Abstract
Fresh fruits and vegetables are potential reservoirs for antimicrobial resistance determinants, but few studies have focused specifically on organic vegetables. The present study aimed to determine the presence of third-generation cephalosporin (3GC)- and carbapenem-resistant Gram-negative bacteria on fresh organic vegetables produced in the city of Valencia (Spain). Main expanded spectrum beta-lactamase (ESBL)- and carbapenemase-encoding genes were also detected in the isolates. One hundred and fifteen samples were analyzed using selective media supplemented with cefotaxime and meropenem. Resistance assays for twelve relevant antibiotics in medical use were performed using a disc diffusion test. A total of 161 isolates were tested. Overall, 33.5% presented multidrug resistance and 16.8% were resistant to all β-lactam antibiotics tested. Imipenem resistance was observed in 18% of isolates, and low resistance levels were found to ceftazidime and meropenem. Opportunistic pathogens such as Acinetobacter baumannii, Enterobacter spp., Raoultella sp., and Stenotrophomonas maltophilia were detected, all presenting high rates of resistance. PCR assays revealed blaVIM to be the most frequently isolated ESBL-encoding gene, followed by blaTEM and blaOXA-48. These results confirm the potential of fresh vegetables to act as reservoirs for 3GC- and carbapenem-producing ARB. Further studies must be carried out to determine the impact of raw organic food on the spread of AMRs into the community.
1. Introduction
Antimicrobial resistance (AMR) is currently one of the most threatening public health issues. According to the European Antimicrobial Resistance Surveillance Network, in 2020, more than 800 000 infections occurred in the European Union (EU) due to bacteria resistant to antibiotics, and more than 35,000 people died as a direct consequence of these infections [1]. In 2021, the most commonly reported bacterial species were E. coli (39.4% of all cases), followed by S. aureus, K. pneumoniae, E. faecalis, E. faecium, P. aeruginosa, Acinetobacter spp., and S. pneumoniae [2]. Between 2020 and 2021, the number of reported antibiotic-resistant bacteria (ARB) cases increased for all pathogens, but especially for Acinetobacter spp. (+43%), E. faecium (+21%), and E. faecalis (+14%).
Among antibiotic-resistant bacteria (ARB), the growing prevalence of Gram-negative extended spectrum ß-lactamase (ESBL)-producing bacteria resistant to third-generation cephalosporins (3GC) and carbapenems is a major concern worldwide, because these antibiotics are frequent last-resort treatment options. Infections with ESBL- and carbapenemase-producing pathogenic bacteria, mainly Enterobacteriaceae, cause high mortality and morbidity [2,3]. Moreover, their prevalence is increasing; in 2021, high percentages of carbapenem-resistant Acinetobacter were found in several countries, reaching 50% of clinical isolates [4,5,6,7]. The treatment options for infections with carbapenem-resistant bacteria are limited, especially because carbapenemase genes are usually localized on mobile genetic elements together with other genes, conferring resistance to other β-lactams, fluoroquinolones, and/or aminoglycosides. This also contributes significantly to their spread [8,9].
However, non-pathogenic ARB are also a matter of concern. Saprophytic Gram-negative species, whose reservoirs are soil and water, are an important source of antibiotic-resistant genes (ARG) in the environment, as they are often resistant to third-generation cephalosporins and carbapenems due to the presence of ESBL genes on their chromosomes or the carriage of plasmids containing ESBL genes [10].
Although the prevalence of ESBLs tends to differ among countries, TEM, SHV, CTX-M, and OXA-type enzymes are the main classes of ESBLs [11,12]. The most common carbapenemases found in Gram-negative bacteria from different sources belong to Ambler class A (blaKPC), class B metallo-b-lactamases (blaIMP, blaVIM, and blaNDM), and class D (blaOXA-48) β-lactamases [9].
The “One Health” concept focuses on the interconnected nature of human, animal, and environmental health and highlights zoonotic diseases, food safety, and antimicrobial resistance as three particularly relevant areas [13]. Considering that ARB can be found in samples from several different origins, including humans, wastewater, foods, animals, and environmental sources such as soil or water, it is widely accepted that the environment is a crucial point for the development and spread of antimicrobial resistance determinants [9,14,15]. Among environmental sources, fruits and vegetables are receiving increasing attention as transmission vehicles for ARB and ARG from the environment to humans because they are usually consumed fresh, without further processing. The presence of 3GC- and carbapenem-resistant bacteria in vegetables has been reported by several authors [10,11,16,17]. Fruits and vegetables can be contaminated by soil, fertilizers, and irrigation water or by cross-contamination during harvesting, distribution, and storage [17,18]; ARB can then be transferred through the food chain from primary production to the final consumer [19]. In this sense, the growing demand for fresh organic vegetables is a matter of concern.
The term “organic agriculture” refers to agricultural methods that aim to limit environmental impact by using natural substances and processes, including organic fertilizers and a predominant reliance on ecosystem services and non-chemical measures for pest prevention and control [20,21]. Globally, organic production has experienced spectacular growth in recent years, and, despite the impact of the pandemic, the global market for organic food strongly increased in 2020, exceeding 120 billion euros [22]. In Comunitat Valenciana (Spain), there was an increase in organic food production of 81.2% between 2016 and 2020 [23].
Consumers are interested in organic vegetables because they subjectively understand that these are healthier and better-quality foods than conventional products. However, there is a knowledge gap about some aspects of organic food microbiological safety [21,24]. One question concerns the potential risk of organic vegetable food products transmitting antibiotic resistance determinants, bacteria (ARB), and genes (ARG). When the prevalence of ARB in organic agriculture is compared with that of conventionally grown vegetables, there is a lack of consensus, mainly due to the fact that very few studies have been performed on organic vegetables. Some authors have reported that the presence of ARB and ARG in soils is increased by animal manure application or the reuse of municipal wastewater for irrigation [25,26,27,28,29]. However, other studies have found no increase or even a lower abundance of antibiotic-resistant bacteria isolated from organic vegetables compared to that of conventionally grown produce [30,31,32,33,34]. It seems that, although the use of human and animal manure as fertilizer is undoubtfully a significant route for ARG to enter agricultural soils, when the appropriate delay or pre-treatment of manure is performed, the impact of biosolids or sewage sludge application does not affect the abundance of pathogenic bacteria or ARB on vegetables at harvest [27,35,36]. However, this is still a matter of controversy.
Thus, further information related to the potential health hazards of organic vegetable products is required as, in the absence of adequate comparative data, generalized conclusions cannot be achieved, and further efforts are needed to understand the risks of ARG spread and ARB infections associated with organic vegetables [37,38,39]. The present study aimed to investigate the prevalence of third-generation cephalosporine- and carbapenem-resistant Gram-negative bacteria on fresh organic fruits and vegetables retailed in the city of Valencia (Eastern Spain). The main ESBL- and carbapenemase-encoding genes of the isolates were also detected.
2. Results
2.1. Isolation and Identification of β-Lactamase-Producing Enterobacterales
After growing in mSuperCARBA and MacConkey media supplemented with cefotaxime, at least one colony was observed from each of the 115 (100%) analyzed vegetable samples. A total of 673 presumptive 3GC- and carbapenem-resistant colonies were obtained (20% from lettuce, 23% from spinach, 40% from cabbage, and 36.6% from strawberries), with 74% being oxidase-positive. Of these 673 colonies, 161 were randomly chosen for further analysis (confidence level > 95% [40]), maintaining both the rate of oxidase-positive/negative isolates and the percentages obtained from each type of vegetable (33 from lettuce, 52 from spinach, 59 from cabbage, and 17 from strawberries) (Table 1).

Table 1.
Representative bacterial species isolated from different fresh vegetables.
Using the API system and 16 rRNA gene partial sequencing, 15 isolates from 12 (10.4%) samples were identified as Acinetobacter sp., with nine of them as Acinetobacter baumannii. Twenty-nine presumptive 3GC- and carbapenem-resistant Enterobacteriaceae were isolated from 15 of the 115 (13.04%) vegetable samples: Enterobacter (n = 10), Pantoea sp. (n = 6), Rahnella sp. (n = 5), Raoutella sp. (n = 3), Serratia sp. (n = 2), Leclercia sp. (n = 1), Buttiauxella agrestis (n = 1), and Proteus sp (n = 1). The other 117 isolates were non-fermenter and oxidase-positive bacteria: Pseudomonas spp. (n = 72, where one isolate was P. aeruginosa, Stenotrophomonas sp. (n = 28, mainly S. maltophilia (n = 12)), Burkholderia sp. (n = 5), Elizabethkingia sp. (n = 2), Ralstonia sp. (n = 3), Pasteurella sp. (n = 3), Sphingobacterium multivorum (n = 2), Achromobacter xylosidans (n = 1), and Ochrobactrum anthropi (n = 1) (Supplementary Material Table S1).
2.2. Antibiotic Susceptibility Patterns
From the results of the disc diffusion test, 157 strains (97.51%) were resistant to at least one antibiotic. Only one Stenotrophomonas isolate was susceptible to all antibiotics tested. Three Pseudomonas isolates presented intermediate susceptibility to amoxicillin (AMC) and were susceptible to the rest of antibiotics. Total resistance levels to AMC and ampicillin (AMP) reached 91.9% and 95.7%, respectively (Table 2).

Table 2.
Incidence of resistance to each antimicrobial compound in the isolates.
The results obtained for different antibiotic classes are shown in Figure 1. All (100%) Enterobacteriaceae, 80% of Acinetobacter, 90.3% of Pseudomonas, and 89.3% of Stenotrophomonas isolates were resistant to AMP. Enterobacteriaceae, Pseudomonas sp., and Stenotrophomonas sp. isolates showed 85.7%, 99%, and 96% resistance to this antibiotic, respectively.
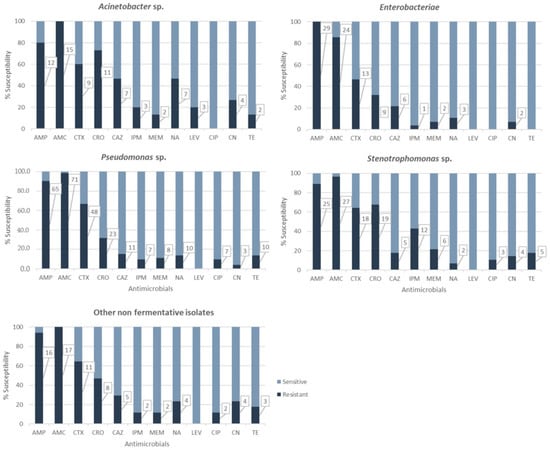
Figure 1.
Antimicrobial resistance for each bacterial group; denotes significant differences between bacterial groups. Numbers in boxes correspond to number of resistant isolates.
Among the Enterobacteriaceae isolates, five were resistant only to AMP and the other five only to AMP and AMC. For carbapenems, only two Enterobacter cloacae isolates were resistant to imipenem (IPM), and one of them was also resistant to meropenem (MEM). One Routella strain was shown to be resistant to imipenem but not to meropenem.
All Acinetobacter isolates (100%) showed resistance to at least three antibiotics, including AMC. Two A. baumannii isolates were resistant to imipenem and meropenem, while the other was only resistant to imipenem.
Regarding third-generation cephalosporins, 116 isolates (72%) showed resistance to at least one of the antibiotics tested. Overall, 61.5% of isolates were resistant to cefotaxime (CTX), 43.5% to ceftriaxone (CRO), and 21.1% to ceftazidime (CAZ). Resistance to cefotaxime was noted for more than 60% of Acinetobacter, Pseudomonas, and Stenotrophomonas isolates. CRO resistance levels in Acinetobacter (73%) and Stenotrophomonas (67.8%) were shown to be statistically significant (χ2 = 18.026, p = 0.0012).
Thirty isolates (18.6%) showed resistance to carbapenems: 15.5% to IMP and 12.4% to MEM. Highly significant IMP resistance levels were observed for Stenotrophomonas (42.8%) (χ2 = 18.026, p = 0.0012).
Resistance to non-β-lactam antibiotics was also studied. The most prevalent resistance was against quinolones (27 (16.1%) isolates resistant to nalidixic acid (NA)), followed by tetracycline (TE) (19 (12.4%) isolates) and gentamicin (CN) (17 (10.6%) isolates). Resistance to the fluoroquinolones ciprofloxacin (CIP) and levofloxacin (LEV) was observed for 12 (7.5%) and three (1.9%) isolates, respectively.
All Enterobacteriaceae were susceptible to CIP, LEV, and TE, while only two isolates showed resistance to CN and three to NA. The 15 Acinetobacter isolates were susceptible to ciprofloxacin. However, resistance rates to NA (46.7%), LEV (three isolates (20%), two A. baumannii and one A. calcoaceticus), and CN (four isolates (26.7%), two being A. baumannii) were significantly higher for Acinetobacter than those observed in the rest of bacterial groups (χ2 = 13.671, p = 0.0084; χ2 = 29.754, p = 0.0000; and χ2 = 11.088, p = 0.0256, respectively).
No significant relationship was found among resistance to any class of antibiotic and the origin of the isolate, except for resistance to CAZ and spinach (χ2 = 9.675, p = 0.0466). However, among the 59 isolates obtained from cabbage, high levels of resistance to CTX (n = 35) were observed. Similarly, 19 and 16 out of the 33 isolates from lettuce were resistant to CTX and CRO, respectively, while isolates from spinach also presented high levels of resistance to CRO (29 out of 52).
Multidrug resistance (MDR) was detected in 36 (22.4%) of the 161 isolates: 11 (33.3%) from lettuce, 13 (25%) from spinach, and 12 (20.3%) from cabbage). No MDR was detected in strawberry isolates (Table 3).

Table 3.
Multidrug resistance patterns among isolates.
Twenty-three MDR isolates showed resistance to 3G-cephaloporins: one Enterobacter sp., one Elizabethkingia sp., one Sphingebacterium sp., two Pasteurella spp., four Burkholderia cepacia., 14 Stenotrophomonas spp., eight Pseudomonas spp. and four Acinetobacter spp. All of these isolates were also resistant to carbapenems, except for one Sphingebacterium, three Stenotrophomonas, one Acinetobacter sp., and three Pseudomonas spp. isolates.
Two isolates of Acinetobacter, 2 A. baumannii and 2 A. calcoaceticus, presented MDR patterns (26.7% of all isolates belonging to this genus). The two A. baumannii were resistant to all antibiotic classes except tetracycline. The only P. aeruginosa strain detected in the samples was resistant to penicillins, 3G-cephalosporins, and tetracycline. One Pseudomonas putida strain, obtained from a spinach sample, was resistant to all types of antibiotics tested. Finally, only one Enterobacteriaceae (Enterobacter sp.) presented an MDR profile.
When comparing the performance of the two selective media used for the isolation of resistant bacteria, similar rates were obtained between MacConkey supplemented with cefotaxime and mSuperCARBA for all the antibiotics tested, except for IMP- and MEM-resistant isolates, which were isolated mainly from the mSuperCARBA medium (93.3%).
2.3. Antibiotic Resistance Genotype Profile
Genes encoding β-lactamases were detected in 64.6% (104/161) of 3GC- and carbapenem-resistant isolates obtained from fresh produce. The most frequently detected gene was blaVIM (n = 43, 26.7%), followed by blaIMP (n = 38, 23.6%), blaTEM (n = 33, 20.5%), blaOXA-48 (n = 28, 17.4%), blaSHV (n = 20, 12.4%), blaCMY-2 (n = 13, 8.1%), and blaKPC (n = 7, 4.3%) (Table 4).

Table 4.
Frequency of ARG detection in the isolates.
Overall, 53 isolates (33%) harbored more than one gene. For Acinetobacter baumannii, at least one gene was detected in six out of nine isolates (66.6%): one gene was detected in three isolates (blaSHV, blaVIM, and blaTEM), one isolate harbored two resistance genes (blaIMP-blaOXA-48), and two isolates from two different samples carried three genes (blaTEM-blaCMY-2-blaKPC).
Out of 29 Enterobacteriaceae isolates, 21 (Enterobacter sp., n = 4; Serratia sp., n = 2; Proteus sp., n = 1; Rahnella sp., n = 5; Pantoea sp., n = 5; Buttiauxella agrestis, n = 1; and Raoutella sp., n = 3) carried resistance genes: one gene was detected in nine isolates (blaOXA-48, n = 5; blaSHV, n = 2; blaIMP, n = 1; and blaVIM, n = 1), seven isolates harbored two genes (blaTEM-blaOXA-48, n = 3; blaTEM-blaVIM, n = 2; blaSHV-blaCMY-2, n = 1; and blaIMP-blaOXA-48, n = 1), and three genes were detected in five isolates (blaSHV-blaIMP-blaVIM, n = 1; blaTEM-blaVIM-blaIMP, n = 1; blaTEM-blaSHV-blaVIM, n = 2; and blaSHV-blaOXA-48-blaVIM, n = 1).
For Stenotrophomonas (28 isolates), no genes were detected in four isolates; one gene was detected in two isolates (blaIMP and blaOXA-48); 14 isolates carried two genes (blaIMP-blaOXA-48, n = 8; blaVIM-blaIMP, n= 4; blaSHV-blaIMP, n = 1; and blaVIM-blaCMY-2, n = 1), and different combinations of three genes were detected in eight isolates (blaTEM-blaCMY-2-blaVIM, n = 4; blaSHV-blaOXA-48-blaIMP, n = 3; and blaTEM-blaVIM-blaIMP, n = 1).
Finally, 24 out of 89 isolates of Pseudomonas sp. and other non-fermenting bacteria carried one resistance gene (blaTEM, n = 7; blaSHV, n = 1; blaIMP, n = 11; blaCMY-2, n = 2; blaVIM, n = 3; and blaKPC, n = 2), while 16 isolates harbored two genes (blaTEM-blaSHV, n = 2; blaTEM-blaCMY-2, n = 2; blaTEM-blaIMP, n = 1; blaTEM-blaKPC, n = 3; blaVIM-blaCMY-2, n = 1; and blaVIM-blaIMP, n = 7), and four carried three genes (blaSHV-blaIMP-blaOXA-48, n = 3 and blaTEM-blaVIM-blaCMY-2, n = 1).
The statistical analysis showed that the presence of blaOXA-48 in Stenotrophomonas and Enterobacteria was significantly greater than in the rest of the bacterial groups (χ2 = 37.933, p = 0.0000). The presence of blaVIM in Stenotrophomonas was also significantly higher than in the rest of bacterial groups (χ2 = 24.365, p = 0.0001).
Among the 27 isolates that showed resistance to all the β-lactam antibiotics in the disc diffusion assay, 16 harbored at least one carbapenemase-encoding gene. In three isolates, only the blaTEM gene was detected, while in eight isolates, we did not detect any of the tested genes.
The statistical correlation between the presence of each gene and the origin of the isolate was investigated: blaTEM was significantly more frequently detected in cabbage (χ2 = 10.405, p = 0.0154), blaKPC in strawberries (χ2 = 18.981, p = 0.0003), blaOXA-48 in lettuce (χ2 = 12.893, p = 0.0049), and blaSHV, blaIMP, and blaVIM in spinach (χ2 = 19.699, p = 0.002, χ2 = 19.358 and 20.359, p = 0.0002 and 0.0001, respectively). No correlation was found for the blaCMY-2 gene.
3. Discussion
The presence of antibiotic-resistant bacteria on fresh organic vegetables is a growing public health concern, due to an increase in the consumption of these products in recent years and evidence that fresh vegetables constitute a source of ARB, in addition to representing a possible route for the dissemination of resistance genes in the community and the environment [34,41]. However, very few studies are available on the actual food safety and environmental risks that occur due to the presence of antibiotic resistance determinants in fresh organic vegetables [16].
Thus, the present study aimed to detect ARB and ARG in organic vegetables grown and distributed in Valencia (Spain) that are usually consumed raw. We focused on Gram-negative bacteria because a large proportion of infections caused by drug-resistant microorganisms in community and healthcare settings are recognized to be caused by these types of bacteria [42]. Due to their special significance in human medicine, ESBL- and carbapenem-producing Gram-negative bacteria were specifically detected.
A total of 115 organic vegetable samples (lettuce, spinach, cabbage, and strawberries) were analyzed. These types of vegetables were selected because they grow near the soil and are usually eaten uncooked. The search for ARB was carried out using selective enrichment and culture media supplemented with β-lactam antibiotics. All the samples were contaminated with ARB, and more than 97% of the 161 selected isolates presented resistance to at least one antibiotic in the disc diffusion test. Moreover, 22.4% presented multidrug resistance and 14.9% were resistant to all β-lactam antibiotics tested, forming 18% of all isolates resistant to carbapenems. Nevertheless, the prevalence of resistance to CAZ and MEM was low among all the isolated bacteria. Among non-β-lactam antibiotics, resistance ranged from 16% for NA to 2% for LE.
Overall, these resistance rates confirm the potential of fresh vegetables to act as a reservoir for AR determinants in the environment. Additionally, the consumption of these raw vegetables may result in the transmission of resistance to other commensal or pathogenic gut microbiota through gene transfer mechanisms, a process previously described as a “silent food safety concern” [34,43], contributing to the dissemination of antibiotic resistance within the community.
In this sense, one of the major threats is the growing prevalence of Enterobacteriaceae resistant to third-generation cephalosporins (3GC) and carbapenemases [44]. In this study, we detected 3GC-resistant Enterobacteriaceae in 13% of the samples; Enterobacter was the most prevalent genus, followed by Pantoea and Rahnella. Our results are similar to those of other studies on conventional vegetables, in which Enterobacter was the most frequently detected genus, followed by Pantoea, Serratia, Citrobacter, and Rahnella [10]. In addition, in a study performed in the same geographical area [34], Pantoea Enterobacter and Serratia were the predominant Enterobacteriaceae genera in organic samples.
Regarding the numbers of ESBL-producing Enterobacteriaceae isolated from raw vegetables, it is difficult to compare our findings with those of other studies due to significant differences in study design, sampling strategy, number, and type of vegetables analyzed, isolation methodologies, and geographical sampling zone. This lack of homogeneity has been also pointed out by other authors [16,31]. However, our results (13%) cannot be considered high and are in the range of those obtained by Reuland et al. [45] (6%), Raphael et al. [42] (12%), Moon et al. [46] (9.09%), and Ritcher et al. [47] (17.4%) for conventional and organic vegetables under different experimental conditions. On the contrary, some authors have found higher rates on conventional vegetables, from 25.4% [41] to 40% [25,27]. Overall, organic vegetables analyzed in this study did not present higher contamination rates than those found by other authors on conventional non-organic produce.
Among the Enterobacteriaceae isolated from organic vegetables, resistance levels to CRO were below 60%. Moreover, only one strain presented an MDR profile. Resistance to carbapenems and CN was also low, reaching 46% for MDR and 70 to 90% for aminoglycoside resistances, far from that obtained by other authors on non-organic vegetables [11,47]. The presence of one MDR, including all carbapenems, Enterobacter strain, is coincident with other studies [16], and may represent a public health concern for immunocompromised patients.
It is also remarkable that we found one Raoultella isolate resistant to IPM. Raoultella organisms are often misidentified, due to their close relatedness to Klebsiella spp. They are commonly found in the natural environment and for many years were considered saprophytic bacteria. However, in recent years, infections in immunocompromised patients have been frequently reported, with high mortality rates. In addition, MDR isolates, including carbapenemase-producing strains, have been described both in clinical settings [48] and in the environment [49].
Acinetobacter isolates were isolated from 9.5% of the analyzed samples, whereas A. baumannii was present in nine samples (5.6%). These rates are similar to those found in conventional non-organic vegetables and fruits [50]. This species is an important hospital-acquired pathogen worldwide, widely found in intensive care units where it can cause severe infections. Moreover, there are increasing reports of multidrug-resistant non-baumannii Acinetobacter that cause infections in healthcare facilities around the world [5,6,51]. Acinetobacter are intrinsically resistant to penicillins and generally present multidrug resistance to third-generation cephalosporins, aminoglycosides, and fluoroquinolones. Their antibiotic-resistant genes are usually harbored in class 1 integrons, which has allowed them to become a major contributor to resistance among clinical and environmental Gram-negative bacteria [51,52]. In this study, we observed worrying resistance rates: 26.7% of Acinetobacter spp. presented MDR patterns, and two A. baumannii isolates were resistant to all antibiotic classes except tetracycline. High levels of antibiotic resistance in Acinetobacter present in conventional vegetables have been previously detected by other authors [5,25,30,53].
Most (73%) of the 161 Gram-negative isolates were oxidase-positive, non-fermenter bacteria, including a high percentage of Pseudomonas and Stenotrophomonas. In contrast to other studies [42,54], most of the isolates were saprophytic, and only one P. aeruginosa isolate was detected. Nonetheless, resistance rates were found to be high, and one Pseudomonas putida isolate was resistant to all antibiotics tested.
Regarding the Stenotrophomonas isolates, they all presented high rates of resistance to CRO and IPM, and almost 40% were resistant to all β-lactam antibiotics tested. Stenotrophomonas are environmental bacteria and are considered to be emergent pathogens, mainly producing severe pulmonary and bloodstream infections in vulnerable populations with mortality rates from 24% to 58%. S. maltophilia was recently described as the most common Gram-negative carbapenem-resistant pathogen isolated from bloodstream infections acquired in community and hospital settings in the USA [55]. Therefore, its presence in vegetables and the high levels of resistance detected cannot be underestimated.
Genotypic characterization using PCR assays revealed blaVIM to be the most frequently isolated carbapenemase-encoding gene, followed by blaTEM and blaOXA-48. Previous studies have also isolated carbapenemase-producing Enterobacteriaceae from fresh vegetables [11,17,56] and pointed out both the great spread of carbapenemase-encoding genes in the environment and the risk of acquisition for this type of ARG through the consumption of fresh vegetables.
The high prevalence of blaOXA-48 in our samples differs from the results of previous studies from other countries [11,17]. Carbapenem-resistant OXA-48 genes are a significant cause of carbapenem resistance, and the emergence of OXA enzymes, particularly in A. baumannii, has transformed these lactamases into a major problem, diminishing the clinical efficacy of carbapenems [57].
Regarding the presence of pathogenic bacteria in the analyzed fruits and vegetables, a relevant result of this study is that we found exclusively opportunistic pathogens. Strict farming regulations in most European and North American countries are aimed at protecting vegetables from contamination with pathogenic microorganisms and, consequently, most antibiotic- and multidrug-resistant bacteria are expected to be predominantly saprophytic and opportunistic organisms [41,58].
Our results confirm that organic vegetables are safe and do not constitute a direct threat to immunocompetent consumers. However, the considerable number of MDR opportunistic bacteria raises concerns about the consumption of this type of food by immunocompromised patients. On the other hand, the fact that all the vegetable samples contained at least one colony of resistant bacteria confirms the role of these foods as reservoirs of antibiotic resistance genes. Saprophytes in fresh produce have been found to harbor drug resistance genes that are also found in internationally circulating strains of pathogens; they may thus serve as a reservoir for drug resistance genes that enter pathogens and affect human health [42]. Further research should thus focus on clarifying and quantifying the involvement of saprophytic bacteria in ARG transfer to pathogens as well as in extraintestinal opportunistic infections after transmission by food [59].
Antimicrobial resistance surveillance programs primarily focus on food from animal origins. However, according to the present work and previous studies [16], monitoring antibiotic-resistant bacterial reservoirs in vegetables seems to be equally important. Some authors have even suggested that including the detection of antibiotic-resistant non-pathogenic Enterobacteriaceae species in fresh vegetables during epidemiological surveillance routines could help to quantify the dissemination of multidrug resistance in non-clinical environments and evaluate the potential role of the consumption of fresh vegetables in spreading resistance in the community [34].
4. Materials and Methods
4.1. Sampling
Between June 2021 and May 2022, a total of 115 fresh vegetable samples with the European organic label were purchased weekly in 12 supermarkets and popular markets in the city of Valencia, Spain. The vegetables included lettuce (Lactuca sativa, n = 30), spinach (Spinacia oleracea, n = 30), cabbage (Brassica oleracea var. sabellica, n = 15; Brassica oleracea var. capitata f. rubra, n = 15), and strawberries (Fragaria, n = 25). Each sample consisted of a batch of 3–4 pieces for leaf vegetables and 750 g for strawberries.
The vegetable samples were handled under aseptic conditions after their arrival at the laboratory. Unwashed leaves were cut with sterile scissors into small portions and the resulting fragments were mixed prior to homogenization. Strawberries were processed by discarding the sepal and pedicle, leaving only the consumable fruit.
The samples were pre-enriched to isolate third-generation cephalosporin (3GC)- and carbapenem-resistant bacteria. From each sample, two portions of 10 g were aseptically placed in separate sterile flasks containing 90 mL of buffered peptone water (BPW) (Scharlau, Barcelona, Spain) with cefotaxime (2.5 mg/L) and 90 mL of trypticase soy broth (TSB, Oxoid, England) supplemented with meropenem (1.0 mg/L). Both broths were supplemented with vancomycin (5 mg/L) to ensure the inhibition of Gram-positive bacterial growth. The samples were homogenized using a Stomacher sample blender and incubated at 37 °C overnight. Thereafter, 50 mL of the sample suspension was centrifuged at 4000 rpm for 10 min for further DNA extraction [14]. For the detection of antibiotic-resistant bacteria, one loopful of each broth was streaked onto mSuperCARBA (CHROMagar™ Paris, France) and MacConkey agar supplemented with cefotaxime (2.5 mg/L), both prepared according to the manufacturer’s instructions. The plates were aerobically incubated for 24 h at 37 °C.
4.2. Isolation and Characterization of Isolates
A maximum of 3 different suspected 3GC- and 3 carbapenem-resistant colonies were picked from each MacConkey and mSuperCARBA plate, respectively, based on the morphological and color-coded distinction of colonies according to the manufacturers’ protocols. Thus, 6 different isolates were recovered from each sample, purified, and identified using Gram, catalase, and oxidase tests. The isolates were stored at −80 °C until use.
The selection of colonies was performed, and isolates were identified using the API phenotypic identification system (API20E and API 20NE strips, BioMèriux, Marcy-l’Étoile, France). When the identity match with the database was less than 99%, identification was confirmed by the partial sequencing of the 16S rDNA gene to amplify the 16S rRNA gene (1500 bp) using the universal bacterial primers 27F (5′-AGAGTTTGATYMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) for the Enterobacteriaceae isolates [60] and 27F and 1525R (5′-AGAAAGGAGGTGATCCAGCC-3′) for the presumptive Acinetobacter (non-fermenter strains) isolates [61]. The PCR conditions for Enterobacteriaceae were: one cycle at 95 °C for 5 min, 35 cycles comprising the denaturation phases at 95 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 1 min, and a final extension cycle at 72 °C for 10 min. For presumptive Acinetobacter strains, the PCR conditions were as follows: one cycle at 95 °C for 2 min, 35 cycles at 94 °C for 30 s, 65 °C for 30 s, 72 °C for 2 min, and a final extension at 72 °C for 10 min. All the reactions were performed in a PTC-100 Thermal Cycler (MJ Research, St. Bruno, QC, Canada).
The PCR products were electrophoresed on a 2% agarose gel (Pronadisa, Madrid, Spain) with 100 mL of Tris-acetate-EDTA buffer and 5 μL of RedSafe™ solution (Intron Biotechnology, Washington, USA) and scanned with a gel monitoring system (Transilluminator Vilber, Lourmat, France). After gel electrophoresis, the amplicons were purified using the GenElute™ PCR Clean-Up Kit (Sigma-Aldrich, Madrid, Spain) following the manufacturer’s instructions. The representative 16S rDNA fragments of the different isolates were sequenced by cycle extension in an ABI 373 DNA sequencer (Applied Biosystems; Foster City, CA, USA). An approximately 800 bp sequence was obtained per fragment, which was compared with sequences deposited in the GenBank database using the online BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/ accessed on 22 October 2022).
4.3. Antibiotic Susceptibility Testing of Isolates
The isolates were subjected to susceptibility testing using the disc diffusion method against 12 antibiotics (Antimicrobial Susceptibility Test Disc, OXOID Ltd., England, United Kingdom), namely ampicillin (AMP, 10 µg), amoxicillin (AMC, 20 µg), cefotaxime (CTX, 30 µg), ceftriaxone (CRO, 30 µg), ceftazidime (CAZ, 30 µg), imipenem (IPM, 10 µg), meropenem (MEM, 10 µg), nalidixic acid (NA, 30 µg), levofloxacin (LEV, 5 µg), ciprofloxacin (CIP, 5 µg), gentamicin (CN, 10 µg), and tetracycline (TE, 30 µg), according to the CLSI guidelines [62].
E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as control strains. Multidrug resistance was defined as nonsusceptibility to at least three antibiotic classes [11,63]. Due to the existence of intrinsic resistance to AMP and AMC in Pseudomonas and Acinetobacter genus, MDR was considered when the isolates presented resistance to 3 classes of antibiotics in addition to penicillins. The results were interpreted according to the European Committee on Antimicrobial Susceptibility Testing [64] and isolates were classified as Susceptible or Resistant [65].
4.4. Antibiotic Resistance Gene Detection
DNA was extracted from all the selected isolates using the GenEluteTM Bacterial Genomic DNA Kit (Sigma-Aldrich, Madrid, Spain), according to the manufacturer’s instructions, and then screened for ESBL genes (blaTEM, blaSHV, and blaCYM-2) and carbapenemase-encoding genes (blaIMP, blaVIM, blaKPC, and blaOXA-48) using the primers described in Table 5.
Three multiplex PCR (mPCR) reactions were performed for detecting ESBL, AmpC, and carbapenemase genes: (1) for the detection of blaSHV, blaTEM, and blaCMY-2; (2) for the detection of blaKPC and blaVIM; and (3) for the detection of blaIPM and blaOXA-48. These genes were selected because of their great prevalence among ESBL- and carbapenemase-producing Gram-negative pathogenic and environmental bacteria in Spain [66,67,68].
The mix for mPCR (1) included 2.5 μL template DNA; 2.5 mM MgCl2; 0.4 μM of each primer for blaSHV, and 0.2 μM for blaTEM and blaCMY-2; 0.2 mM of each dNTP; 5 U of Taq polymerase; and 1X PCR buffer, reaching a final reaction volume of 25 μL. The thermal cycler conditions for this assay were as described by [69]: 15 min at 94 °C, 30 cycles of amplification consisting of 1 min at 94 °C, 1 min at 55 °C, and 1 min at 72 °C, with 10 min for the final extension at 72 °C. As positive controls, we used Klebsiella pneumoniae subsp. pneumoniae ATCC 700603 for blaSHV, Escherichia coli ATCC 35218 for blaTEM, and our own positive E. coli strain (M1A mec8) for blaCMY-2. MilliQ water was used as a negative control.
For mPCR (2) and (3), the mix consisted of 1X PCR buffer; 1.5 mM MgCl2; 0.4 μM of each primer; 0.125 mM of dNTPs; 2 U of Taq polymerase; and 2.5 μL of extracted DNA, reaching a final reaction volume of 25 μL. Amplification was carried out according to Poirel et al. [70] with slight modifications: 10 min at 94 °C and 36 cycles of amplification consisting of 30 s at 94 °C, 40 s at 55 °C, and 50 s at 72 °C, finishing with 5 min at 72 °C for the final extension. As positive controls, K. pneumoniae NCTC 13442, K. pneumoniae NCTC 13438, K. pneumoniae NCTC 13440, and E. coli NCTC 13476 were used for blaOXA-48, blaKPC, blaVIM, and blaIMP, respectively. MilliQ water was used as a negative control.
All amplifications were developed in a Mastercycler®Pro (Eppendorf, Hamburg, Germany) thermal cycler. PCR products were detected by electrophoresis on 1.2% (w/v) agarose gel in TAE 1X (Tris 40 mM, acetic acid 20 mM, EDTA 1 mM) buffer with RedSafeTM (iNtRON Biotechnology, Sungnam, Korea) at 90 V for about 75 min and visualized under UV light.

Table 5.
PCR primer sequences, targets, and conditions.
Table 5.
PCR primer sequences, targets, and conditions.
| Target Gene | Sequence | Conditions | Reference |
|---|---|---|---|
| blaTEM | 5′-TTAACTGGCGAACTACTTAC-3′ 5′-GTCTATTTCGTTCATCCATA-3′ | 94 °C 15 min (1 cycle); 94 °C 1 min, 55 °C 1 min (36 cycles); 72 °C 1 min; 72 °C 10 min (elongation) | Kozak et al. (2009) [69] |
| blaSHV | 5′-AGGATTGACTGCCTTTTTG-3′ 5′-ATTTGCTGATTTCGCTCG-3′ | ||
| blaCMY-2 | 5′-GACAGCCTCTTTCTCCACA-3′ 5′-TGGACACGAAGGCTACGTA-3′ | ||
| blaKPC | 5′-CGTCTAGTTCTGCTGTCTTG-3′ 5′-CTTGTCATCCTTGTTAGGCG-3′ | 94 °C 10 min (1 cycle); 94 °C 30 s, 55 °C 40 s (36 cycles); 72 °C 55 s; 72 °C 5 min (elongation) | Poirel et al. (2011) [70] |
| blaOXA-48 | 5′-GCGTGGTTAAGGATGAACAC-3′ 5′-CATCAAGTTCAACCCAACCG-3′ | ||
| blaIMP | 5′-GGAATAGAGTGGCTTAAYTCTC-3′ 5′-GGTTTAAYAAAACAACCACC-3′ | ||
| blaVIM | 5′-GATGGTGTTTGGTCGCATA-3′ 5′-CGAATGCGCAGCACCAG-3′ |
4.5. Statistical Analysis
Statistical analysis was performed using the Statgraphics (Centurion XVII) software (Statpoint Technologies, Inc. Warrenton, VA, USA). Antibiotic resistance and ARG detection were analyzed via a χ2 test, using contingency tables, to establish any possible dependent correlation with vegetable type and/or bacterial group. A probability value of less than 5% was considered significant.
5. Conclusions
The results of this study demonstrated a high abundance of relatively non-pathogenic ESBL- and carbapenemase-producing Gram-negative bacteria on fresh organic vegetables retailed in Valencia, Spain. Our results indicate that organic vegetables represent reservoirs and possible routes for the spread of ARB and ARG into the community. Although the absence of enteric pathogens was encouraging, saprophytic bacteria can carry antibiotic resistance determinants to consumers via the food chain and transfer them to gut commensal or pathogenic microorganisms. Moreover, the prevalence of opportunistic bacteria presenting high resistance levels was relevant in all the analyzed vegetables. Exposure to these opportunistic pathogens may result in the colonization of immunocompromised patients, causing severe and difficult-to-treat infections. Thus, the presence of ARB and ARGs on fresh organic vegetables that are consumed raw poses potential public health risks whose magnitude has yet to be established.
Our study points out the need for more research to determine the actual impact of raw organic food on the spread of ARGs, to understand the effects of human colonization with ARBs originating from fresh vegetables, and to qualitatively and quantitatively assess the health risks associated with the ARB/ARGs in these foods.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12020387/s1, Table S1: Identification of isolates.
Author Contributions
A.I.J.-B. and M.Á.C. equally participated in the development of the experimental design, supervision of research, and working on and preparing the manuscript draft. M.Á.C. performed the formal data analysis. M.A.F. participated in the conceptualization of the research as the main leader, in funding acquisition and project administration, as well as in manuscript redaction. M.H., J.G.-H. and Y.M. participated in the development and application of microbiological and molecular techniques and validation of procedures. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministerio de Ciencia e Innovación, Spain, Grant number PID2019-105691RB-I00.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- European Centre for Disease Prevention and Control. Assessing the Health Burden of Infections with Antibiotic-Resistant Bacteria in the EU/EEA, 2016–2020. 2022. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/Health-burden-infections-antibiotic-resistant-bacteria.pdf (accessed on 23 December 2022).
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2021. 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2021 (accessed on 23 December 2022).
- CDC. Antibiotic Resistance Threats in the United States. Department of Health and Human Services, CDC 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 23 December 2022).
- European Centre for Disease Prevention and Control and World Health Organization Surveillance of Antimicrobial Resistance in Europe, 2021 Data. Executive Summary. 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2021-data (accessed on 23 December 2022).
- Al Atrouni, A.; Hamze, M.; Rafei, R.; Eveillard, M.; Joly-Guillou, M.-L.; Kempf, M. Diversity of Acinetobacter species isolated from different environments in Lebanon: A nationwide study. Future Microbiol. 2016, 11, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Ababneh, Q.; Abu Laila, S.; Jaradat, Z. Prevalence, genetic diversity, antibiotic resistance and biofilm formation of Acinetobacter baumannii isolated from urban environments. J. Appl. Microbiol. 2022, 133, 3617–3633. [Google Scholar] [CrossRef] [PubMed]
- Amudhan, M.S.; Sekar, U.; Kamalanathan, A.; Balaraman, S. bla(IMP) and bla(VIM) mediated carbapenem resistance in Pseudomonas and Acinetobacter species in India. J. Infect. Dev. Ctries. 2012, 6, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Dortet, L.; Poirel, L. Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 2012, 18, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Taggar, G.; Rehman, M.A.; Boerlin, P.; Diarra, M. Molecular Epidemiology of Carbapenemases in Enterobacteriales from Humans, Animals, Food and the Environment. Antibiotics 2020, 9, 693. [Google Scholar] [CrossRef]
- Blaak, H.; van Hoek, A.H.; Veenman, C.; van Leeuwen, A.E.D.; Lynch, G.; van Overbeek, W.M.; de Roda Husman, A.M. Extended spectrum β-lactamase-and constitutively AmpC-producing Enterobacteriaceae on fresh produce and in the agricultural environment. Int. J. Food Microbiol. 2014, 168, 8–16. [Google Scholar] [CrossRef]
- Colosi, I.A.; Baciu, A.M.; Opriș, R.V.; Peca, L.; Gudat, T.; Simon, L.M.; Colosi, H.A.; Costache, C. Prevalence of ESBL, AmpC and Carbapenemase-Producing Enterobacterales Isolated from Raw Vegetables Retailed in Romania. Foods 2020, 9, 1726. [Google Scholar] [CrossRef]
- Pishtiwan, A.H.; Khadija, K.M. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumoniae and Escherichia coli isolated from thalassemia patients in Erbil, Iraq. Mediterr. J. Hematol. Infect. Dis. 2019, 11, e2019041. [Google Scholar] [CrossRef]
- European Commission. A European One Health Action Plan Against Antimicrobial Resistance (AMR). 2017. Available online: https://health.ec.europa.eu/system/files/2020-01/amr_2017_action-plan_0.pdf (accessed on 23 December 2022).
- Sung, G.-H.; Kim, S.H.; Park, E.H.; Hwang, S.N.; Kim, J.-D.; Kim, G.R.; Kim, E.-Y.; Jeong, J.; Kim, S.; Shin, J.H. Association of Carbapenemase-Producing Enterobacterales Detected in Stream and Clinical Samples. Front. Microbiol. 2022, 13, 923979. [Google Scholar] [CrossRef]
- Wang, F.; Fu, Y.-H.; Sheng, H.-J.; Topp, E.; Jiang, X.; Zhu, Y.-G.; Tiedje, J.M. Antibiotic resistance in the soil ecosystem: A One Health perspective. Curr. Opin. Environ. Sci. Health 2021, 20, 100230. [Google Scholar] [CrossRef]
- Rahman, M.; Alam, M.-U.; Luies, S.K.; Kamal, A.; Ferdous, S.; Lin, A.; Sharior, F.; Khan, R.; Rahman, Z.; Parvez, S.M.; et al. Contamination of Fresh Produce with Antibiotic-Resistant Bacteria and Associated Risks to Human Health: A Scoping Review. Int. J. Environ. Res. Public Health 2022, 19, 360. [Google Scholar] [CrossRef]
- Liu, B.-T.; Zhang, X.-Y.; Wan, S.-W.; Hao, J.-J.; Jiang, R.-D.; Song, F.-J. Characteristics of Carbapenem-Resistant Enterobacteriaceae in Ready-to-Eat Vegetables in China. Front. Microbiol. 2018, 9, 1147. [Google Scholar] [CrossRef]
- Machado-Moreira, B.; Richards, K.; Brennan, F.; Abram, F.; Burgess, C.M. Microbial Contamination of Fresh Produce: What, Where, and How? Compr. Rev. Food Sci. Food Saf. 2019, 18, 1727–1750. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2016. EFSA J. 2018, 16, e05182. [Google Scholar] [CrossRef]
- European Union (EU). Legislation for the Organics Sectors. 2019. Available online: https://ec.europa.eu/info/food-farming-fisheries/farming/organic-farming/legislation_en (accessed on 23 December 2022).
- Mie, A.; Andersen, H.R.; Gunnarsson, S.; Kahl, J.; Kesse-Guyot, E.; Rembiałkowska, E.; Quaglio, G.; Grandjean, P. Human health implications of organic food and organic agriculture: A comprehensive review. Environ. Health 2017, 16, 111. [Google Scholar] [CrossRef] [PubMed]
- Research Institute of Organic Agriculture FIBL, IFOAM-Organics International. The World of Organic Agriculture Statistics and Emerging Trends 2022; Willer, H., Trávníček, J., Meier, C., Schlatter, B., Eds.; Druckerei Hachenburg: Hachenburg, Germany, 2022; Available online: https://www.fibl.org/fileadmin/documents/shop/1344-organic-world-2022.pdf (accessed on 23 December 2022).
- CAECV. Informe del Sector Ecológico de la Comunitat Valenciana. 2020. Available online: https://www.caecv.com/wp-content/uploads/2021/05/Informe.pdf (accessed on 23 December 2022).
- Nguyen-The, C.; Bardin, M.; Berard, A.; Berge, O.; Brillard, J.; Broussolle, V.; Carlin, F.; Renault, P.; Tchamitchian, M.; Morris, C.E. Agrifood systems and the microbial safety of fresh produce: Trade-offs in the wake of increased sustainability. Sci. Total Environ. 2016, 562, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Rahube, T.O.; Marti, R.; Scott, A.; Tien, Y.-C.; Murray, R.; Sabourin, L.; Zhang, Y.; Duenk, P.; Lapen, D.R.; Topp, E. Impact of Fertilizing with Raw or Anaerobically Digested Sewage Sludge on the Abundance of Antibiotic-Resistant Coliforms, Antibiotic Resistance Genes, and Pathogenic Bacteria in Soil and on Vegetables at Harvest. Appl. Environ. Microbiol. 2014, 80, 6898–6907. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Reynnells, R. Importance of Soil Amendments: Survival of Bacterial Pathogens in Manure and Compost Used as Organic Fertilizers. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Salmanov, A.G.; Ushkalov, V.O.; Shunko, Y.Y.; Piven, N.; Vygovska, L.M.; Verner, O.M.; Kushnirenko, S. One health: Antibiotic-resistant bacteria contamination in fresh vegetables sold at a retail markets in Kyiv, Ukraine. Wiad. Lek. 2021, 74, 83–89. [Google Scholar] [CrossRef]
- Marano, R.B.; Zolti, A.; Jurkevitch, E.; Cytryn, E. Antibiotic resistance and class 1 integron gene dynamics along effluent, reclaimed wastewater irrigated soil, crop continua: Elucidating potential risks and ecological constraints. Water Res. 2019, 164, 114906. [Google Scholar] [CrossRef]
- Li, H.; Zheng, X.; Tan, L.; Shao, Z.; Cao, H.; Xu, Y. The vertical migration of antibiotic-resistant genes and pathogens in soil and vegetables after the application of different fertilizers. Environ. Res. 2022, 203, 111884. [Google Scholar] [CrossRef] [PubMed]
- Ruimy, R.; Brisabois, A.; Bernede, C.; Skurnik, D.; Barnat, S.; Arlet, G.; Momcilovic, S.; Elbaz, S.; Moury, F.; Vibet, M.-A.; et al. Organic and conventional fruits and vegetables contain equivalent counts of Gram-negative bacteria expressing resistance to antibacterial agents. Environ. Microbiol. 2010, 12, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Marti, R.; Scott, A.; Tien, Y.-C.; Murray, R.; Sabourin, L.; Zhang, Y.; Topp, E. Impact of Manure Fertilization on the Abundance of Antibiotic-Resistant Bacteria and Frequency of Detection of Antibiotic Resistance Genes in Soil and on Vegetables at Harvest. Appl. Environ. Microbiol. 2013, 79, 5701–5709. [Google Scholar] [CrossRef] [PubMed]
- van Hoek, A.H.; Veenman, C.; van Overbeek, W.M.; Lynch, G.; de Roda Husman, A.M.; Blaak, H. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 2015, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Guarddon, M.; Miranda, J.M.; Vázquez, B.I.; Cepeda, A.; Franco, C.M. Assessment of Tetracyclines Residues and Tetracycline Resistant Bacteria in Conventional and Organic Baby Foods. Foods 2015, 4, 306–317. [Google Scholar] [CrossRef]
- Rico, H.; Falomir, P. Comparison of the Antibiotic-Resistant Enterobacteriaceae Content in Conventional, Organic and Fresh-Cut Vegetables Sold in Valencia (Spain). AIMS Agric. Food 2020, 5, 233–244. [Google Scholar] [CrossRef]
- Zhang, Y.-J.; Hu, H.-W.; Chen, Q.L.; Yan, H.; Wang, J.-T.; Chen, D.; He, J.-Z. Manure Application Did Not Enrich Antibiotic Resistance Genes in Root Endophytic Bacterial Microbiota of Cherry Radish Plants. Appl. Environ. Microbiol. 2020, 86, e02106–e02119. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, Q.; Chen, S.; Zhu, Y.-G. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 2017, 98, 152–159. [Google Scholar] [CrossRef]
- Maffei, D.F.; Batalha, E.Y.; Landgraf, M.; Schaffner, D.W.; Franco, B.D. Microbiology of organic and conventionally grown fresh produce. Braz. J. Microbiol. 2016, 47, 99–105. [Google Scholar] [CrossRef]
- Sun, Y.; Qiu, T.; Gao, M.; Shi, M.; Zhang, H.; Wang, X. Inorganic and organic fertilizers application enhanced antibiotic resistome in greenhouse soils growing vegetables. Ecotoxicol. Environ. Saf. 2019, 179, 24–30. [Google Scholar] [CrossRef]
- Gudda, F.O.; Waigi, M.G.; Odinga, E.S.; Yang, B.; Carter, L.; Gao, Y. Antibiotic-contaminated wastewater irrigated vegetables pose resistance selection risks to the gut microbiome. Environ. Pollut. 2020, 264, 114752. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.P.; Verma, P. Determining Sample Size in Experimental Studies. In Determining Sample Size and Power in Research Studies; Verma, J.P., Verma, P., Eds.; Springer: Singapore, 2020. [Google Scholar] [CrossRef]
- Zurfluh, K.; Nüesch-Inderbinen, M.; Morach, M.; Zihler Berner, A.; Hächler, H.; Stephan, R. Extended-spectrum-β-lactamase-producing Enterobacteriaceae isolated from vegetables imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 2015, 81, 3115–3120. [Google Scholar] [CrossRef] [PubMed]
- Raphael, E.; Wong, L.K.; Riley, L.W. Extended-Spectrum Beta-Lactamase Gene Sequences in Gram-Negative Saprophytes on Retail Organic and Nonorganic Spinach. Appl. Environ. Microbiol. 2011, 77, 1601–1607. [Google Scholar] [CrossRef] [PubMed]
- Iseppi, R.; De Niederhäusern, S.; Bondi, M.; Messi, P.; Sabia, C. Extended-Spectrum β-Lactamase, AmpC, and MBL-Producing Gram-Negative Bacteria on Fresh Vegetables and Ready-to-Eat Salads Sold in Local Markets. Microb. Drug Resist. 2018, 24, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Reuland, E.A.; Al Naiemi, N.; Raadsen, S.A.; Savelkoul, P.H.M.; Kluytmans, J.A.J.W.; Vandenbroucke-Grauls, C.M.J.E. Prevalence of ESBL-producing Enterobacteriaceae in raw vegetables. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.H.; Udaondo, Z.; Abram, K.Z.; Li, X.; Yang, X.; DiCaprio, E.L.; Jun, S.-R.; Huang, E. Isolation of AmpC- and extended spectrum β-lactamase-producing Enterobacterales from fresh vegetables in the United States. Food Control. 2022, 132, 108559. [Google Scholar] [CrossRef]
- Richter, L.; Du Plessis, E.M.; Duvenage, S.; Korsten, L. Occurrence, Identification, and Antimicrobial Resistance Profiles of Extended-Spectrum and AmpC β-Lactamase-Producing Enterobacteriaceae from Fresh Vegetables Retailed in Gauteng Province, South Africa. Foodborne Pathog. Dis. 2019, 16, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Appel, T.M.; Quijano-Martínez, N.; De La Cadena, E.; Mojica, M.F.; Villegas, M.V. Microbiological and Clinical Aspects of Raoultella spp. Front. Public Health 2021, 9, 686789. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Berglund, B.; Wang, S.; Zhou, Z.; Gu, C.; Zhao, L.; Meng, C.; Li, X. Emergence of blaNDM-1, blaNDM-5, blaKPC-2 and blaIMP-4 carrying plasmids in Raoultella spp. in the environment. Environ. Pollut. 2022, 306, 119437. [Google Scholar] [CrossRef] [PubMed]
- Ababneh, Q.; Al-Rousan, E.; Jaradat, Z. Fresh produce as a potential vehicle for transmission of Acinetobacter baumannii. Int. J. Food Contam. 2022, 9, 5. [Google Scholar] [CrossRef]
- Hou, C.; Yang, F. Drug-resistant gene of blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58 in Acinetobacter baumannii. Int. J. Clin. Exp. Med. 2015, 8, 13859–13863. [Google Scholar]
- Rafei, R.; Hamze, M.; Pailhoriès, H.; Eveillard, M.; Marsollier, L.; Joly-Guillou, M.-L.; Dabboussi, F.; Kempf, M. Extrahuman Epidemiology of Acinetobacter baumannii in Lebanon. Appl. Environ. Microbiol. 2015, 81, 2359–2367. [Google Scholar] [CrossRef] [PubMed]
- Carvalheira, A.; Silva, J.; Teixeira, P. Lettuce and fruits as a source of multidrug resistant Acinetobacter spp. Food Microbiol. 2017, 64, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Allydice-Francis, K.; Brown, P.D. Diversity of Antimicrobial Resistance and Virulence Determinants in Pseudomonas aeru-ginosa Associated with Fresh Vegetables. Int. J. Microbiol. 2012, 2012, 426241. [Google Scholar] [CrossRef] [PubMed]
- Mojica, M.F.; Humphries, R.; Lipuma, J.J.; Mathers, A.J.; Rao, G.G.; Shelburne, S.A.; Fouts, D.E.; Van Duin, D.; Bonomo, R.A. Clinical challenges treating Stenotrophomonas maltophilia infections: An update. JAC-Antimicrob. Resist. 2022, 4, dlac040. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yao, X.; Luo, J.; Lv, L.; Zeng, Z.; Liu, J.H. Emergence of Escherichia coli coproducing NDM-1 and KPC-2 carbapenemases from a retail vegetable, China. J. Antimicrob. Chemother. 2018, 73, 252–254. [Google Scholar] [CrossRef]
- Evans, B.; Amyes, S.G.B. OXA β-Lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef]
- Karumathil, D.P.; Yin, H.-B.; Kollanoor-Johny, A.; Venkitanarayanan, K. Prevalence of Multidrug-Resistant Bacteria on Fresh Vegetables Collected from Farmers’ Markets in Connecticut. J. Food Prot. 2016, 79, 1446–1451. [Google Scholar] [CrossRef]
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the Role of Vegetables in Spreading Antimicrobial-Resistant Bacteria: A Need for Quantitative Risk Assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- Cherif-Antar, A.; Moussa–Boudjemâa, B.; Didouh, N.; Medjahdi, K.; Mayo, B.; Flórez, A.B. Diversity and biofilm-forming capability of bacteria recovered from stainless steel pipes of a milk-processing dairy plant. Dairy Sci. Technol. 2016, 96, 27–38. [Google Scholar] [CrossRef]
- Khosravi, A.D.; Sadeghi, P.; Shahraki, A.H.; Heidarieh, P.; Sheikhi, N. Molecular Methods for Identi fi cation of Acinetobacter Species by Partial Sequencing of the rpoB and 16S rRNA Genes. J. Clin. Diagn. Res. 2015, 9, DC09–DC13. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. In Twenty-Fifth Informational Supplement; CLSI Document M100-S25; Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2015. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- EUCAST. EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. Available online: http://www.eucast.org (accessed on 21 November 2022).
- EUCAST. New Definitions of S, I and R from 2019. Available online: https://www.eucast.org/newsiandr (accessed on 23 January 2023).
- Mateos, M.; Hernández-García, M.; del Campo, R.; Martínez-García, L.; Gijón, D.; Morosini, M.I.; Ruiz-Garbajosa, P.; Cantón, R. Emergence and Persistence over Time of Carbapenemase-Producing Enterobacter Isolates in a Spanish University Hospital in Madrid, Spain (2005–2018). Microb. Drug Resist. 2021, 27, 895–903. [Google Scholar] [CrossRef] [PubMed]
- González, D.; Gallagher, E.; Zúñiga, T.; Leiva, J.; Vitas, A.I. Prevalence and characterization of β-lactamase-producing Enterobacteriaceae in healthy human carriers. Int. Microbiol. 2020, 23, 171–177. [Google Scholar] [CrossRef]
- Pérez, C.D.-A.; López-Fresneña, N.; Carlavilla, A.L.R.; Garcia, M.H.; Ruiz-Garbajosa, P.; Aranaz-Andrés, J.M.; Maechler, F.; Gastmeier, P.; Bonten, M.J.M.; Canton, R. Local prevalence of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae intestinal carriers at admission and co-expression of ESBL and OXA-48 carbapenemase in Klebsiella pneumoniae: A prevalence survey in a Spanish University Hospital. BMJ Open 2019, 9, e024879. [Google Scholar] [CrossRef]
- Kozak, G.K.; Boerlin, P.; Janecko, N.; Reid-Smith, R.J.; Jardine, C. Antimicrobial Resistance in Escherichia coli Isolates from Swine and Wild Small Mammals in the Proximity of Swine Farms and in Natural Environments in Ontario, Canada. Appl. Environ. Microbiol. 2009, 75, 559–566. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).