Mutations Related to Antibiotics Resistance in Helicobacter pylori Clinical Isolates from Bangladesh
Abstract
1. Introduction
2. Results
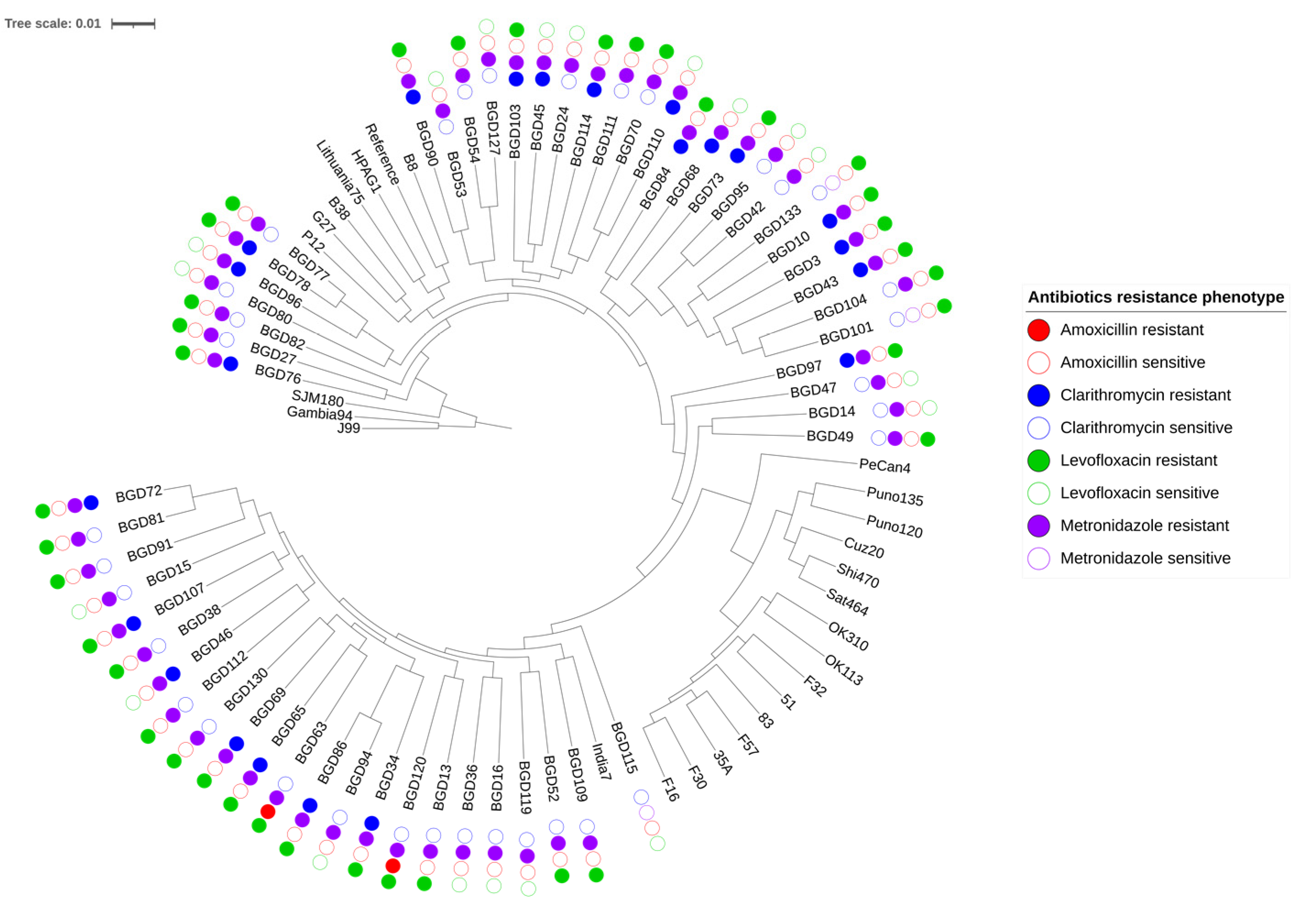
2.1. Genome Relatedness and Antibiotic Resistance
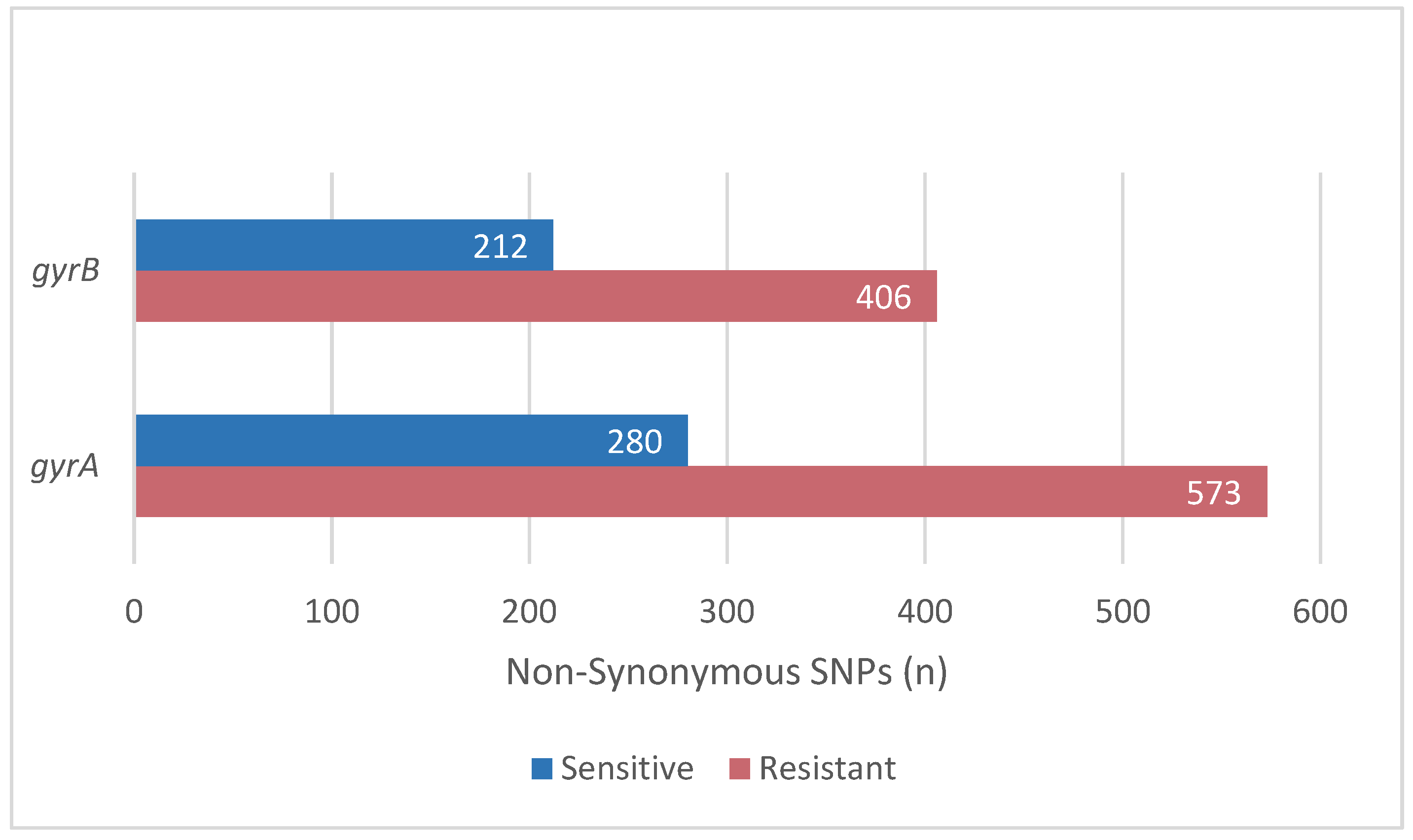
2.2. Mutations Related to Resistance to Levofloxacin
2.3. Mutations Related to Resistance to Amoxicillin
2.4. Mutations Related to Resistance to Metronidazole
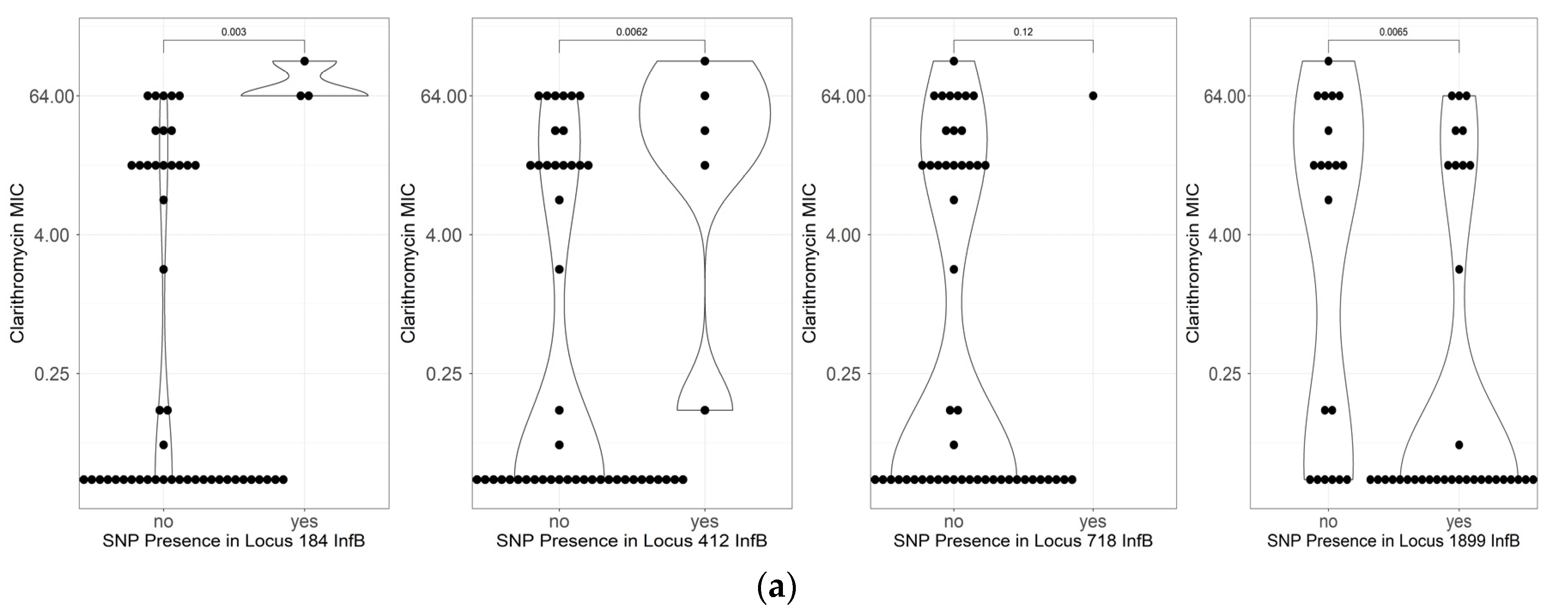
2.5. Mutations Related to Resistance to Clarithromycin
3. Discussion
4. Materials and Methods
4.1. Patient Sampling and H. pylori Isolates
4.2. Antibiotics Susceptibility Test
4.3. Genome Sequencing
4.4. Analysis of the SNP and Protein Modelling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeo, Y.H.; Shiu, S.-I.; Ho, H.J.; Zou, B.; Lin, J.-T.; Wu, M.-S.; Liou, J.-M.; Wu, C.-Y. First-line Helicobacter pylori eradication therapies in countries with high and low clarithromycin resistance: A systematic review and network meta-analysis. Gut 2018, 67, 20–27. [Google Scholar] [CrossRef]
- Kuo, Y.-T.; Liou, J.-M.; El-Omar, E.M.; Wu, J.-Y.; Leow, A.H.R.; Goh, K.L.; Das, R.; Lu, H.; Lin, J.-T.; Tu, Y.-K.; et al. Primary antibiotic resistance in Helicobacter pylori in the Asia-Pacific region: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017, 2, 707–715. [Google Scholar] [CrossRef]
- Correa, P.; Houghton, J. Carcinogenesis of Helicobacter pylori. Gastroenterology 2007, 133, 659–672. [Google Scholar] [CrossRef]
- Malfertheiner, P.; Megraud, F.; O’Morain, C.A.; Gisbert, J.P.; Kuipers, E.J.; Axon, A.T.; Bazzoli, F.; Gasbarrini, A.; Atherton, J.; Graham, D.Y.; et al. Management ofHelicobacter pyloriinfection—The Maastricht V/Florence Consensus Report. Gut 2017, 66, 6–30. [Google Scholar] [CrossRef] [PubMed]
- Gingold-Belfer, R.; Niv, Y.; Schmilovitz-Weiss, H.; Levi, Z.; Boltin, D. Susceptibility-guided versus empirical treatment for Helicobacter pylori infection: A systematic review and meta-analysis. J. Gastroenterol. Hepatol. 2021, 36, 2649–2658. [Google Scholar] [CrossRef] [PubMed]
- Mannion, A.; Dzink-Fox, J.; Shen, Z.; Piazuelo, M.B.; Wilson, K.T.; Correa, P.; Peek, R.M.; Camargo, M.C.; Fox, J.G. Helicobacter pylori Antimicrobial Resistance and Gene Variants in High- and Low-Gastric-Cancer-Risk Populations. J. Clin. Microbiol. 2021, 59, e03203-20. [Google Scholar] [CrossRef] [PubMed]
- Tshibangu-Kabamba, E.; Yamaoka, Y. Helicobacter pylori infection and antibiotic resistance—From biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 613–629. [Google Scholar] [CrossRef] [PubMed]
- Mahant, S.; Sharma, A.K.; Gehlot, V.; Mukhopadhyay, A.K.; Chhawchharia, A.; Dutta, S.; Agarwal, A.; Som, A.; Das, K.; Das, R. Geographically Distinct North-East Indian Helicobacter pylori Strains are Highly Sensitive to Clarithromycin but are Levofloxacin Resistant. Indian J. Med. Microbiol. 2019, 37, 337–344. [Google Scholar] [CrossRef]
- Subsomwong, P.; Doohan, D.; Fauzia, K.A.; Akada, J.; Matsumoto, T.; Yee, T.T.; Htet, K.; Waskito, L.A.; Tuan, V.P.; Uchida, T.; et al. Next-Generation Sequencing-Based Study of Helicobacter pylori Isolates from Myanmar and Their Susceptibility to Antibiotics. Microorganisms 2022, 10, 196. [Google Scholar] [CrossRef]
- Binh, T.T.; Shiota, S.; Suzuki, R.; Matsuda, M.; Trang, T.T.H.; Kwon, D.H.; Iwatani, S.; Yamaoka, Y. Discovery of novel mutations for clarithromycin resistance in Helicobacter pylori by using next-generation sequencing. J. Antimicrob. Chemother. 2014, 69, 1796–1803. [Google Scholar] [CrossRef]
- Rimbara, E.; Noguchi, N.; Kawai, T.; Sasatsu, M. Mutations in penicillin-binding proteins 1, 2 and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J. Antimicrob. Chemother. 2008, 61, 995–998. [Google Scholar] [CrossRef]
- Rimbara, E.; Noguchi, N.; Kawai, T.; Sasatsu, M. Fluoroquinolone Resistance in Helicobacter pylori: Role of Mutations at Position 87 and 91 of GyrA on the Level of Resistance and Identification of a Resistance Conferring Mutation in GyrB. Helicobacter 2012, 17, 36–42. [Google Scholar] [CrossRef]
- Suerbaum, S.; Smith, J.M.; Bapumia, K.; Morelli, G.; Smith, N.H.; Kunstmann, E.; Dyrek, I.; Achtman, M. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 1998, 95, 12619–12624. [Google Scholar] [CrossRef]
- Suerbaum, S.; Achtman, M. Population Genetics. In Helicobacter pylori: Physiology and Genetics; Mobley, H.L.T., Mendz, G.L., Hazell, S.L., Eds.; ASM Press: Washington, DC, USA, 2001. [Google Scholar]
- Aftab, H.; Miftahussurur, M.; Subsomwong, P.; Ahmed, F.; Khan, A.A.; Yamaoka, Y. Helicobacter pylori antibiotic susceptibility patterns in Bangladesh: Emerging levofloxacin resistance. J. Infect. Dev. Ctries. 2016, 10, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Hallur, V.; Panigrahi, M.; Sable, M.; Ghosh, M.; Mohanty, S.; Purkait, S.; Praharaj, A. Low clarithromycin resistance in virulent Helicobacter pylori from dyspeptic patients at a tertiary care centre in Odisha. Indian J. Med. Microbiol. 2022, 40, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Salehi, N.; Attaran, B.; Eskini, N.; Esmaeili, M.; Sharifirad, A.; Sadeghi, M.; Mohammadi, M. New insights into resistance ofHelicobacter pyloriagainst third- and fourth-generation fluoroquinolones: A molecular docking study of prevalent GyrA mutations. Helicobacter 2019, 24, e12628. [Google Scholar] [CrossRef] [PubMed]
- Ziver-Sarp, T.; Yuksel-Mayda, P.; Saribas, S.; Demiryas, S.; Gareayaghi, N.; Ergin, S.; Tasci, I.; Ozbey, D.; Bal, K.; Erzin, Y.; et al. Point Mutations at gyrA and gyrB Genes of Levofloxacin Resistant Helicobacter pylori Strains and Dual Resistance with Clarithromycin. Clin. Lab. 2021, 67, 10. [Google Scholar] [CrossRef] [PubMed]
- Hanafiah, A.; Binmaeil, H.; Ali, R.A.R.; Rose, I.M.; Lopes, B.S. Molecular characterization and prevalence of antibiotic resistance in Helicobacter pylori isolates in Kuala Lumpur, Malaysia. Infect. Drug Resist. 2019, 12, 3051–3061. [Google Scholar] [CrossRef]
- Vianna, J.S.; Ramis, I.B.; Ramos, D.F.; Gastal, O.L.; Da Silva, R.A.; Gonçalves, C.V.; Da Silva, P.E.A. The interplay between mutations in cagA, 23S rRNA, gyrA and drug resistance in Helicobacter pylori. Rev. Inst. Med. Trop. São Paulo 2018, 60, e25. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Suzuki, H.; Matsuzaki, J.; Masaoka, T.; Kanai, T. Acquisition of double mutation in gyrA caused high resistance to sitafloxacin in Helicobacter pylori after unsuccessful eradication with sitafloxacin-containing regimens. United Eur. Gastroenterol. J. 2018, 6, 391–397. [Google Scholar] [CrossRef]
- Zapun, A.; Contreras-Martel, C.; Vernet, T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 2008, 32, 361–385. [Google Scholar] [CrossRef] [PubMed]
- Eng, N.F.; Ybazeta, G.; Chapman, K.; Fraleigh, N.L.; Letto, R.; Altman, E.; Diaz-Mitoma, F. Antimicrobial susceptibility of Canadian isolates of Helicobacter pylori in Northeastern Ontario. Can. J. Infect. Dis. Med. Microbiol. 2015, 26, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Sukri, A.; Hanafiah, A.; Yusoff, H.; Nizam, N.A.S.; Nameyrra, Z.; Wong, Z.; Ali, R.A.R. Multidrug-Resistant Helicobacter pylori Strains: A Five-Year Surveillance Study and Its Genome Characteristics. Antibiotics 2022, 11, 1391. [Google Scholar] [CrossRef] [PubMed]
- Metwally, M.; Ragab, R.; Hamid, H.S.A.; Emara, N.; Elkholy, H. Helicobacter pylori Antibiotic Resistance in Egypt: A Single-Center Study. Infect. Drug Resist. 2022, 15, 5905–5913. [Google Scholar] [CrossRef] [PubMed]
- Albert, T.J.; Dailidiene, D.; Dailide, G.; Norton, J.E.; Kalia, A.; Richmond, T.A.; Molla, M.; Singh, J.; Green, R.D.; Berg, D.E. Mutation discovery in bacterial genomes: Metronidazole resistance in Helicobacter pylori. Nat. Methods 2005, 2, 951–953. [Google Scholar] [CrossRef] [PubMed]
- Wani, F.A.; Bashir, G.; Khan, M.A.; Zargar, S.A.; Rasool, Z.; Qadri, Q. Antibiotic Resistance in Helicobacter pylori: A Mutational Analysis from a Tertiary Care Hospital in Kashmir, India. Indian J. Med Microbiol. 2018, 36, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Gehlot, V.; Mahant, S.; Mukhopadhyay, A.K.; Das, K.; Alam, J.; Ghosh, P.; Das, R. Low prevalence of clarithromycin-resistant Helicobacter pylori isolates with A2143G point mutation in the 23S rRNA gene in North India. J. Glob. Antimicrob. Resist. 2016, 6, 39–43. [Google Scholar] [CrossRef]
- Qumar, S.; Nguyen, T.H.; Nahar, S.; Sarker, N.; Baker, S.; Bulach, D.; Ahmed, N.; Rahman, M. A comparative whole genome analysis of Helicobacter pylori from a human dense South Asian setting. Helicobacter 2021, 26, e12766. [Google Scholar] [CrossRef]
- Khan, R.; Nahar, S.; Sultana, J.; Ahmad, M.M.; Rahman, M. T2182C Mutation in 23S rRNA Is Associated with Clarithromycin Resistance in Helicobacter pylori Isolates Obtained in Bangladesh. Antimicrob. Agents Chemother. 2004, 48, 3567–3569. [Google Scholar] [CrossRef]
- Hoshiya, S.; Watanabe, K.; Tokunaga, K.; Tanaka, A.; Ninomiya, H.; Shingaki, M.; Itoh, T.; Saito, S.; Ishida, H.; Takahashi, S. Relationship between eradication therapy and clarithromycin-resistant Helicobacter pylori in Japan. J. Gastroenterol. 2000, 35, 10–14. [Google Scholar]
- Liou, J.-M.; Chang, C.-Y.; Sheng, W.-H.; Wang, Y.-C.; Chen, M.-J.; Lee, Y.-C.; Hung, H.-W.; Chian, H.; Chang, S.-C.; Wu, M.-S.; et al. Genotypic Resistance in Helicobacter pylori Strains Correlates with Susceptibility Test and Treatment Outcomes after Levofloxacin- and Clarithromycin-Based Therapies. Antimicrob. Agents Chemother. 2011, 55, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Xuan, S.-H.; Wu, L.-P.; Zhou, Y.-G.; Xiao, M.-B. Detection of clarithromycin-resistant Helicobacter pylori in clinical specimens by molecular methods: A review. J. Glob. Antimicrob. Resist. 2016, 4, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Yahara, K.; Furuta, Y.; Oshima, K.; Yoshida, M.; Azuma, T.; Hattori, M.; Uchiyama, I.; Kobayashi, I. Chromosome painting in silico in a bacterial species reveals fine population structure. Mol. Biol. Evol. 2013, 30, 1454–1464. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Argimón, S.; Abudahab, K.; Goater, R.J.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.; Yeats, C.A.; Grundmann, H. Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2005; pp. 571–607. [Google Scholar]


| SNP | Resistant (n = 37) | Sensitive (n = 19) | Total | p-Value | ||
|---|---|---|---|---|---|---|
| Absent | Present | Absent | Present | |||
| gyrA | ||||||
| D91G | 32 | 5 | 19 | 0 | 56 | 0.15 |
| D91N | 24 | 13 | 19 | 0 | 56 | 0.002 |
| D91Y | 36 | 1 | 19 | 0 | 56 | 1 |
| N87K | 24 | 13 | 19 | 0 | 56 | 0.002 |
| N87Y | 36 | 1 | 19 | 0 | 56 | 1 |
| gyrB | ||||||
| A343V | 29 | 8 | 19 | 0 | 56 | 0.041 |
| Genes | AMX Resistant (n = 2) | AMX Susceptible (n = 54) | p-Value | ||
|---|---|---|---|---|---|
| Present | Absent | Present | Absent | ||
| pbp1a | |||||
| T30S | 1 | 1 | 0 | 54 | 0.0357 |
| V45I | 2 | 0 | 6 | 48 | 0.0182 |
| K306R | 1 | 1 | 0 | 54 | 0.0357 |
| V374L | 1 | 1 | 0 | 54 | 0.0357 |
| N562Y | 2 | 0 | 0 | 54 | 0.0006 |
| G594fs | 1 | 1 | 0 | 54 | 0.0357 |
| pbp2 | |||||
| S79A | 1 | 1 | 0 | 54 | 0.0357 |
| F99S | 1 | 1 | 0 | 54 | 0.0357 |
| E312G | 1 | 1 | 0 | 54 | 0.0357 |
| G529V | 1 | 1 | 0 | 54 | 0.0357 |
| P582T | 1 | 1 | 0 | 54 | 0.0357 |
| pbp3 | |||||
| I14V | 2 | 0 | 4 | 50 | 0.0097 |
| I191V | 2 | 0 | 8 | 46 | 0.0292 |
| V223M | 2 | 0 | 7 | 47 | 0.0234 |
| pbp4 | |||||
| Q226R | 1 | 1 | 0 | 54 | 0.0357 |
| T272A | 1 | 1 | 0 | 54 | 0.0357 |
| Genes | Frameshift | Truncated | SNP | Resistant | Susceptible | ||
|---|---|---|---|---|---|---|---|
| n Total | Average | n Total | Average | ||||
| ribF | 2 | 0 | 67 | 615 | 11.6 | 38 | 12.7 |
| frxA | 14 | 5 | 81 | 231 | 4.4 | 14 | 4.7 |
| rdxA | 8 | 5 | 95 | 353 | 6.7 | 14 | 4.7 |
| mdaB | 10 | 1 | 40 | 289 | 5.5 | 11 | 3.7 |
| omp11 | 1 | 0 | 17 | 52 | 1.0 | 0 | 0.0 |
| fur | 0 | 0 | 14 | 42 | 0.8 | 2 | 0.7 |
| Row Labels | Resistant (n = 22) | Susceptible (n = 34) | p-Value | ||
|---|---|---|---|---|---|
| Yes | Percentage | Yes | Percentage | ||
| 23S rRNA | |||||
| A2146C | 1 | 4.5% | 0 | 0.0% | 0.39 |
| A2146G | 3 | 13.6% | 0 | 0.0% | 0.05 |
| A2147G | 10 | 45.5% | 0 | 0.0% | 1.8 × 10−5 |
| infB | |||||
| C718T | 8 | 36.4% | 3 | 8.8% | 0.024239 |
| A1899G | 10 | 45.5% | 26 | 76.5% | 0.016799 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fauzia, K.A.; Aftab, H.; Tshibangu-Kabamba, E.; Alfaray, R.I.; Saruuljavkhlan, B.; Cimuanga-Mukanya, A.; Matsumoto, T.; Subsomwong, P.; Akada, J.; Miftahussurur, M.; et al. Mutations Related to Antibiotics Resistance in Helicobacter pylori Clinical Isolates from Bangladesh. Antibiotics 2023, 12, 279. https://doi.org/10.3390/antibiotics12020279
Fauzia KA, Aftab H, Tshibangu-Kabamba E, Alfaray RI, Saruuljavkhlan B, Cimuanga-Mukanya A, Matsumoto T, Subsomwong P, Akada J, Miftahussurur M, et al. Mutations Related to Antibiotics Resistance in Helicobacter pylori Clinical Isolates from Bangladesh. Antibiotics. 2023; 12(2):279. https://doi.org/10.3390/antibiotics12020279
Chicago/Turabian StyleFauzia, Kartika Afrida, Hafeza Aftab, Evariste Tshibangu-Kabamba, Ricky Indra Alfaray, Batsaikhan Saruuljavkhlan, Alain Cimuanga-Mukanya, Takashi Matsumoto, Phawinee Subsomwong, Junko Akada, Muhammad Miftahussurur, and et al. 2023. "Mutations Related to Antibiotics Resistance in Helicobacter pylori Clinical Isolates from Bangladesh" Antibiotics 12, no. 2: 279. https://doi.org/10.3390/antibiotics12020279
APA StyleFauzia, K. A., Aftab, H., Tshibangu-Kabamba, E., Alfaray, R. I., Saruuljavkhlan, B., Cimuanga-Mukanya, A., Matsumoto, T., Subsomwong, P., Akada, J., Miftahussurur, M., & Yamaoka, Y. (2023). Mutations Related to Antibiotics Resistance in Helicobacter pylori Clinical Isolates from Bangladesh. Antibiotics, 12(2), 279. https://doi.org/10.3390/antibiotics12020279










