Phage Therapy as an Alternative Treatment Modality for Resistant Staphylococcus aureus Infections
Abstract
1. Introduction
2. Narrative Review
2.1. Staphylococci
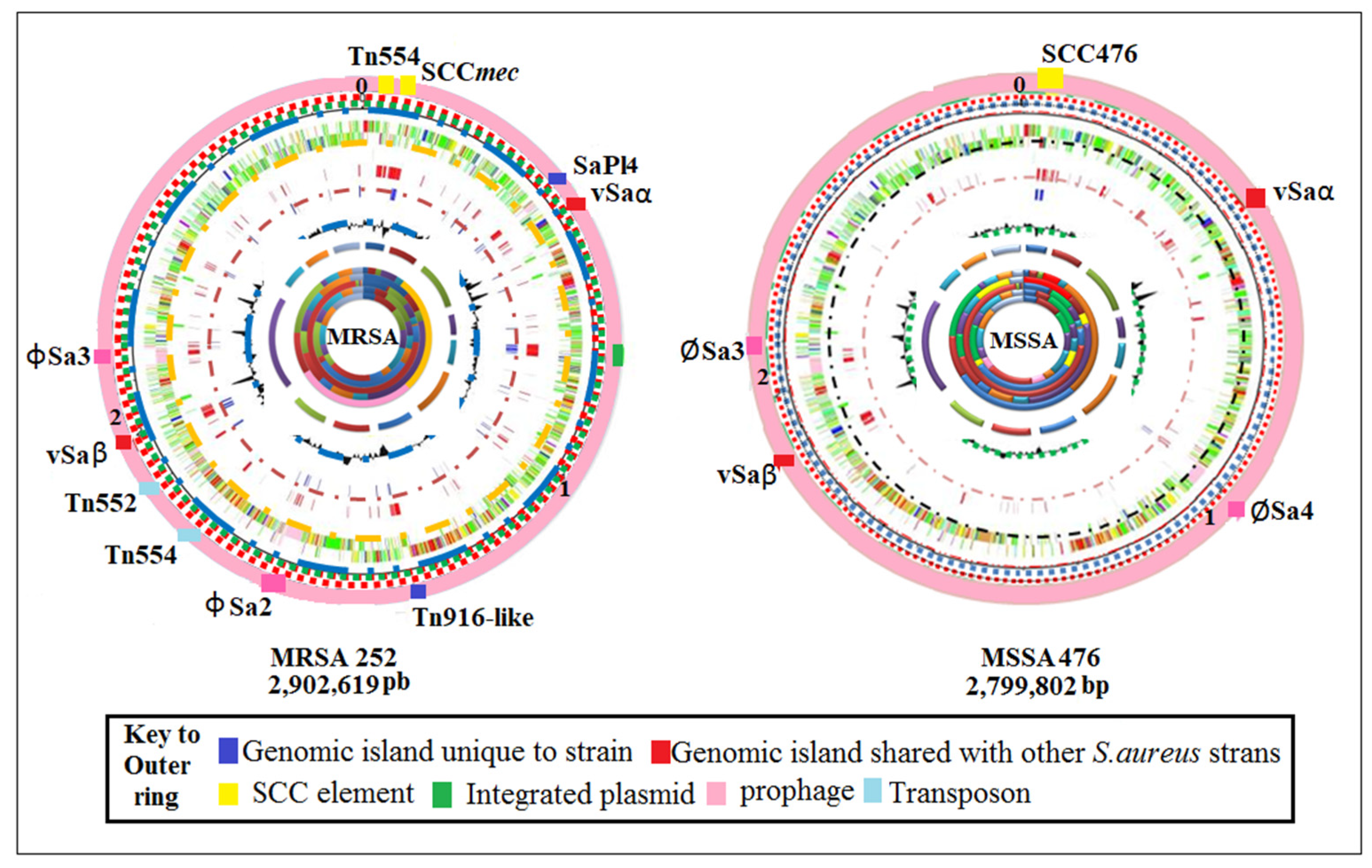
2.2. Genetic Structure of Staphylococcus aureus
2.3. Staphylococcal Cassette Chromosome (SCC)
2.4. Update on Treatment of S. aureus Infections and Its Associated Challenges
2.5. Resistance to Antimicrobial Therapy
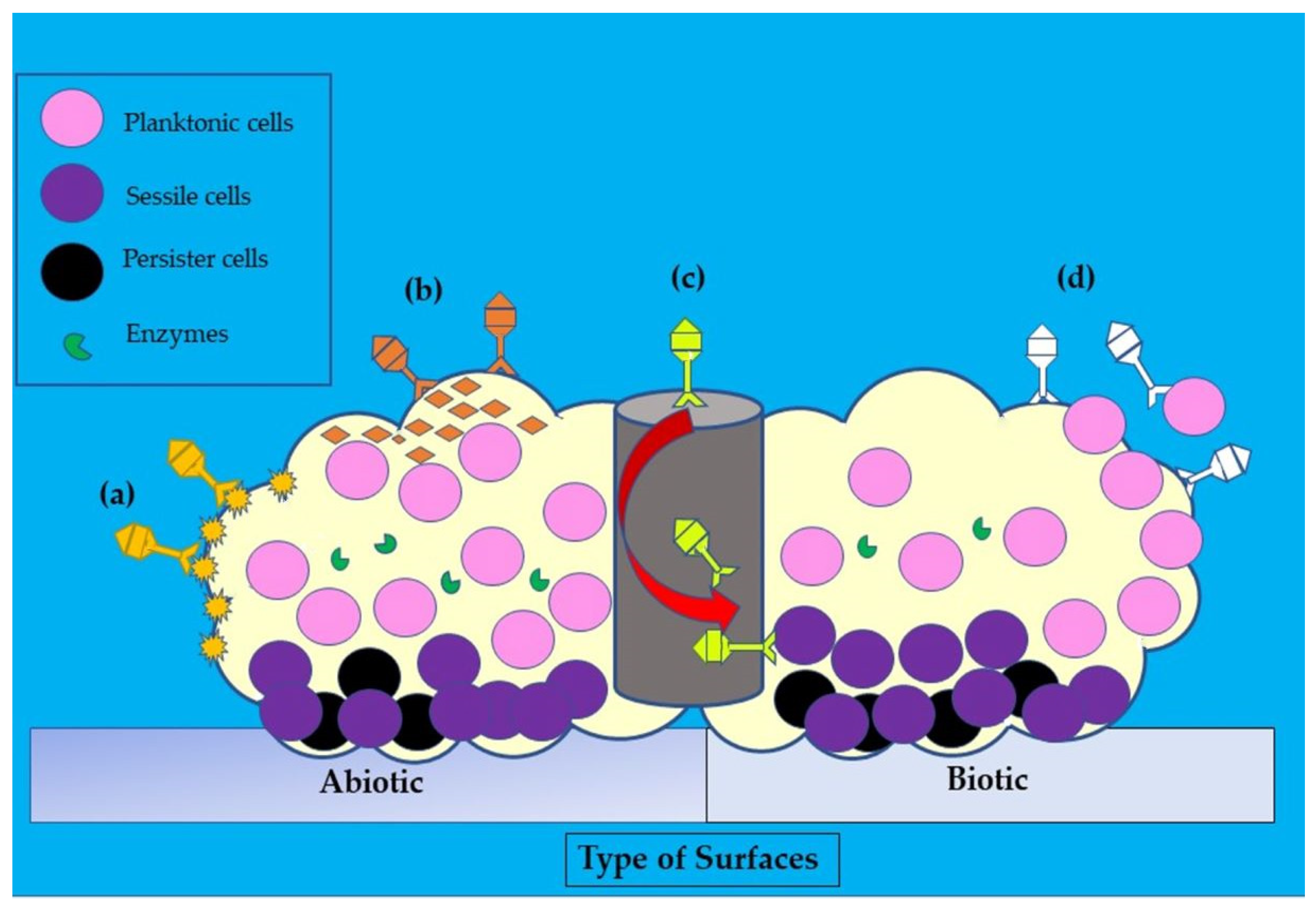
2.6. Role of Biofilm Formation in S. aureus Infections
2.7. Phage Therapy
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019, 57, e01405–e01418. [Google Scholar] [CrossRef] [PubMed]
- Trend, S.; Fonceca, A.M.; Ditcham, W.G.; Kicic, A.; Cf, A. The potential of phage therapy in cystic fibrosis: Essential human-bacterial-phage interactions and delivery considerations for use in Pseudomonas aeruginosa-infected airways. J. Cyst. Fibros. 2017, 16, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Kodong, F.R.; Shanono, N.M.; AL-Jaberi, M.A.A. The monitoring infectious diseases diffusion through GIS. SciTech Framew. 2020, 2, 23–33. [Google Scholar]
- Pincus, N.B.; Reckhow, J.D.; Saleem, D.; Jammeh, M.L.; Datta, S.K.; Myles, I.A. Strain Specific Phage Treatment for Staphylococcus aureus Infection Is Influenced by Host Immunity and Site of Infection. PLoS ONE 2015, 10, e0124280. [Google Scholar] [CrossRef] [PubMed]
- Breederveld, R.S. Phage therapy 2.0: Where do we stand? Lancet Infect. Dis. 2019, 19, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Fish, R.; Kutter, E.; Bryan, D.; Wheat, G.; Kuhl, S. Resolving Digital Staphylococcal Osteomyelitis Using Bacteriophage—A Case Report. Antibiotics 2018, 7, 87. [Google Scholar] [CrossRef]
- Morozova, V.V.; Vlassov, V.V.; Tikunova, N.V. Applications of Bacteriophages in the Treatment of Localized Infections in Humans. Front. Microbiol. 2018, 9, 1696. [Google Scholar] [CrossRef]
- Petrovic Fabijan, A.; Lin, R.C.Y.; Ho, J.; Maddocks, S.; Ben Zakour, N.L.; Iredell, J.R.; Khalid, A.; Venturini, C.; Chard, R.; Morales, S.; et al. Safety of bacteriophage therapy in severe Staphylococcus aureus infection. Nat. Microbiol. 2020, 5, 465–472. [Google Scholar] [CrossRef]
- Oduor, J.M.O.; Kadija, E.; Nyachieo, A.; Mureithi, M.W.; Skurnik, M. Bioprospecting Staphylococcus Phages with Therapeutic and Bio-Control Potential. Viruses 2020, 12, 133. [Google Scholar] [CrossRef]
- Kornienko, M.; Kuptsov, N.; Gorodnichev, R.; Bespiatykh, D.; Guliaev, A.; Letarova, M.; Kulikov, E.; Veselovsky, V.; Malakhova, M.; Letarov, A.; et al. Contribution of Podoviridae and Myoviridae bacteriophages to the effectiveness of anti-staphylococcal therapeutic cocktails. Sci. Rep. 2020, 10, 18612. [Google Scholar] [CrossRef]
- McCallin, S.; Sarker, S.A.; Sultana, S.; Oechslin, F.; Brüssow, H. Metagenome analysis of Russian and Georgian Pyophage cocktails and a placebo-controlled safety trial of single phage versus phage cocktail in healthy Staphylococcus aureus carriers. Environ. Microbiol. 2018, 20, 3278–3293. [Google Scholar] [CrossRef] [PubMed]
- Romero-Calle, D.; Guimarães Benevides, R.; Góes-Neto, A.; Billington, C. Bacteriophages as Alternatives to Antibiotics in Clinical Care. Antibiotics 2019, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Taylor, T.A.; Unakal, C.G. Staphylococcus aureus. In StatPearls [Internet]; StatPearls Publishing: Tampa, FL, USA, 2022. [Google Scholar]
- Brock, T.D.; Madigan, M.T.; Martinko, J.M.; Parker, J. Brock Biology of Microorganisms, 10th ed.; Prentice-Hall: Upper Saddle River, NJ, USA, 2003. [Google Scholar]
- Kållberg, C.; Årdal, C.; Salvesen Blix, H.; Klein, E.; Martinez, E.M.; Lindbæk, M.; Outterson, K.; Røttingen, J.-A.; Laxminarayan, R. Introduction and geographic availability of new antibiotics approved between 1999 and 2014. PLoS ONE 2018, 13, e0205166. [Google Scholar] [CrossRef]
- Molina-Manso, D.; del Prado, G.; Ortiz-Pérez, A.; Manrubia-Cobo, M.; Gómez-Barrena, E.; Cordero-Ampuero, J.; Esteban, J. In vitro susceptibility to antibiotics of staphylococci in biofilms isolated from orthopaedic infections. Int. J. Antimicrob. Agents 2013, 41, 521–523. [Google Scholar] [CrossRef] [PubMed]
- Kreiswirth, B.; Kornblum, J.; Arbeit, R.D.; Eisner, W.; Maslow, J.N.; McGeer, A.; Low, D.E.; Novick, R.P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 1993, 259, 227–230. [Google Scholar] [CrossRef]
- Rad, E.G. Molecular Epidemiology and Therapeutic Potential of Persian Shallot (Allium ascalonicum L.) in the Management of Methicillin Resistant Staphylococcus Aureus Infection. Ph.D. Thesis, Universiti Putra Malaysia, Selangor, Malaysia, 2010. [Google Scholar]
- Dickmann, P.; Keeping, S.; Döring, N.; Schmidt, A.E.; Binder, C.; Ariño-Blasco, S.; Gil, J. Communicating the Risk of MRSA: The Role of Clinical Practice, Regulation and Other Policies in Five European Countries. Front. Public Health 2017, 5, 44. [Google Scholar] [CrossRef]
- Palavecino, E. Clinical, Epidemiological, and Laboratory Aspects of Methicillin-Resistant Staphylococcus aureus (MRSA) Infections. In Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols; Ji, Y., Ed.; Humana Press: Totowa, NJ, USA, 2007; pp. 1–19. [Google Scholar]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef] [PubMed]
- Howden Benjamin, P.; Seemann, T.; Harrison Paul, F.; McEvoy Chris, R.; Stanton Jo-Ann, L.; Rand Christy, J.; Mason Chris, W.; Jensen Slade, O.; Firth, N.; Davies John, K.; et al. Complete Genome Sequence of Staphylococcus aureus Strain JKD6008, an ST239 Clone of Methicillin-Resistant Staphylococcus aureus with Intermediate-Level Vancomycin Resistance. J. Bacteriol. 2010, 192, 5848–5849. [Google Scholar] [CrossRef] [PubMed]
- Pang, R.; Wu, S.; Zhang, F.; Huang, J.; Wu, H.; Zhang, J.; Li, Y.; Ding, Y.; Zhang, J.; Chen, M.; et al. The Genomic Context for the Evolution and Transmission of Community-Associated Staphylococcus aureus ST59 Through the Food Chain. Front. Microbiol. 2020, 11, 422. [Google Scholar] [CrossRef]
- El Garch, F.; Hallin, M.; De Mendonça, R.; Denis, O.; Lefort, A.; Struelens, M.J. StaphVar-DNA microarray analysis of accessory genome elements of community-acquired methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2009, 63, 877–885. [Google Scholar] [CrossRef]
- Slott Jensen, M.L.; Nielsine Skov, M.; Pries Kristiansen, H.; Toft, A.; Lundgaard, H.; Gumpert, H.; Westh, H.; Holm, A.; Kolmos, H.J.; Kemp, M. Core genome multi-locus sequence typing as an essential tool in a high-cost livestock-associated meticillin-resistant Staphylococcus aureus CC398 hospital outbreak. J. Hosp. Infect. 2020, 104, 574–581. [Google Scholar] [CrossRef]
- Balaky, S. The Effect of Antibiotics on Toxin Gene Expression in PVL-positive Staphylococcus aureus Strains. Ph.D. Thesis, Durham University, Durham, UK, 2011. [Google Scholar]
- Holden, M.T.G.; Feil, E.J.; Lindsay, J.A.; Peacock, S.J.; Day, N.P.J.; Enright, M.C.; Foster, T.J.; Moore, C.E.; Hurst, L.; Atkin, R.; et al. Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 2004, 101, 9786–9791. [Google Scholar] [CrossRef]
- Ioanas, M.; Lode, H. Linezolid in VAP by MRSA: A better choice? Intensive Care Med. 2004, 30, 343–346. [Google Scholar] [CrossRef]
- Livermore, D.M. Linezolid in vitro: Mechanism and antibacterial spectrum. J. Antimicrob. Chemother. 2003, 51, ii9–ii16. [Google Scholar] [CrossRef]
- Ito, T.; Ma Xiao, X.; Takeuchi, F.; Okuma, K.; Yuzawa, H.; Hiramatsu, K. Novel Type V Staphylococcal Cassette Chromosome mec Driven by a Novel Cassette Chromosome Recombinase, ccrC. Antimicrob. Agents Chemother. 2004, 48, 2637–2651. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.-A.; Elsayed, S.; Conly John, M. Novel Staphylococcal Cassette Chromosome mec Type, Tentatively Designated Type VIII, Harboring Class A mec and Type 4 ccr Gene Complexes in a Canadian Epidemic Strain of Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 531–540. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Chongtrakool, P.; Ito, T.; Ma Xiao, X.; Kondo, Y.; Trakulsomboon, S.; Tiensasitorn, C.; Jamklang, M.; Chavalit, T.; Song, J.-H.; Hiramatsu, K. Staphylococcal Cassette Chromosome mec (SCCmec) Typing of Methicillin-Resistant Staphylococcus aureus Strains Isolated in 11 Asian Countries: A Proposal for a New Nomenclature for SCCmec Elements. Antimicrob. Agents Chemother. 2006, 50, 1001–1012. [Google Scholar] [CrossRef]
- Rasmussen, R.V.; Fowler Jr, V.G.; Skov, R.; Bruun, N.E. Future challenges and treatment of Staphylococcus aureus bacteremia with emphasis on MRSA. Future Microbiol. 2010, 6, 43–56. [Google Scholar] [CrossRef]
- Kavanagh, K.T.; Abusalem, S.; Calderon, L.E. View point: Gaps in the current guidelines for the prevention of Methicillin-resistant Staphylococcus aureus surgical site infections. Antimicrob. Resist. Infect. Control 2018, 7, 112. [Google Scholar] [CrossRef]
- Brown, N.M.; Goodman, A.L.; Horner, C.; Jenkins, A.; Brown, E.M. Treatment of methicillin-resistant Staphylococcus aureus (MRSA): Updated guidelines from the UK. JAC-Antimicrob. Resist. 2021, 3, dlaa114. [Google Scholar] [CrossRef] [PubMed]
- Vonberg, R.P.; Weitzel-Kage, D.; Behnke, M.; Gastmeier, P. Worldwide Outbreak Database: The largest collection of nosocomial outbreaks. Infection 2011, 39, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Rubin, I.M.; Hansen, T.A.; Klingenberg, A.M.; Petersen, A.M.; Worning, P.; Westh, H.; Bartels, M.D. A Sporadic Four-Year Hospital Outbreak of a ST97-IVa MRSA With Half of the Patients First Identified in the Community. Front. Microbiol. 2018, 9, 1494. [Google Scholar] [CrossRef] [PubMed]
- Talan, D.A.; Mower, W.R.; Krishnadasan, A.; Abrahamian, F.M.; Lovecchio, F.; Karras, D.J.; Steele, M.T.; Rothman, R.E.; Hoagland, R.; Moran, G.J. Trimethoprim–Sulfamethoxazole versus Placebo for Uncomplicated Skin Abscess. N. Engl. J. Med. 2016, 374, 823–832. [Google Scholar] [CrossRef]
- Talan, D.A.; Moran, G.J.; Krishnadasan, A.; Abrahamian, F.M.; Lovecchio, F.; Karras, D.J.; Steele, M.T.; Rothman, R.E.; Mower, W.R. Subgroup Analysis of Antibiotic Treatment for Skin Abscesses. Ann. Emerg. Med. 2018, 71, 21–30. [Google Scholar] [CrossRef]
- Nathwani, D.; Morgan, M.; Masterton, R.G.; Dryden, M.; Cookson, B.D.; French, G.; Lewis, D.; on behalf of the British Society for Antimicrobial Chemotherapy Working Party on Community-onset MRSA Infections. Guidelines for UK practice for the diagnosis and management of methicillin-resistant Staphylococcus aureus (MRSA) infections presenting in the community. J. Antimicrob. Chemother. 2008, 61, 976–994. [Google Scholar] [CrossRef]
- Tanus, T.; Scangarella-Oman, N.E.; Dalessandro, M.; Li, G.; Breton, J.J.; Tomayko, J.F. A Randomized, Double-blind, Comparative Study to Assess the Safety and Efficacy of Topical Retapamulin Ointment 1% Versus Oral Linezolid in the Treatment of Secondarily Infected Traumatic Lesions and Impetigo Due to Methicillin-Resistant Staphylococcus aureus. Adv. Ski. Wound Care 2014, 27, 548–559. [Google Scholar]
- Noel, G.J.; Bush, K.; Bagchi, P.; Ianus, J.; Strauss, R.S. A Randomized, Double-Blind Trial Comparing Ceftobiprole Medocaril with Vancomycin plus Ceftazidime for the Treatment of Patients with Complicated Skin and Skin-Structure Infections. Clin. Infect. Dis. 2008, 46, 647–655. [Google Scholar] [CrossRef]
- Boucher, H.W.; Wilcox, M.; Talbot, G.H.; Puttagunta, S.; Das, A.F.; Dunne, M.W. Once-Weekly Dalbavancin versus Daily Conventional Therapy for Skin Infection. New Engl. J. Med. 2014, 370, 2169–2179. [Google Scholar] [CrossRef]
- Moran, G.J.; Fang, E.; Corey, G.R.; Das, A.F.; De Anda, C.; Prokocimer, P. Tedizolid for 6 days versus linezolid for 10 days for acute bacterial skin and skin-structure infections (ESTABLISH-2): A randomised, double-blind, phase 3, non-inferiority trial. Lancet Infect. Dis. 2014, 14, 696–705. [Google Scholar] [CrossRef]
- John, J., Jr. The treatment of resistant staphylococcal infections. F1000Research 2020, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Gould, F.K.; Brindle, R.; Chadwick, P.R.; Fraise, A.P.; Hill, S.; Nathwani, D.; Ridgway, G.L.; Spry, M.J.; Warren, R.E.; on behalf of the Joint Working Party of the British Society for Antimicrobial Chemotherapy, Hospital Infection Society and Infection Control Nurses Association. Guidelines (2008) for the prophylaxis and treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in the United Kingdom. J. Antimicrob. Chemother. 2009, 63, 849–861. [Google Scholar] [CrossRef]
- Rubinstein, E.; Lalani, T.; Corey, G.R.; Kanafani, Z.A.; Nannini, E.C.; Rocha, M.G.; Rahav, G.; Niederman, M.S.; Kollef, M.H.; Shorr, A.F.; et al. Telavancin versus Vancomycin for Hospital-Acquired Pneumonia due to Gram-positive Pathogens. Clin. Infect. Dis. 2011, 52, 31–40. [Google Scholar] [CrossRef]
- Silverman, J.A.; Mortin, L.I.; VanPraagh, A.D.G.; Li, T.; Alder, J. Inhibition of Daptomycin by Pulmonary Surfactant: In Vitro Modeling and Clinical Impact. J. Infect. Dis. 2005, 191, 2149–2152. [Google Scholar] [CrossRef] [PubMed]
- Thitiananpakorn, K.; Aiba, Y.; Tan, X.-E.; Watanabe, S.; Kiga, K.; Sato’o, Y.; Boonsiri, T.; Li, F.-Y.; Sasahara, T.; Taki, Y.; et al. Association of mprF mutations with cross-resistance to daptomycin and vancomycin in methicillin-resistant Staphylococcus aureus (MRSA). Sci. Rep. 2020, 10, 16107. [Google Scholar] [CrossRef] [PubMed]
- Dunne Michael, W.; Puttagunta, S.; Sprenger Craig, R.; Rubino, C.; Van Wart, S.; Baldassarre, J. Extended-Duration Dosing and Distribution of Dalbavancin into Bone and Articular Tissue. Antimicrob. Agents Chemother. 2015, 59, 1849–1855. [Google Scholar] [CrossRef] [PubMed]
- Filippone, E.J.; Kraft, W.K.; Farber, J.L. The Nephrotoxicity of Vancomycin. Clin. Pharmacol. Ther. 2017, 102, 459–469. [Google Scholar] [CrossRef]
- Liu, C.; Bayer, A.; Cosgrove, S.E.; Daum, R.S.; Fridkin, S.K.; Gorwitz, R.J.; Kaplan, S.L.; Karchmer, A.W.; Levine, D.P.; Murray, B.E.; et al. Clinical Practice Guidelines by the Infectious Diseases Society of America for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections in Adults and Children. Clin. Infect. Dis. 2011, 52, e18–e55. [Google Scholar] [CrossRef]
- Choo, E.J.; Chambers, H.F. Treatment of Methicillin-Resistant Staphylococcus aureus Bacteremia. Infect. Chemother. 2016, 48, 267–273. [Google Scholar] [CrossRef]
- Demir, A.; Çamlar, M.; Kuşçu, G.C.; Gürel, Ç.; Oltulu, F.; Oren, M.; Aldağ, C.; Karabay Yavaşoğlu, N.Ü.; Sandal, E.; Özer, F. How Safe Is the Use of Intrathecal Vancomycin? World Neurosurg. 2022, 160, e55–e60. [Google Scholar] [CrossRef]
- Gould, F.K.; Denning, D.W.; Elliott, T.S.J.; Foweraker, J.; Perry, J.D.; Prendergast, B.D.; Sandoe, J.A.T.; Spry, M.J.; Watkin, R.W. Guidelines for the diagnosis and antibiotic treatment of endocarditis in adults: A report of the Working Party of the British Society for Antimicrobial Chemotherapy. J. Antimicrob. Chemother. 2012, 67, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Morrisette, T.; Alosaimy, S.; Abdul-Mutakabbir, J.C.; Kebriaei, R.; Rybak, M.J. The Evolving Reduction of Vancomycin and Daptomycin Susceptibility in MRSA—Salvaging the Gold Standards with Combination Therapy. Antibiotics 2020, 9, 762. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2020, 21, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.A.; Enright Mark, C. Evolutionary Models of the Emergence of Methicillin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 3926–3934. [Google Scholar] [CrossRef]
- Rolo, J.; Worning, P.; Boye Nielsen, J.; Sobral, R.; Bowden, R.; Bouchami, O.; Damborg, P.; Guardabassi, L.; Perreten, V.; Westh, H.; et al. Evidence for the evolutionary steps leading to mecA-mediated β-lactam resistance in staphylococci. PLOS Genet. 2017, 13, e1006674. [Google Scholar] [CrossRef]
- Cafini, F.; Romero, V.M.; Morikawa, K. Mechanisms of horizontal gene transfer. In The Rise of Virulence Antibiotic Resistance in Staphylococcus aureus; Intech Open: Rijeka, Croatia, 2017; Volume 61. [Google Scholar]
- Babakhani, S.; Oloomi, M. Transposons: The agents of antibiotic resistance in bacteria. J. Basic Microbiol. 2018, 58, 905–917. [Google Scholar] [CrossRef]
- Pillar, C.M.; Draghi, D.C.; Sheehan, D.J.; Sahm, D.F. Prevalence of multidrug-resistant, methicillin-resistant Staphylococcus aureus in the United States: Findings of the stratified analysis of the 2004 to 2005 LEADER Surveillance Programs. Diagn. Microbiol. Infect. Dis. 2008, 60, 221–224. [Google Scholar] [CrossRef]
- National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2003, issued August 2003. Am. J. Infect. Control 2003, 31, 481–498. [Google Scholar] [CrossRef]
- National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 2004, 32, 470–485. [Google Scholar] [CrossRef]
- Ehsanollah, G.; Chong, P.; Zamberi, S.; Mariana, N. Anti-adhesive and anti-invasive activities of an oil based di-herbal extract against methicillin resistant Staphylococcus aureus. Afr. J. Microbiol. Res. 2009, 3, 272–275. [Google Scholar]
- Appelbaum, P.C.; Bozdogan, B. Vancomycin resistance in Staphylococcus aureus. Clin. Lab. Med. 2004, 24, 381–402. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; McDonald, L.C. Vancomycin-resistant staphylococci and enterococci: Epidemiology and control. Curr. Opin. Infect. Dis. 2005, 18, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.F.J.; Edwards, D.I.; Hawkey, P.M.; Morrison, D.; Ridgway, G.L.; Towner, K.J.; Wren, M.W.D.; on behalf of the Joint Working Party of the British Society for Antimicrobial Chemotherapy, Hospital Infection Society and Infection Control Nurses Association. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J. Antimicrob. Chemother. 2005, 56, 1000–1018. [Google Scholar] [CrossRef] [PubMed]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2011, 2, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef]
- Chain, E.; Florey, H.W.; Gardner, A.D.; Heatley, N.G.; Jennings, M.A.; Orr-Ewing, J.; Sanders, A.G. Penicillin as a chemotherapeutic agent. Lancet 1940, 236, 226–228. [Google Scholar] [CrossRef]
- Bauer, A. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966, 45, 149–158. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef]
- Jevons, M.P. Celebin-resistant staphylococci. Br Med. J. 1961, 1, 124–125. [Google Scholar] [CrossRef]
- Kowalski, T.J.; Berbari, E.F.; Osmon, D.R. Epidemiology, Treatment, and Prevention of Community-Acquired Methicillin-Resistant Staphylococcus aureus Infections. Mayo Clin. Proc. 2005, 80, 1201–1208. [Google Scholar] [CrossRef]
- Martínez-Meléndez, A.; Morfín-Otero, R.; Villarreal-Treviño, L.; González-González, G.; Llaca-Díaz, J.; Rodríguez-Noriega, E.; Camacho-Ortíz, A.; Garza-González, E. Staphylococcal Cassette Chromosome mec (SCCmec) in coagulase negative staphylococci. Med. Univ. 2015, 17, 229–233. [Google Scholar] [CrossRef]
- Antignac, A.; Tomasz, A. Reconstruction of the Phenotypes of Methicillin-Resistant Staphylococcus aureus by Replacement of the Staphylococcal Cassette Chromosome mec with a Plasmid-Borne Copy of Staphylococcus sciuri pbpD Gene. Antimicrob. Agents Chemother. 2009, 53, 435–441. [Google Scholar] [CrossRef] [PubMed]
- Lee John, H. Methicillin (Oxacillin)-Resistant Staphylococcus aureus Strains Isolated from Major Food Animals and Their Potential Transmission to Humans. Appl. Environ. Microbiol. 2003, 69, 6489–6494. [Google Scholar] [CrossRef]
- Howden Benjamin, P.; Davies John, K.; Johnson Paul, D.R.; Stinear Timothy, P.; Grayson, M.L. Reduced Vancomycin Susceptibility in Staphylococcus aureus, Including Vancomycin-Intermediate and Heterogeneous Vancomycin-Intermediate Strains: Resistance Mechanisms, Laboratory Detection, and Clinical Implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef] [PubMed]
- Katzif, S.; Lee, E.-H.; Law Anthony, B.; Tzeng, Y.-L.; Shafer William, M. CspA Regulates Pigment Production in Staphylococcus aureus through a SigB-Dependent Mechanism. J. Bacteriol. 2005, 187, 8181–8184. [Google Scholar] [CrossRef] [PubMed]
- Périchon, B.; Courvalin, P. VanA-Type Vancomycin-Resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 4580–4587. [Google Scholar] [CrossRef]
- Mussa, E.A.M.; Alsalahi, A.; Aljaberi, M.A.; Jasni, A.S.; Desa, M.N.M.; Al-Mahdi, A.Y.M.; Hamat, R.A. Acquired tetracycline resistance genes by transposons and virulence factors in enterococci recovered from overland and aquatic animals: A systematic review. Rev. Aquac. 2022, 14, 399–413. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Monroe, D. Looking for Chinks in the Armor of Bacterial Biofilms. PLOS Biol. 2007, 5, e307. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Hsu, L.-Y.; Koh, T.-H.; Singh, K.; Kang, M.-L.; Kurup, A.; Tan, B.-H. Dissemination of Multisusceptible Methicillin-Resistant Staphylococcus aureus in Singapore. J. Clin. Microbiol. 2005, 43, 2923–2925. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Stenz, L. The GTP-Dependant Pleiotropic Repressor” CodY” Regulates Biofilm Formation in Staphylococcus aureus. Ph.D. Thesis, University of Geneva, Geneva, Switzerland, 2011. [Google Scholar]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef]
- Akturk, E.; Oliveira, H.; Santos, S.B.; Costa, S.; Kuyumcu, S.; Melo, L.D.R.; Azeredo, J. Synergistic Action of Phage and Antibiotics: Parameters to Enhance the Killing Efficacy Against Mono and Dual-Species Biofilms. Antibiotics 2019, 8, 103. [Google Scholar] [CrossRef]
- Dickey, J.; Perrot, V. Adjunct phage treatment enhances the effectiveness of low antibiotic concentration against Staphylococcus aureus biofilms in vitro. PLoS ONE 2019, 14, e0209390. [Google Scholar] [CrossRef]
- Wang, L.; Tkhilaishvili, T.; Trampuz, A.; Gonzalez Moreno, M. Evaluation of Staphylococcal Bacteriophage Sb-1 as an Adjunctive Agent to Antibiotics Against Rifampin-Resistant Staphylococcus aureus Biofilms. Front. Microbiol. 2020, 11, 602057. [Google Scholar] [CrossRef]
- Pires, D.P.; Meneses, L.; Brandão, A.C.; Azeredo, J. An overview of the current state of phage therapy for the treatment of biofilm-related infections. Curr. Opin. Virol. 2022, 53, 101209. [Google Scholar] [CrossRef]
- Mitchell, A.A. The Development and Spread of Antibiotic Resistant Strains of Staphylococcus aureus in a General Hospital; University of Glasgow: Glasgow, UK, 1970. [Google Scholar]
- Atshan, S.S.; Hamat, R.A.; Coolen, M.J.L.; Dykes, G.; Sekawi, Z.; Mullins, B.J.; Than, L.T.; Abduljaleel, S.A.; Kicic, A. The Role of Subinhibitory Concentrations of Daptomycin and Tigecycline in Modulating Virulence in Staphylococcus aureus. Antibiotics 2021, 10, 39. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Reza, A.; Farid, A.J.; Zamberi, S.; Amini, R.; Sajedeh, K.; Ahmad, N.; Hassan, A.; Norkhoda, S.; Morovat, T.; Iraj, P. Dynamics of bacteriophages as a promising antibiofilm agents. J. Pure Appl. Microbiol. 2014, 8, 1015–1019. [Google Scholar]
- Miernikiewicz, P.; Dąbrowska, K.; Piotrowicz, A.; Owczarek, B.; Wojas-Turek, J.; Kicielińska, J.; Rossowska, J.; Pajtasz-Piasecka, E.; Hodyra, K.; Macegoniuk, K.; et al. T4 Phage and Its Head Surface Proteins Do Not Stimulate Inflammatory Mediator Production. PLoS ONE 2013, 8, e71036. [Google Scholar] [CrossRef] [PubMed]
- Van Belleghem, J.D.; Dąbrowska, K.; Vaneechoutte, M.; Barr, J.J.; Bollyky, P.L. Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Letarov, A.V.; Kulikov, E.E. Adsorption of bacteriophages on bacterial cells. Biochem. 2017, 82, 1632–1658. [Google Scholar] [CrossRef]
- Twort, F.W. An investigation on the nature of ultra-microscopic viruses. Acta Kravsi 1961, 189, 1241–1243. [Google Scholar] [CrossRef]
- d’Herelle, F. On an invisible microbe antagonistic to dysentery bacilli. CR Acad. Sci. 1917, 165, 373–375. [Google Scholar]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef]
- Międzybrodzki, R.; Borysowski, J.; Weber-Dąbrowska, B.; Fortuna, W.; Letkiewicz, S.; Szufnarowski, K.; Pawełczyk, Z.; Rogóż, P.; Kłak, M.; Wojtasik, E.; et al. Chapter 3—Clinical Aspects of Phage Therapy. In Advances in Virus Research; Łobocka, M., Szybalski, W., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 83, pp. 73–121. [Google Scholar]
- Chanishvili, N. Literature Review of the Practical Application of Bacteriophage Research; Nova Science Publishers, Incorporated: New York, NY, USA, 2012. [Google Scholar]
- Chan, B.K.; Abedon, S.T. Chapter 1—Phage Therapy Pharmacology: Phage Cocktails. In Advances in Applied Microbiology; Laskin, A.I., Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 78, pp. 1–23. [Google Scholar]
- Levin, B.R.; Bull, J.J. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2004, 2, 166–173. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Gannon, B.W.; Halfhide, D.E.; Santos, S.B.; Hayes, C.M.; Roe, J.M.; Azeredo, J. The in vivo efficacy of two administration routes of a phage cocktail to reduce numbers of Campylobacter coli and Campylobacter jejuni in chickens. BMC Microbiol. 2010, 10, 232. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.-P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef]
- Viazis, S.; Akhtar, M.; Feirtag, J.; Diez-Gonzalez, F. Reduction of Escherichia coli O157:H7 viability on hard surfaces by treatment with a bacteriophage mixture. Int. J. Food Microbiol. 2011, 145, 37–42. [Google Scholar] [CrossRef]
- Geredew Kifelew, L.; Mitchell, J.G.; Speck, P. Mini-review: Efficacy of lytic bacteriophages on multispecies biofilms. Biofouling 2019, 35, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Li, J.; Nazir, A.; Tong, Y. Bacteriophage—A Promising Alternative Measure for Bacterial Biofilm Control. Infect. Drug Resist. 2021, 14, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; García, P. Bacteriophages as Weapons Against Bacterial Biofilms in the Food Industry. Front. Microbiol. 2016, 7, 825. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.M.; Pereira, M.O. Pseudomonas aeruginosa Diversification during Infection Development in Cystic Fibrosis Lungs—A Review. Pathogens 2014, 3, 680–703. [Google Scholar] [CrossRef]
- Wright, A.; Hawkins, C.H.; Änggård, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef]
- Gu, J.; Liu, X.; Li, Y.; Han, W.; Lei, L.; Yang, Y.; Zhao, H.; Gao, Y.; Song, J.; Lu, R.; et al. A Method for Generation Phage Cocktail with Great Therapeutic Potential. PLoS ONE 2012, 7, e31698. [Google Scholar] [CrossRef]
- Chaudhry, W.N.; Concepción-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and Order Effects of Antibiotics and Phages in Killing Pseudomonas aeruginosa Biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef]
- Tagliaferri, T.L.; Jansen, M.; Horz, H.-P. Fighting Pathogenic Bacteria on Two Fronts: Phages and Antibiotics as Combined Strategy. Front. Cell. Infect. Microbiol. 2019, 9, 22. [Google Scholar] [CrossRef]
- Trend, S.; Chang, B.J.; O’Dea, M.; Stick, S.M.; Kicic, A.; Waerp; WAERP; AusREC; AREST CF. Use of a Primary Epithelial Cell Screening Tool to Investigate Phage Therapy in Cystic Fibrosis. Front. Pharmacol. 2018, 9, 1330. [Google Scholar] [CrossRef]
- Estrella, L.A.; Quinones, J.; Henry, M.; Hannah, R.M.; Pope, R.K.; Hamilton, T.; Teneza-mora, N.; Hall, E.; Biswajit, B. Characterization of novel Staphylococcus aureus lytic phage and defining their combinatorial virulence using the OmniLog® system. Bacteriophage 2016, 6, e1219440. [Google Scholar] [CrossRef]
- Wunderink, R.G.; Niederman, M.S.; Kollef, M.H.; Shorr, A.F.; Kunkel, M.J.; Baruch, A.; McGee, W.T.; Reisman, A.; Chastre, J. Linezolid in Methicillin-Resistant Staphylococcus aureus Nosocomial Pneumonia: A Randomized, Controlled Study. Clin. Infect. Dis. 2012, 54, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Kirby Amy, E.; Garner, K.; Levin Bruce, R. The Relative Contributions of Physical Structure and Cell Density to the Antibiotic Susceptibility of Bacteria in Biofilms. Antimicrob. Agents Chemother. 2012, 56, 2967–2975. [Google Scholar] [CrossRef] [PubMed]
- Lehman, S.M.; Mearns, G.; Rankin, D.; Cole, R.A.; Smrekar, F.; Branston, S.D.; Morales, S. Design and Preclinical Development of a Phage Product for the Treatment of Antibiotic-Resistant Staphylococcus aureus Infections. Viruses 2019, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Chhibber, S.; Shukla, A.; Kaur, S. Transfersomal Phage Cocktail Is an Effective Treatment against Methicillin-Resistant Staphylococcus aureus-Mediated Skin and Soft Tissue Infections. Antimicrob. Agents Chemother. 2017, 61, e02146-16. [Google Scholar] [CrossRef]
- Prazak, J.; Iten, M.; Cameron, D.R.; Save, J.; Grandgirard, D.; Resch, G.; Goepfert, C.; Leib, S.L.; Takala, J.; Jakob, S.M.; et al. Bacteriophages Improve Outcomes in Experimental Staphylococcus aureus Ventilator-associated Pneumonia. Am. J. Respir. Crit. Care Med. 2019, 200, 1126–1133. [Google Scholar] [CrossRef]
- Rahman, M.; Kim, S.; Kim, S.M.; Seol, S.Y.; Kim, J. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 2011, 27, 1087–1093. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, D.; Taha, M.; Yi, Q.; Ramirez-Arcos, S.; Diallo, J.-S.; Carli, A.; Abdelbary, H. Does Treatment Order Matter? Investigating the Ability of Bacteriophage to Augment Antibiotic Activity against Staphylococcus aureus Biofilms. Front. Microbiol. 2018, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Shimamori, Y.; Mitsunaka, S.; Yamashita, H.; Suzuki, T.; Kitao, T.; Kubori, T.; Nagai, H.; Takeda, S.; Ando, H. Staphylococcal Phage in Combination with Staphylococcus epidermidis as a Potential Treatment for Staphylococcus aureus-Associated Atopic Dermatitis and Suppressor of Phage-Resistant Mutants. Viruses 2020, 13, 7. [Google Scholar] [CrossRef]
- Brives, C.; Pourraz, J. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. 2020, 6, 100. [Google Scholar] [CrossRef]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Aslam, S.; Pretorius, V.; Lehman, S.M.; Morales, S.; Schooley, R.T. Novel bacteriophage therapy for treatment of left ventricular assist device infection. J. Heart Lung Transplant. 2019, 38, 475–476. [Google Scholar] [CrossRef]
- Doub, J.B.; Ng, V.Y.; Johnson, A.J.; Slomka, M.; Fackler, J.; Horne, B.A.; Brownstein, M.J.; Henry, M.; Malagon, F.; Biswas, B. Salvage Bacteriophage Therapy for a Chronic MRSA Prosthetic Joint Infection. Antibiotics 2020, 9, 241. [Google Scholar] [CrossRef]
- Gilbey, T.; Ho, J.; Cooley, L.A.; Petrovic Fabijan, A.; Iredell, J.R. Adjunctive bacteriophage therapy for prosthetic valve endocarditis due to Staphylococcus aureus. Med. J. Aust. 2019, 211, 142–143.e141. [Google Scholar] [CrossRef]
- Ferry, T.; Batailler, C.; Petitjean, C.; Chateau, J.; Fevre, C.; Forestier, E.; Brosset, S.; Leboucher, G.; Kolenda, C.; Laurent, F.; et al. The Potential Innovative Use of Bacteriophages Within the DAC® Hydrogel to Treat Patients With Knee Megaprosthesis Infection Requiring “Debridement Antibiotics and Implant Retention” and Soft Tissue Coverage as Salvage Therapy. Front. Med. 2020, 7, 342. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Kolenda, C.; Batailler, C.; Gustave, C.-A.; Lustig, S.; Malatray, M.; Fevre, C.; Josse, J.; Petitjean, C.; Chidiac, C.; et al. Phage Therapy as Adjuvant to Conservative Surgery and Antibiotics to Salvage Patients With Relapsing S. aureus Prosthetic Knee Infection. Front. Med. 2020, 7, 570572. [Google Scholar] [CrossRef]
- Rubalskii, E.; Ruemke, S.; Salmoukas, C.; Boyle, E.C.; Warnecke, G.; Tudorache, I.; Shrestha, M.; Schmitto, J.D.; Martens, A.; Rojas, S.V.; et al. Bacteriophage Therapy for Critical Infections Related to Cardiothoracic Surgery. Antibiotics 2020, 9, 232. [Google Scholar] [CrossRef]
- Ramirez-Sanchez, C.; Gonzales, F.; Buckley, M.; Biswas, B.; Henry, M.; Deschenes, M.V.; Horne, B.A.; Fackler, J.; Brownstein, M.J.; Schooley, R.T.; et al. Successful Treatment of Staphylococcus aureus Prosthetic Joint Infection with Bacteriophage Therapy. Viruses 2021, 13, 1182. [Google Scholar] [CrossRef] [PubMed]
- Rojas, S.V.; Junghans, S.; Fox, H.; Lazouski, K.; Schramm, R.; Morshuis, M.; Gummert, J.F.; Gross, J. Bacteriophage-Enriched Galenic for Intrapericardial Ventricular Assist Device Infection. Antibiotics 2022, 11, 602. [Google Scholar] [CrossRef]
- Püschel, A.; Skusa, R.; Bollensdorf, A.; Gross, J. Local Treatment of Driveline Infection with Bacteriophages. Antibiotics 2022, 11, 1310. [Google Scholar] [CrossRef] [PubMed]
- Doub, J.B.; Ng, V.Y.; Lee, M.; Chi, A.; Lee, A.; Würstle, S.; Chan, B. Salphage: Salvage Bacteriophage Therapy for Recalcitrant MRSA Prosthetic Joint Infection. Antibiotics 2022, 11, 616. [Google Scholar] [CrossRef]
- Schoeffel, J.; Wang, E.W.; Gill, D.; Frackler, J.; Horne, B.A.; Manson, T.; Doub, J.B. Successful Use of Salvage Bacteriophage Therapy for a Recalcitrant MRSA Knee and Hip Prosthetic Joint Infection. Pharmaceuticals 2022, 15, 177. [Google Scholar] [CrossRef]
- Grambow, E.; Junghans, S.; Kröger, J.C.; Reisinger, E.C.; Krause, B.J.; Groß, J. Treatment of an Infected TEVAR with Extra- and Endovascular Bacteriophage Application. EJVES Vasc. Forum 2022, 56, 20–23. [Google Scholar] [CrossRef]
- Onsea, J.; Soentjens, P.; Djebara, S.; Merabishvili, M.; Depypere, M.; Spriet, I.; De Munter, P.; Debaveye, Y.; Nijs, S.; Vanderschot, P.; et al. Bacteriophage Application for Difficult-To-Treat Musculoskeletal Infections: Development of a Standardized Multidisciplinary Treatment Protocol. Viruses 2019, 11, 891. [Google Scholar] [CrossRef] [PubMed]
- Nadareishvili, L.; Hoyle, N.; Nakaidze, N.; Nizharadze, D.; Kutateladze, M.; Balarjishvili, N.; Kutter, E.; Pruidze, N. Bacteriophage Therapy as a Potential Management Option for Surgical Wound Infections. PHAGE 2020, 1, 158–165. [Google Scholar] [CrossRef]
- Van Nieuwenhuyse, B.; Galant, C.; Brichard, B.; Docquier, P.-L.; Djebara, S.; Pirnay, J.-P.; Van der Linden, D.; Merabishvili, M.; Chatzis, O. A Case of In Situ Phage Therapy against Staphylococcus aureus in a Bone Allograft Polymicrobial Biofilm Infection: Outcomes and Phage-Antibiotic Interactions. Viruses 2021, 13, 1898. [Google Scholar] [CrossRef]
- Gupta, P.; Singh, H.S.; Shukla, V.K.; Nath, G.; Bhartiya, S.K. Bacteriophage Therapy of Chronic Nonhealing Wound: Clinical Study. Int. J. Low. Extrem. Wounds 2019, 18, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M.; Woodworth, B.A.; Horne, B.A.; Fackler, J.; Brownstein, M.J. Case Report: Successful use of phage therapy in refractory MRSA chronic rhinosinusitis. Int. J. Infect. Dis. 2022, 121, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Bleibtreu, A.; Fevre, C.; Robert, J.; Haddad, E.; Caumes, E.; Lantieri, L.; Peyre, M. Combining bacteriophages and dalbavancin for salvage therapy of complex Staphylococcus aureus extradural empyema. Médecine Mal. Infect. 2020, 50, 458–459. [Google Scholar] [CrossRef] [PubMed]
- Johri, A.V.; Johri, P.; Hoyle, N.; Pipia, L.; Nadareishvili, L.; Nizharadze, D. Case Report: Chronic Bacterial Prostatitis Treated With Phage Therapy After Multiple Failed Antibiotic Treatments. Front. Pharmacol. 2021, 12, 692614. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, G.S.; Greenberg, R.G.; Hornik, C.D.; Cassino, C.; Ghahramani, P.; Kumar, K.R.; Fowler, V.G.; Cohen-Wolkowiez, M. Safety and Pharmacokinetics of Exebacase in an Infant With Disseminated Staphylococcus aureus Infection Clin. Infect. Dis. 2022, 75, 338–341. [Google Scholar] [CrossRef] [PubMed]
- Schuch, R.; Khan Babar, K.; Raz, A.; Rotolo Jimmy, A.; Wittekind, M. Bacteriophage Lysin CF-301, a Potent Antistaphylococcal Biofilm Agent. Antimicrob. Agents Chemother. 2017, 61, e02666-16. [Google Scholar] [CrossRef]
- Khullar, L.; Harjai, K.; Chhibber, S. Exploring the therapeutic potential of staphylococcal phage formulations: Current challenges and applications in phage therapy. J. Appl. Microbiol. 2022, 132, 3515–3532. [Google Scholar] [CrossRef]
- Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application. Antibiotics 2021, 10, 1497. [Google Scholar] [CrossRef]
- Krylov, V.N. Phagotherapy in terms of bacteriophage genetics: Hopes, perspectives, safety, limitations. Genetika 2001, 37, 869–887. [Google Scholar]
- Huh, H.; Wong, S.; St. Jean, J.; Slavcev, R. Bacteriophage interactions with mammalian tissue: Therapeutic applications. Adv. Drug Deliv. Rev. 2019, 145, 4–17. [Google Scholar] [CrossRef]
- Guerin, E.; Hill, C. Shining Light on Human Gut Bacteriophages. Front. Cell. Infect. Microbiol. 2020, 10, 481. [Google Scholar] [CrossRef] [PubMed]
- Barrow, P.A.; Soothill, J.S. Bacteriophage therapy and prophylaxis: Rediscovery and renewed assessment of potential. Trends Microbiol. 1997, 5, 268–271. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Cha, K.E.; Myung, H. Observation of inflammatory responses in mice orally fed with bacteriophage T7. J. Appl. Microbiol. 2014, 117, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.-P.; De Vos, D.; Verbeken, G.; Merabishvili, M.; Chanishvili, N.; Vaneechoutte, M.; Zizi, M.; Laire, G.; Lavigne, R.; Huys, I.; et al. The Phage Therapy Paradigm: Prêt-à-Porter or Sur-mesure? Pharm. Res. 2011, 28, 934–937. [Google Scholar] [CrossRef]
- Torres-Barceló, C. Phage Therapy Faces Evolutionary Challenges. Viruses 2018, 10, 323. [Google Scholar] [CrossRef]
- Kolenda, C.; Medina, M.; Bonhomme, M.; Laumay, F.; Roussel-Gaillard, T.; Martins-Simoes, P.; Tristan, A.; Pirot, F.; Ferry, T.; Laurent, F.; et al. Phage Therapy against Staphylococcus aureus: Selection and Optimization of Production Protocols of Novel Broad-Spectrum Silviavirus Phages. Pharmaceutics 2022, 14, 1885. [Google Scholar] [CrossRef]
- Sanmukh, S.G.; Santos, N.J.; Barquilha, C.N.; dos Santos, S.A.; Duran, B.O.; Delella, F.K.; Moroz, A.; Justulin, L.A.; Carvalho, H.F.; Felisbino, S.L. Exposure to Bacteriophages T4 and M13 Increases Integrin Gene Expression and Impairs Migration of Human PC-3 Prostate Cancer Cells. Antibiotics 2021, 10, 1202. [Google Scholar] [CrossRef]
- Vellon, L.; Menendez, J.A.; Liu, H.; Lupu, R. Up-regulation of αVβ3 integrin expression is a novel molecular response to chemotherapy-induced cell damage in a heregulin-dependent manner. Differentiation 2007, 75, 819–830. [Google Scholar] [CrossRef]
- Bichet, M.C.; Chin, W.H.; Richards, W.; Lin, Y.-W.; Avellaneda-Franco, L.; Hernandez, C.A.; Oddo, A.; Chernyavskiy, O.; Hilsenstein, V.; Neild, A.; et al. Bacteriophage uptake by mammalian cell layers represents a potential sink that may impact phage therapy. iScience 2021, 24, 102287. [Google Scholar] [CrossRef]
- Hsiao, I.-L.; Gramatke, A.M.; Joksimovic, R.; Sokolowski, M.; Gradzielski, M.; Haase, A. Size and cell type dependent uptake of silica nanoparticles. J. Nanomed. Nanotechnol. 2014, 5, 1. [Google Scholar]
- Ackermann, H.-W.; Tremblay, D.; Moineau, S. Long-term bacteriophage preservation. WFCC Newsl. 2004, 38, 35–40. [Google Scholar]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Chapter 7—Endolysins as Antimicrobials. In Advances in Virus Research; Łobocka, M., Szybalski, W., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 83, pp. 299–365. [Google Scholar]
- Brzozowska, E.; Bazan, J.; Gamian, A. The functions of bacteriophage proteins. Postep. Hig. I Med. Dosw. 2011, 65, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]



| Year | Clinical Condition | Bacterial Pathogen | Phage Source | Phage Preparation | Phage Dose (PFU/mL) | Number of Doses | Route of Administration | Combined Therapy | Treatment Duration | Clinical Outcome | Adverse Events | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2018 | Diabetic foot osteomyelitis | MSSA | Eliava Institute | Single Phage | NR | Multi-dose | Local | Levofloxacin (stopped on Day 7) | 7w | Resolved | NR | [6] |
| 2019 | Device-related infection | MSSA | AmpliPhi Biosciences | Phage Cocktail | 3.0 × 109 | Multi-dose | Intravenous | Cefazolin and minocycline | 4w | Resolved | No | [137] |
| 2019 | Osteomyelitis | S. aureus * | Queen Astrid Military Hospital | Phage cocktail | 1.0 × 107 | Multi-dose | Local | Amoxicillin, vancomycin, rifampicin, moxifloxacin, colistin, fosfomycin and clindamycin, and surgery | 7–10 d | Resolved | No | [149] |
| 2019 | Chronic wound | S. aureus | Banaras Hindu University | Phage cocktail | 1.0 × 109 | Multi-dose | Local | No | 13 d | Resolved | No | [152] |
| 2019 | Device-related infection (prosthetic valve endocarditis) | MSSA | AmpliPhi Biosciences | Phage cocktail | 1.0 × 109 | Multi-dose | Intravenous | Flucloxacillin, ciprofloxacin and rifampicin | 2w | Resolved | No | [139] |
| 2020 | Cranial empyema | MSSA | Pherecydes Pharma | Phage Cocktail | NR | Single-dose | Local | Dalbavancin | NA | Resolved | No | [154] |
| 2020 | Device-related infection (knee prosthetic) | MSSA | Pherecydes Pharma | Phage Cocktail | 1.0 × 109 | Single-dose | Local | Daptomycin, cloxacillin, levofloxacin and rifampicin, and surgery | NA | Resolved | NR | [141] |
| 2020 | Device-related infection (knee prosthetic) | MRSA | Adaptive Phage Therapeutics | Single Phage | 5.4 × 109 | Multi-dose | Dual routes (local and intravenous) | Daptomycin and surgery | 3 d | Resolved | Reversible transaminitis | [138] |
| 2020 | Osteomyelitis | MSSA | LTD Eliava Biopreparations | Phage Cocktail | 1.0 × 107 | Multi-dose | Dual routes (local and oral) | No | 5w | Resolved | No | [150] |
| Osteomyelitis | MSSA | LTD Eliava Biopreparations | Phage Cocktail | 1.0 × 107 | Multi-dose | Dual routes (local and oral) | No | 5w | Resolved | No | ||
| Diabetic foot osteomyelitis | S. aureus * | LTD Eliava Biopreparations | Phage Cocktail | 1.0 × 107 | Multi-dose | Dual routes (local and oral) | No | 6w | Resolved | No | ||
| 2020 | Device-related infection (knee prosthetic) | MSSA | Pherecydes Pharma | Phage Cocktail | 1.0 × 109 | Single-dose | Local | Daptomycin, levofloxacin, ofloxacin and doxycycline, and surgery | NA | Resolved | NR | [140] |
| Device-related infection (knee prosthetic) | MSSA | Pherecydes Pharma | Phage Cocktail | 1.0 × 1010 | Single-dose | Local | Daptomycin, ceftazidime, ciprofloxacin and rifampin, and surgery | NA | Improved clinically | No | ||
| Device-related infection (knee prosthetic) | MSSA | Pherecydes Pharma | Phage Cocktail | 1.0 × 109 | Single-dose | Local | Daptomycin, cefepime, rifampin and levofloxacin, and surgery | NA | Improved clinically | NR | ||
| 2020 | Device-related infection | S. aureus * | Gabrichevsky Institute | Phage cocktail | 1.0 × 108 | Multi-dose | Dual routes (local and oral) | Cefepime, daptomycin, linezolid and tobramycin | NA | Resolved but died of other complications | No | [142] |
| Device-related infection | S. aureus | Gabrichevsky Institute | Single Phage | 1.0 × 109 | Multi-dose | Dual routes (local and oral) | Rifampicin, flucloxacillin | 2 d | Resolved | No | ||
| Device-related infection | S. aureus | Gabrichevsky Institute | Single Phage | 1.0 × 109 | Multi-dose | Dual routes (local and oral) | Daptomycin | 12 d | Resolved but died of other complications | No | ||
| Device-related infection | S. aureus | Gabrichevsky Institute | Phage cocktail | 1.0 × 109 | Multi-dose | Multiple (local, inhaled, and oral) | Daptomycin | 8 d | Not resolved | No | ||
| Device-related infection | S. aureus | Gabrichevsky Institute | Single Phage | 4.0 × 1010 | Single-dose | Local | Sultamicillin | NA | Resolved | No | ||
| 2021 | Chronic bacterial prostatitis | MRSA * | Eliava Institute | Phage Cocktail | 1.0 × 105–1.0 × 107 | Multi-dose | Multiple (oral, intra-rectal, and intra-urethral) | No | NA | Resolved | No | [155] |
| 2021 | Osteomyelitis | MSSA * | Queen Astrid Military Hospital | Phage Cocktail | 1.0 × 107 | Multi-dose | Local | Clindamycin, rifampin and ciprofloxacin, and surgery | 2w | Clinical improvement | NR | [151] |
| 2021 | Device-related infection (knee prosthetic) | MSSA | Adaptive PT | Single Phage | 2.9 × 1010 | Multi-dose | Dual routes (local and intravenous) | Cefazolin and surgery | 6w | Resolved | No | [143] |
| 2022 | Device-related infection | S. aureus | Sanubiom GmbH, Fritzens, Austria | Phage Cocktail | 1.0 × 107 | Single-dose | Local | Piperacillin/tazobactam and surgery | NA | Resolved | NR | [144] |
| 2022 | Chronic rhinosinusitis | MRSA | Adaptive Phage Therapeutics | Single Phage | 1.0 × 109–1.0 × 1010 | Multi-dose | Local | Oritavancin | 3w | Resolved | NR | [153] |
| 2022 | Device-related infection | MSSA * | Sanubiom GmbH, Fritzens, Austria, Phage 24.com | Phage Cocktail | 1.0 × 107 | Single-dose | Local | Piperacillin/tazobactam and surgery | NA | Not resolved | NR | [145] |
| 2022 | Device-related infection | MSSA | Phage24.com, Austria | Phage Cocktail | NA | Single-dose | Local (endovascular grafts coated with phage) | Surgery | NA | Resolved | NR | [148] |
| 2022 | Device-related infection | MRSA | NA | Single Phage | 1.0 × 109–1.0 × 1010; 2.0 × 108 | Multi-dose | Intravenous | Daptomycin and ceftaroline and surgery | 3 d | Resolved | Reversible transaminitis | [146] |
| 2022 | Device-related infection | MRSA | Adaptive PT (Gaithersburg, MD, USA) | Single Phage | 1.2 × 109; 2.4 × 107–2.4 × 108; 3.0 × 108–4.0 × 108 | Multi-dose | Dual routes (local and intravenous) | Daptomycin and Bactrim and surgery | 3 d | Resolved | Reversible transaminitis | [147] |
| 2022 | Disseminated S. aureus infections: meningitis, retropharyngeal abscess, cranial empyema, and endocarditis | MRSA | ContraFect Corp | exebacase | 3 mg | Single-dose | Intravenous | Vancomycin, linezolid, daptomycin and ceftaroline, and surgery | NA | Resolved | NR | [156] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atshan, S.S.; Hamat, R.A.; Aljaberi, M.A.; Chen, J.-S.; Huang, S.-W.; Lin, C.-Y.; Mullins, B.J.; Kicic, A. Phage Therapy as an Alternative Treatment Modality for Resistant Staphylococcus aureus Infections. Antibiotics 2023, 12, 286. https://doi.org/10.3390/antibiotics12020286
Atshan SS, Hamat RA, Aljaberi MA, Chen J-S, Huang S-W, Lin C-Y, Mullins BJ, Kicic A. Phage Therapy as an Alternative Treatment Modality for Resistant Staphylococcus aureus Infections. Antibiotics. 2023; 12(2):286. https://doi.org/10.3390/antibiotics12020286
Chicago/Turabian StyleAtshan, Salman Sahab, Rukman Awang Hamat, Musheer A. Aljaberi, Jung-Sheng Chen, Shih-Wei Huang, Chung-Ying Lin, Benjamin J. Mullins, and Anthony Kicic. 2023. "Phage Therapy as an Alternative Treatment Modality for Resistant Staphylococcus aureus Infections" Antibiotics 12, no. 2: 286. https://doi.org/10.3390/antibiotics12020286
APA StyleAtshan, S. S., Hamat, R. A., Aljaberi, M. A., Chen, J.-S., Huang, S.-W., Lin, C.-Y., Mullins, B. J., & Kicic, A. (2023). Phage Therapy as an Alternative Treatment Modality for Resistant Staphylococcus aureus Infections. Antibiotics, 12(2), 286. https://doi.org/10.3390/antibiotics12020286









