A. Removal of antibiotic resistance genes in two tertiary level municipal wastewater treatment plants. Sci. Total Environ. 2018, 643, 292–300. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, M. Effects of Advanced Treatment Systems on the Removal of Antibiotic Resistance Genes in Wastewater Treatment Plants from Hangzhou, China. Environ. Sci. Technol. 2013, 47, 8157–8163. [Google Scholar] [CrossRef] [PubMed]
- Amos, G.C.A.; Hawkey, P.M.; Gaze, W.H.; Wellington, E.M. Wastewater effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 2014, 69, 1785–1791. [Google Scholar] [CrossRef]
- Hiller, C.; Hübner, U.; Fajnorova, S.; Schwartz, T.; Drewes, J. Antibiotic microbial resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review. Sci. Total Environ. 2019, 685, 596–608. [Google Scholar] [CrossRef]
- Liu, L.; Xin, Y.; Huang, X.; Liu, C. Response of antibiotic resistance genes in constructed wetlands during treatment of livestock wastewater with different exogenous inducers: Antibiotic and antibiotic-resistant bacteria. Bioresour. Technol. 2020, 314, 123779. [Google Scholar] [CrossRef]
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of Antibiotic Resistance Genes and Bacterial Community Composition in a River Influenced by a Wastewater Treatment Plant. PLoS ONE 2013, 8, e78906. [Google Scholar] [CrossRef]
- Sousa, J.M.; Macedo, G.; Pedrosa, M.; Becerra-Castro, C.; Castro-Silva, S.; Pereira, M.F.R.; Silva, A.M.; Nunes, O.C.; Manaia, C.M. Ozonation and UV254nm radiation for the removal of microorganisms and antibiotic resistance genes from urban wastewater. J. Hazard. Mater. 2017, 323, 434–441. [Google Scholar] [CrossRef]
- Lee, J.; Jeon, J.H.; Shin, J.; Jang, H.M.; Kim, S.; Song, M.S.; Kim, Y.M. Quantitative and qualitative changes in antibiotic resistance genes after passing through treatment processes in municipal wastewater treatment plants. Sci. Total Environ. 2017, 605, 906–914. [Google Scholar] [CrossRef]
- Umar, M. From Conventional Disinfection to Antibiotic Resistance Control—Status of the Use of Chlorine and UV Irradiation during Wastewater Treatment. Int. J. Environ. Res. Public Health 2022, 19, 1636. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Hammarén, R.; Pal, C.; Östman, M.; Björlenius, B.; Flach, C.-F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712. [Google Scholar] [CrossRef]
- Ferreira da Silva, M.; Tiago, I.; Veríssimo, A.; Boaventura, R.A.R.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance of enterococci and related bacteria in an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2006, 55, 322–329. [Google Scholar] [CrossRef]



| Sampling Times | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Urban WWTPs | Parameters | 06/2020 | 07/2020 | 08/2020 | 09/2020 | 10/2020 | 11/2020 | 12/2020 | 01/2021 | 02/2021 | 03/2021 | 04/2021 | 05/2021 |
| A | Treatment | UV | UV | UV | Cl + UV | Cl + UV | UV | UV | UV | UV | PAA + UV | PAA + UV | PAA + UV |
| Effluent Flow (m3/day) | _____ | _____ | 2916 | 2600 | 2430 | 2029 | 2065 | 2126 | 1897 | 2900 | 2441 | 2441 | |
| B | Treatment | Cl + UV | Cl + UV | Cl + UV | Cl + UV | PAA + UV | PAA + UV | PAA + UV | PAA + UV | PAA + UV | PAA + UV | PAA + UV | PAA + UV |
| Effluent Flow (m3/day) | 4479 | 4180 | 4050 | 4065 | 3921 | 4150 | 3894 | _____ | _____ | _____ | _____ | _____ | |
| C | Treatment | UV | Cl | UV | Cl + UV | UV | UV | PAA + UV | PAA + UV | PAA + UV | UV | UV | UV |
| Effluent Flow (m3/day) | 1700 | 1740 | 1533 | 1641 | 1673 | 1520 | 1529 | 1616 | 1714 | 2560 | 4284 | 2769 | |
| D | Treatment | Cl | UV | UV | Cl + UV | UV | UV | UV | UV | UV | UV | UV | UV |
| Effluent Flow (m3/day) | 1115 | 3366 | _____ | 2272 | 1015 | 1895 | 2156 | 1914 | _____ | 2504 | 2838 | _____ | |
| Target Gene | Primer Sequence (5′ → 3′) | Concentration (µM) | Cycling Parameters | Amplicon Size (bp) | Reference | |
|---|---|---|---|---|---|---|
| blaCTX-M-G1 | FW | TTAGGAARTGTGCCGCTGYA | 0.4 | 94 °C for 10 min; 35 cycles (94 °C for 40 s, 60 °C for 40 s, 72 °C for 1 min); 72 °C for 10 min | 688 | [39] |
| RV | CGATATCGTTGGTGGTRCCAT | |||||
| blaTEM | FW | CATTTCCGTGTCGCCCTTATTC | 0.4 | 94 °C for 10 min; 30 cycles (94 °C for 40 s, 60 °C for 40 s, 72 °C for 1 min); 72 °C for 10 min | 800 | [39] |
| RV | CGTTCATCCATAGTTGCCTGAC | |||||
| catA1 | FW | GGTGATATGGGATAGTGTT | 0.4 | 95 °C for 7 min; 34 cycles (95 °C for 40 s, 55 °C for 1 min, 72 °C for 40 s); 72 °C for 5 min | 349 | [40] |
| RV | CCATCACATACTGCATGATG | |||||
| cmlA | FW | GCCAGCAGTGCCGTTTAT | 1 | 95 °C for 5 min; 35 cycles (95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s); 72 °C for 7 min | 158 | [41] |
| RV | GGCCACCTCCCAGTAGAA | |||||
| qnrA | FW | GATAAAGTTTTTCAGCAAGAGG | 0.5 | 95 °C for 7 min; 34 cycles (95 °C for 40 s, 55 °C for 1 min, 72 °C for 40 s); 72 °C for 5 min | 543 | [42] |
| RV | ATCCAGATCGGCAAAGGTTA | |||||
| qnrB | FW | GATCGTGAAAGCCAGAAAGG | 0.5 | 95 °C for 5 min; 38 cycles (95 °C for 40 s, 50 °C for 40 s, 72 °C for 40 s); 72 °C for 7 min | 476 | [42] |
| RV | ATGAGCAACGATGCCTGGTA | |||||
| sul1 | FW | TGAGATCAGACGTATTGCGC | 1 | 95 °C for 4 min; 32 cycles (95 °C for 2 min, 52,7 °C for 2 min, 72 °C for 2 min); 72 °C for 10 min | 400 | [43] |
| RV | TTGAAGGTTCGACAGCACGT | |||||
| sul2 | FW | GCGCTCAAGGCAGATGGCATT | 1 | 94 °C for 5 min; 30 cycles (94 °C for 15 s, 69 °C for 30 s, 72 °C for 60 s); 72 °C for 7 min | 293 | [44] |
| RV | GCGTTTGATACCGGCACCCGT | |||||
| tetA | FW | GTAATTCTGAGCACTGTCGC | 0.4 | 94 °C for 3 min; 25 cycles (94 °C for 1 min, 57 °C for 1 min, 72 °C for 1 min); 72 °C for 10 min | 956 | [45] |
| RV | CTGCCTGGACAACATTGCTT | |||||
| tetB | FW | AAAACTTATTATATTATAGTG | 0.4 | 94 °C for 3 min; 30 cycles (94 °C for 1 min, 52 °C for 1 min, 72 °C for 1 min); 72 °C for 10 min | 169 | [46] |
| RV | TGGAGTATCAATAATATTCAC | |||||
| Gene (nº Positive Samples/nº Total Samples) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Urban WWTPs | Sampling Site | blaCTX-M-G1 | blaTEM | sul1 | sul2 | catI | cmlA | qnrA | qnrB | tetA | tetB |
| A | Influent | 83.33 (10/12) | 91.67 (11/12) | 83.33 (10/12) | 100 (12/12) | 100 (12/12) | 100 (12/12) | 50.00 (6/12) | 66.67 (8/12) | 100 (12/12) | 100 (12/12) |
| Effluent | 66.67 (8/12) | 91.67 (11/12) | 75.00 (9/12) | 91.67 (11/12) | 91.67 (11/12) | 91.67 (11/12) | 25.00 (3/12) | 58.33 (7/12) | 66.67 (8/12) | 83.33 (10/12) | |
| B | Influent | 75.00 (9/12) | 100 (12/12) | 91.67 (11/12) | 100 (12/12) | 83.33 (10/12) | 100 (12/12) | 66.67 (8/12) | 66.67 (8/12) | 100 (12/12) | 91.67 (11/12) |
| Effluent | 58.33 (7/12) | 83.33 (10/12) | 75.00 (9/12) | 83.33 (10/12) | 83.33 (10/12) | 100 (12/12) | 33.33 (4/12) | 58.33 (7/12) | 33.33 (4/12) | 75.00 (9/12) | |
| C | Influent | 75.00 (9/12) | 100 (12/12) | 100 (12/12) | 100 (12/12) | 100 (12/12) | 100 (12/12) | 50.00 (6/12) | 75.00 (9/12) | 100 (12/12) | 75.00 (9/12) |
| Effluent | 41.67 (5/12) | 91.67 (11/12) | 75.00 (9/12) | 100 (12/12) | 91.67 (11/12) | 100 (12/12) | 0 (0/12) | 66.67 (8/12) | 75.00 (9/12) | 75.00 (9/12) | |
| D | Influent | 75.00 (9/12) | 91.67 (11/12) | 91.67 (11/12) | 100 (12/12) | 100 (12/12) | 100 (12/12) | 41.67 (5/12) | 66.67 (8/12) | 83.33 (10/12) | 58.33 (7/12) |
| Effluent | 16.67 (2/12) | 50.00 (6/12) | 83.33 (10/12) | 100 (12/12) | 66.67 (8/12) | 100 (12/12) | 8.33 (1/12) | 58.33 (7/12) | 33.33 (4/12) | 58.33 (7/12) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, M.; Truchado, P.; Cordero-García, R.; Gil, M.I.; Soler, M.A.; Rancaño, A.; García, F.; Álvarez-Ordóñez, A.; Allende, A. Surveillance on ESBL-Escherichia coli and Indicator ARG in Wastewater and Reclaimed Water of Four Regions of Spain: Impact of Different Disinfection Treatments. Antibiotics 2023, 12, 400. https://doi.org/10.3390/antibiotics12020400
Oliveira M, Truchado P, Cordero-García R, Gil MI, Soler MA, Rancaño A, García F, Álvarez-Ordóñez A, Allende A. Surveillance on ESBL-Escherichia coli and Indicator ARG in Wastewater and Reclaimed Water of Four Regions of Spain: Impact of Different Disinfection Treatments. Antibiotics. 2023; 12(2):400. https://doi.org/10.3390/antibiotics12020400
Chicago/Turabian StyleOliveira, Márcia, Pilar Truchado, Rebeca Cordero-García, María I. Gil, Manuel Abellán Soler, Amador Rancaño, Francisca García, Avelino Álvarez-Ordóñez, and Ana Allende. 2023. "Surveillance on ESBL-Escherichia coli and Indicator ARG in Wastewater and Reclaimed Water of Four Regions of Spain: Impact of Different Disinfection Treatments" Antibiotics 12, no. 2: 400. https://doi.org/10.3390/antibiotics12020400
APA StyleOliveira, M., Truchado, P., Cordero-García, R., Gil, M. I., Soler, M. A., Rancaño, A., García, F., Álvarez-Ordóñez, A., & Allende, A. (2023). Surveillance on ESBL-Escherichia coli and Indicator ARG in Wastewater and Reclaimed Water of Four Regions of Spain: Impact of Different Disinfection Treatments. Antibiotics, 12(2), 400. https://doi.org/10.3390/antibiotics12020400







