Prevalence of Plasmid-Associated Tetracycline Resistance Genes in Multidrug-Resistant Escherichia coli Strains Isolated from Environmental, Animal and Human Samples in Panama
Abstract
1. Introduction
2. Results
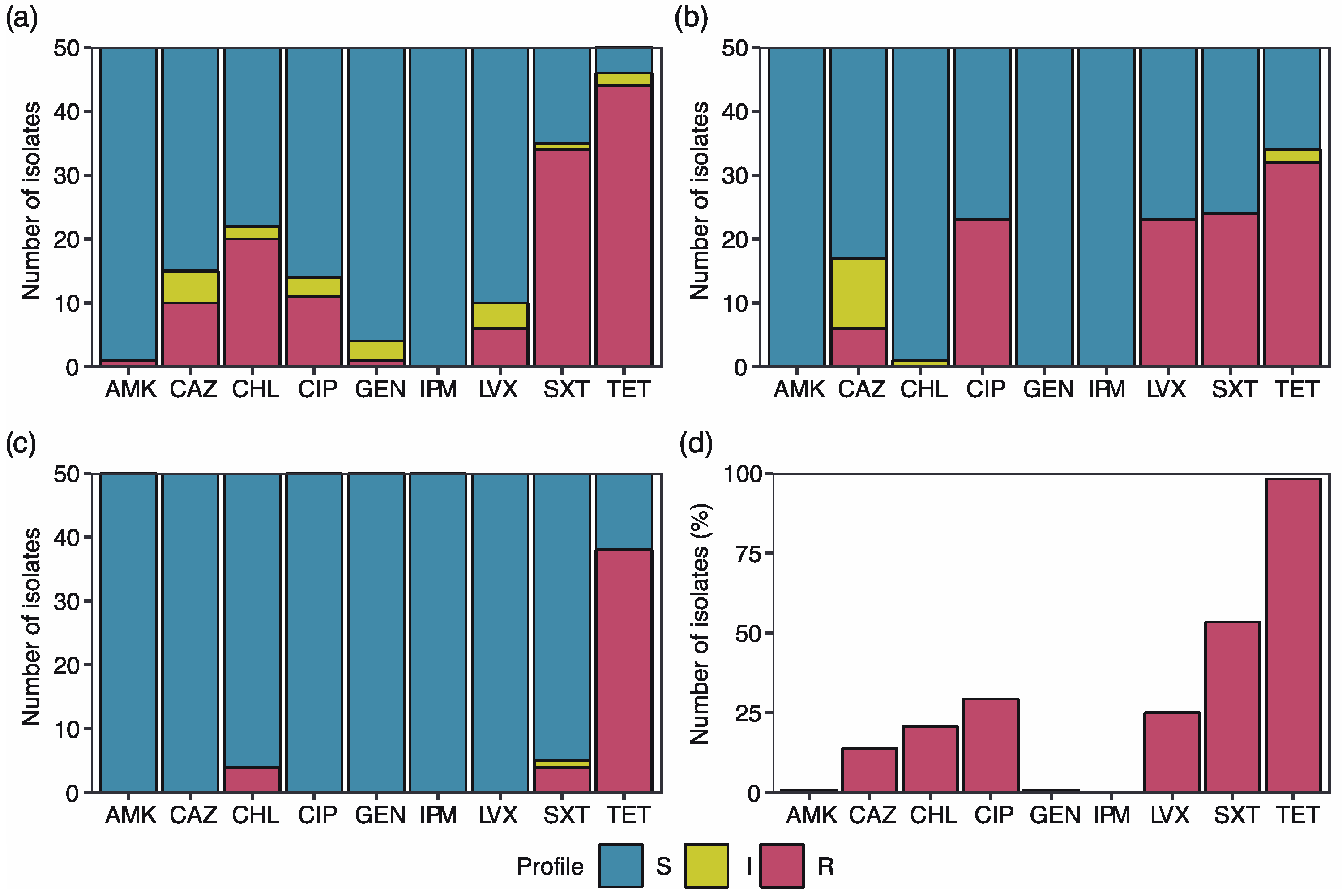
2.1. High Percentage of the E. coli Isolates Exhibit Antibiotic Resistance
2.2. Multidrug Resistance Profiles
2.3. Tetracycline Resistance
2.4. tetA and tetB Are Present in Nearly 45% of the Samples Analyzed
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. E. coli Isolation and Confirmation
4.3. Antibiotic Susceptibility Testing
4.4. Plasmid DNA Extraction
4.5. Detection of tetA and tetB Genes by Multiplex PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Al-Bahry, S.N.; Al-Mashani, B.M.; Al-Ansari, A.S.; Elshafie, A.E.; Mahmoud, I.Y. Escherichia Coli Tetracycline Efflux Determinants in Relation to Tetracycline Residues in Chicken. Asian Pac. J. Trop. Med. 2013, 6, 718–722. [Google Scholar] [CrossRef]
- Frye, J.G.; Jackson, C.R. Genetic Mechanisms of Antimicrobial Resistance Identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. Isolated from U.S. Food Animals. Front. Microbiol. 2013, 4, 135. [Google Scholar] [CrossRef] [PubMed]
- Wasyl, D.; Hoszowski, A.; Zając, M.; Szulowski, K. Antimicrobial Resistance in Commensal Escherichia Coli Isolated from Animals at Slaughter. Front. Microbiol. 2013, 4, 221. [Google Scholar] [CrossRef]
- Karami, N.; Nowrouzian, F.; Adlerberth, I.; Wold, A.E. Tetracycline Resistance in Escherichia Coli and Persistence in the Infantile Colonic Microbiota. Antimicrob. Agents Chemother. 2006, 50, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology Drives a Global Network of Gene Exchange Connecting the Human Microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological Effects of Antibiotics on Natural Ecosystems: A Review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria—A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Jones, C.H.; Tuckman, M.; Murphy, E.; Bradford, P.A. Identification and Sequence of a Tet(M) Tetracycline Resistance Determinant Homologue in Clinical Isolates of Escherichia Coli. J. Bacteriol. 2006, 188, 7151–7164. [Google Scholar] [CrossRef]
- Kaesbohrer, A.; Schroeter, A.; Tenhagen, B.-A.; Alt, K.; Guerra, B.; Appel, B. Emerging Antimicrobial Resistance in Commensal Escherichia Coli with Public Health Relevance. Zoonoses Public Health 2012, 59 (Suppl. S2), 158–165. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Salvatore, D.; Sakhria, S.; Catelli, E.; Lupini, C.; Abbassi, M.S.; Bessoussa, G.; Ben Yahia, S.; Ben Chehida, N. High Frequency and Diversity of Tetracycline Resistance Genes in the Microbiota of Broiler Chickens in Tunisia. Animals 2021, 11, 377. [Google Scholar] [CrossRef] [PubMed]
- Thaker, M.; Spanogiannopoulos, P.; Wright, G.D. The Tetracycline Resistome. Cell. Mol. Life Sci. 2010, 67, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Laurent, P.; Jean-Yves, M.; Agnese, L.; Anne-Kathrin, S.; Nicolas, K.; Patrice, N.; Stefan, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 6-4. [Google Scholar] [CrossRef]
- Mejía, F.; Castro-del Campo, N.; García, A.; Rodríguez, K.; Cornejo, H.; Ahumada-Ruiz, S.; Soto-Beltrán, M.; Castillo, M.; Querol-Audi, J.; Chaidez-Quiroz, C.; et al. Prevalence and Characterization of Antibiotic-Resistant Staphylococcus Aureus Recovered from Pasteurized Cheese Commercialized in Panama City Markets. J. Food Qual. 2021, 2021, 9923855. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic Resistance in the Environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Hou, J.; Long, X.; Wang, X.; Li, L.; Mao, D.; Luo, Y.; Ren, H. Global Trend of Antimicrobial Resistance in Common Bacterial Pathogens in Response to Antibiotic Consumption. J. Hazard. Mater. 2023, 442, 130042. [Google Scholar] [CrossRef]
- Österberg, J.; Wingstrand, A.; Nygaard Jensen, A.; Kerouanton, A.; Cibin, V.; Barco, L.; Denis, M.; Aabo, S.; Bengtsson, B. Antibiotic Resistance in Escherichia Coli from Pigs in Organic and Conventional Farming in Four European Countries. PLoS ONE 2016, 11, e0157049. [Google Scholar] [CrossRef]
- Nhung, N.T.; Cuong, N.V.; Campbell, J.; Hoa, N.T.; Bryant, J.E.; Truc, V.N.T.; Kiet, B.T.; Jombart, T.; Trung, N.V.; Hien, V.B.; et al. High Levels of Antimicrobial Resistance among Escherichia Coli Isolates from Livestock Farms and Synanthropic Rats and Shrews in the Mekong Delta of Vietnam. Appl. Environ. Microbiol. 2015, 81, 812–820. [Google Scholar] [CrossRef]
- Muloi, D.; Kiiru, J.; Ward, M.; Hassell, J.; Bettridge, J.; Robinson, T.; van Bunnik, B.; Chase-Topping, M.; Robertson, G.; Pedersen, A.; et al. Epidemiology of Antimicrobial Resistant Escherichia Coli Carriage in Sympatric Humans and Livestock in a Rapidly Urbanising City. Int. J. Antimicrob. Agents 2019, 54, 531–537. [Google Scholar] [CrossRef]
- Azabo, R.R.; Mshana, S.E.; Matee, M.I.; Kimera, S.I. Antimicrobial Resistance Pattern of Escherichia Coli Isolates from Small Scale Dairy Cattle in Dar Es Salaam, Tanzania. Animals 2022, 12, 1853. [Google Scholar] [CrossRef] [PubMed]
- Al Azad, M.A.R.; Rahman, M.M.; Amin, R.; Begum, M.I.A.; Fries, R.; Husna, A.; Khairalla, A.S.; Badruzzaman, A.T.M.; El Zowalaty, M.E.; Lampang, K.N.; et al. Susceptibility and Multidrug Resistance Patterns of Escherichia Coli Isolated from Cloacal Swabs of Live Broiler Chickens in Bangladesh. Pathogens 2019, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Adefisoye, M.A.; Okoh, A.I. Identification and Antimicrobial Resistance Prevalence of Pathogenic Escherichia Coli Strains from Treated Wastewater Effluents in Eastern Cape, South Africa. Microbiologyopen 2016, 5, 143–151. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. NPJ Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Noah, R.-F. The Landscape of Antibiotic Resistance. Environ. Health Perspect. 2009, 117, A244–A250. [Google Scholar] [CrossRef]
- Belaynehe, K.M.; Shin, S.W.; Yoo, H.S. Interrelationship between Tetracycline Resistance Determinants, Phylogenetic Group Affiliation and Carriage of Class 1 Integrons in Commensal Escherichia Coli Isolates from Cattle Farms. BMC Vet. Res. 2018, 14, 340. [Google Scholar] [CrossRef]
- Seifi, S.; Khoshbakht, R. Prevalence of Tetracycline Resistance Determinants in Broiler Isolated Escherichia Coli in Iran. Br. Poult. Sci. 2016, 57, 729–733. [Google Scholar] [CrossRef]
- Rolain, J.-M. Food and Human Gut as Reservoirs of Transferable Antibiotic Resistance Encoding Genes. Front. Microbiol. 2013, 4, 173. [Google Scholar] [CrossRef] [PubMed]
- Akingbade, O.; Balogun, S.; Ojo, D.; Akinduti, P.; Okerentugba, P.O.; Nwanze, J.C.; Okonko, I.O. Resistant Plasmid Profile Analysis of Multidrug Resistant Escherichia Coli Isolated from Urinary Tract Infections in Abeokuta, Nigeria. Afr. Health Sci. 2014, 14, 821–828. [Google Scholar] [CrossRef]
- Capkin, E.; Terzi, E.; Altinok, I. Occurrence of Antibiotic Resistance Genes in Culturable Bacteria Isolated from Turkish Trout Farms and Their Local Aquatic Environment. Dis. Aquat. Organ. 2015, 114, 127–137. [Google Scholar] [CrossRef]
- Xi, X.; Zhang, J.; Kwok, L.; Huo, D.; Feng, S.; Zhang, H.; Sun, T. Microbial Pollution Tracking of Dairy Farm with a Combined PCR-DGGE and QPCR Approach. Curr. Microbiol. 2015, 71, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.-J.; Woo, G.-J. Distribution and Transferability of Tetracycline Resistance Determinants in Escherichia Coli Isolated from Meat and Meat Products. Int. J. Food Microbiol. 2011, 145, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Adesoji, A.T.; Ogunjobi, A.A.; Olatoye, I.O.; Call, D.R. Prevalence of Tetracycline Resistance Genes among Multi-Drug Resistant Bacteria from Selected Water Distribution Systems in Southwestern Nigeria. Ann. Clin. Microbiol. Antimicrob. 2015, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Yue, D.; Peng, Y.; Liu, Y.; Xiao, L. Occurrence and Distribution of Antibiotic-Resistant Bacteria and Transfer of Resistance Genes in Lake Taihu. Microbes Environ. 2013, 28, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. The Role of Antibiotics and Antibiotic Resistance in Nature. Environ. Microbiol. 2009, 11, 2970–2988. [Google Scholar] [CrossRef] [PubMed]
- Csuros, M. Microbiological Examination of Water and Wastewater; CRC Press: Boca Raton, FL, USA, 1999; ISBN 1-56670-179-1. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th ed.; CLSI Standard M02 Wayne; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2018. [Google Scholar]
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN1 9781936113415. ISBN2 1936113414. ISBN3 9781936113422. ISBN4 1936113422. [Google Scholar]
- Bailey, J.K.; Pinyon, J.L.; Anantham, S.; Hall, R.M. Commensal Escherichia Coli of Healthy Humans: A Reservoir for Antibiotic-Resistance Determinants. J. Med. Microbiol. 2010, 59, 1331–1339. [Google Scholar] [CrossRef]



| Source | Site (%) | Total Antibiotic Resistance Strains (%) | ||
|---|---|---|---|---|
| Ciudad del Niño | El Arado | Escobal | ||
| Chicken | 10 (6.67) | 9 (6.00) | 8 (5.33) | 27 (18.00) |
| Cow | 10 (6.67) | 5 (3.33) | 10 (6.67) | 25 (16.67) |
| Human | 5 (3.33) | 1 (0.67) | 10 (6.67) | 16 (10.67) |
| Swine | 10 (6.67) | 10 (6.67) | 10 (6.67) | 30 (20.00) |
| Water | 10 (6.67) | 8 (5.33) | 0 (0) | 18 (12.00) |
| Total (%) | 45 (30.00) | 33 (22.00) | 38 (25.33) | 116 (77.33) |
| tetA (%) | tetB (%) | tetA and tetB (%) | Total (%) | |
|---|---|---|---|---|
| Chicken | 7 (15.22) | 1 (2.17) | 3 (6.52) | 11 (23.91) |
| Cow | 6 (13.04) | 0 (0) | 4 (8.70) | 10 (21.74) |
| Human | 3 (6.52) | 1 (2.17) | 2 (4.35) | 6 (13.04) |
| Swine | 6 (13.04) | 1 (2.17) | 4 (8.70) | 11 (23.91) |
| Water | 5 (10.87) | 0 (0) | 3 (6.52) | 8 (17.39) |
| Ciudad del Niño | 9 (19.57) | 3 (6.52) | 6 (13.04) | 18 (39.13) |
| El Arado | 10 (21.74) | 0 (0) | 6 (13.04) | 16 (34.78) |
| Escobal | 8 (17.39) | 0 (0) | 4 (8.70) | 12 (26.09) |
| Positive (%) | 27 (58.70) | 3 (6.52) | 16 (34.78) | 46 (100) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Bayard, I.E.; Mejía, F.; Medina-Sánchez, J.R.; Cornejo-Reyes, H.; Castillo, M.; Querol-Audi, J.; Martínez-Torres, A.O. Prevalence of Plasmid-Associated Tetracycline Resistance Genes in Multidrug-Resistant Escherichia coli Strains Isolated from Environmental, Animal and Human Samples in Panama. Antibiotics 2023, 12, 280. https://doi.org/10.3390/antibiotics12020280
Ramírez-Bayard IE, Mejía F, Medina-Sánchez JR, Cornejo-Reyes H, Castillo M, Querol-Audi J, Martínez-Torres AO. Prevalence of Plasmid-Associated Tetracycline Resistance Genes in Multidrug-Resistant Escherichia coli Strains Isolated from Environmental, Animal and Human Samples in Panama. Antibiotics. 2023; 12(2):280. https://doi.org/10.3390/antibiotics12020280
Chicago/Turabian StyleRamírez-Bayard, I. E., F. Mejía, J. R. Medina-Sánchez, H. Cornejo-Reyes, M. Castillo, J. Querol-Audi, and A. O. Martínez-Torres. 2023. "Prevalence of Plasmid-Associated Tetracycline Resistance Genes in Multidrug-Resistant Escherichia coli Strains Isolated from Environmental, Animal and Human Samples in Panama" Antibiotics 12, no. 2: 280. https://doi.org/10.3390/antibiotics12020280
APA StyleRamírez-Bayard, I. E., Mejía, F., Medina-Sánchez, J. R., Cornejo-Reyes, H., Castillo, M., Querol-Audi, J., & Martínez-Torres, A. O. (2023). Prevalence of Plasmid-Associated Tetracycline Resistance Genes in Multidrug-Resistant Escherichia coli Strains Isolated from Environmental, Animal and Human Samples in Panama. Antibiotics, 12(2), 280. https://doi.org/10.3390/antibiotics12020280







