Regulatory Landscape of the Pseudomonas aeruginosa Phosphoethanolamine Transferase Gene eptA in the Context of Colistin Resistance
Abstract
1. Introduction
2. Results
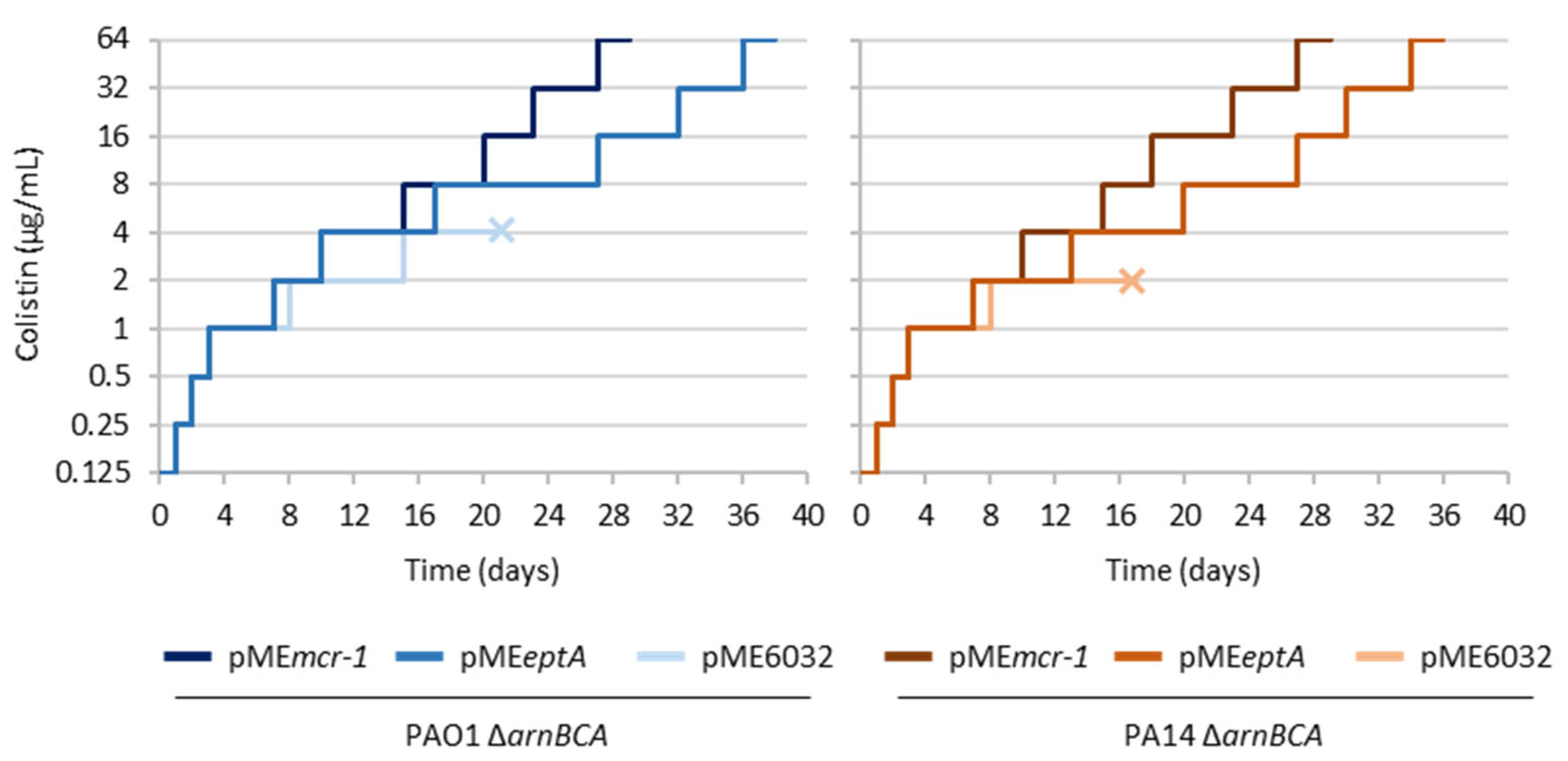
2.1. PEtN Transferases Support the Evolution of P. aeruginosa towards High Levels of Colistin Resistance
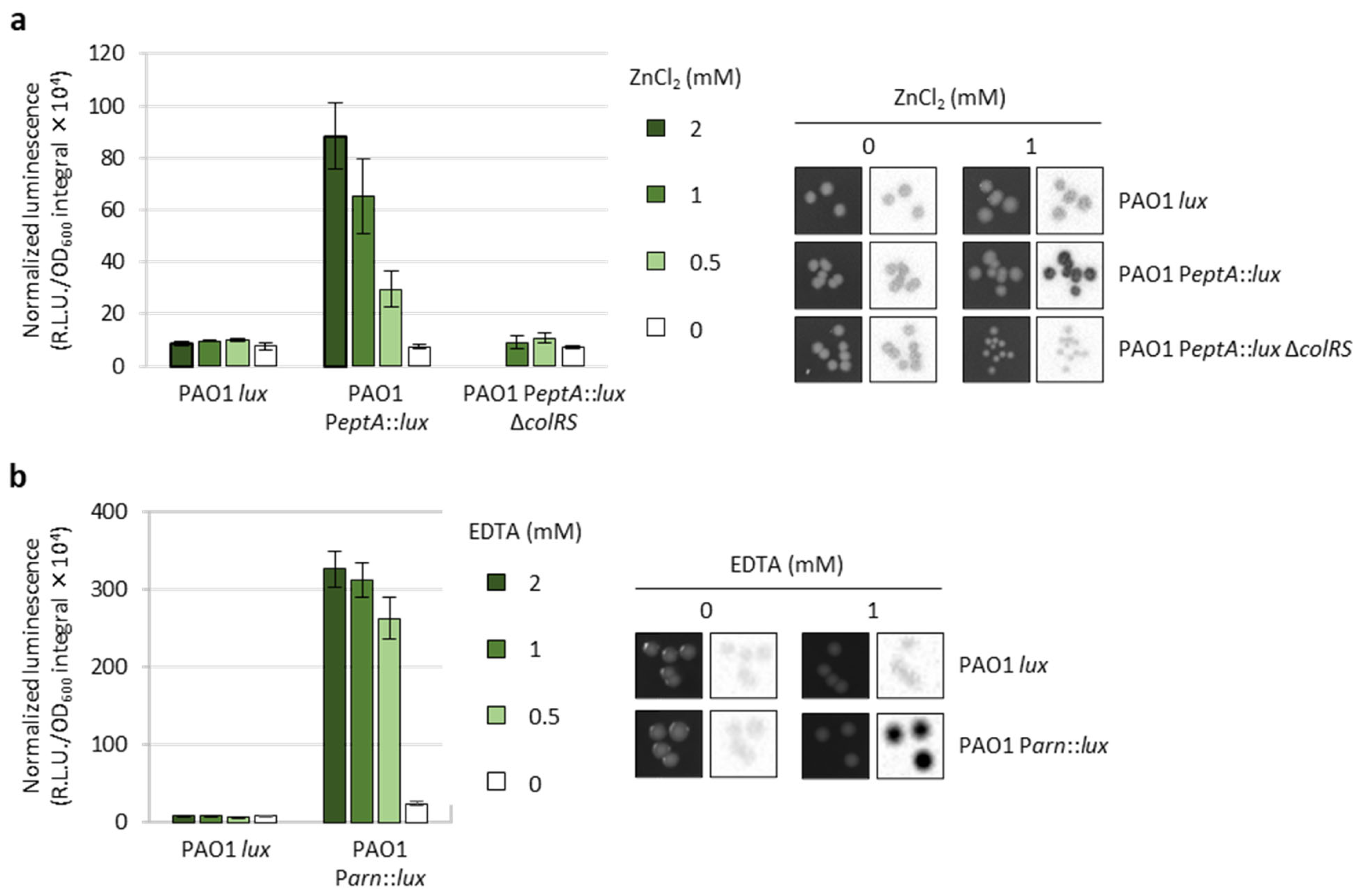
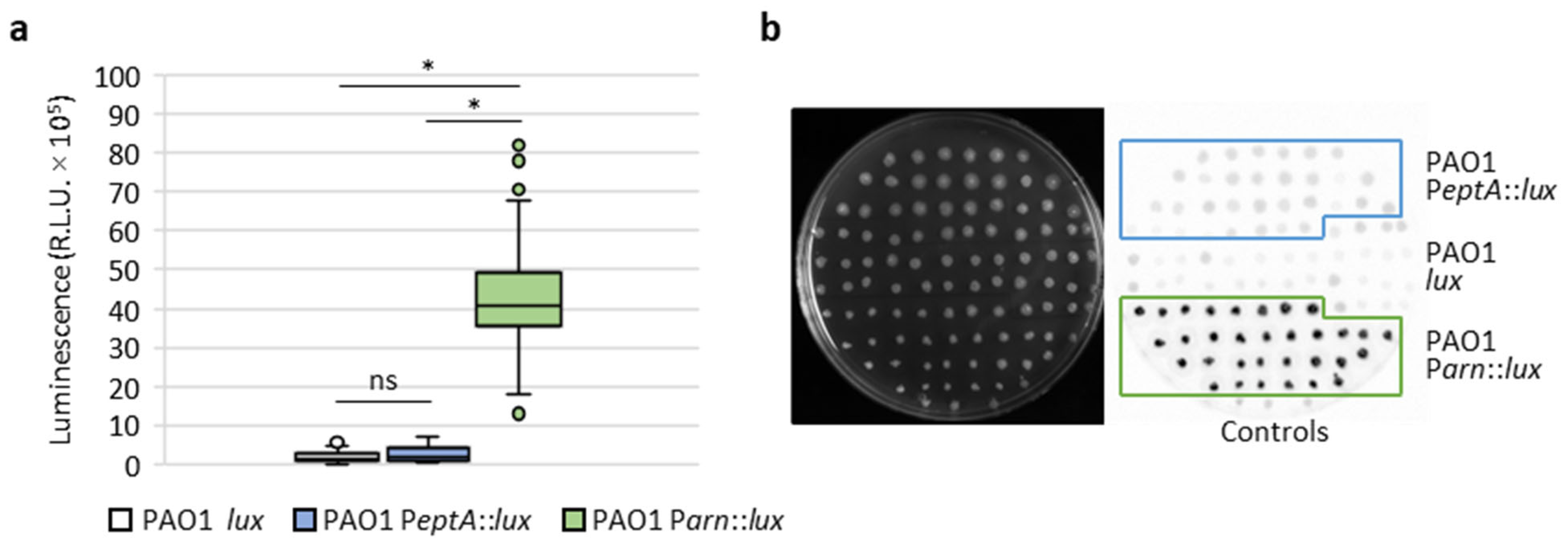
2.2. Generation and Validation of Reporter Strains for PeptA and Parn Activity
2.3. Identification of Genes Influencing eptA Promoter Activity and Evaluation of Their Impact on Colistin Resistance
2.4. Further Characterization of Genes Influencing eptA Promoter Activity
2.5. The eptA Gene Is Not Induced in Colistin-Resistant Spontaneous Mutants
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids and Growth Media
4.2. In Vitro Evolution Assays
4.3. Generation of Luminescent Reporter Strains, Deletion Mutants and Complementing Plasmids
4.4. Growth and Luminescence Assays
4.5. Transposon-Mediated Random Mutagenesis
4.6. MIC Assays
4.7. Selection of Colistin-Resistant Spontaneous Mutants
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Ledger, E.V.K.; Sabnis, A.; Edwards, A.M. Polymyxin and lipopeptide antibiotics: Membrane-targeting drugs of last resort. Microbiology 2022, 168, 001136. [Google Scholar] [CrossRef] [PubMed]
- Binsker, U.; Käsbohrer, A.; Hammerl, J.A. Global colistin use: A review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 2022, 46, fuab049. [Google Scholar] [CrossRef] [PubMed]
- Jeannot, K.; Bolard, A.; Plésiat, P. Resistance to polymyxins in Gram-negative organisms. Int. J. Antimicrob. Agents 2017, 49, 526–535. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Velkov, T.; Thompson, P.E.; Nation, R.L.; Li, J. Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 2010, 53, 1898–1916. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymyxins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmids or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Sabnis, A.; Hagart, K.L.; Klöckner, A.; Becce, M.; Evans, L.E.; Furniss, R.C.D.; Mavridou, D.A.; Murphy, R.; Stevens, M.M.; Davies, J.C.; et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. eLife 2021, 10, e65836. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Morand, S.; Rolain, J.M. Mechanisms of polymyxin resistance: Acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef]
- Baron, S.; Hadjadj, L.; Rolain, J.M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [Google Scholar] [CrossRef]
- Lichtenberg, M.; Jakobsen, T.H.; Kühl, M.; Kolpen, M.; Jensen, P.Ø.; Bjarnsholt, T. The structure-function relationship of Pseudomonas aeruginosa in infections and its influence on the microenvironment. FEMS Microbiol. Rev. 2022, 46, fuac018. [Google Scholar] [CrossRef]
- Schurek, K.N.; Sampaio, J.L.; Kiffer, C.R.; Sinto, S.; Mendes, C.M.; Hancock, R.E. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 4345–4351. [Google Scholar] [CrossRef]
- Moskowitz, S.M.; Brannon, M.K.; Dasgupta, N.; Pier, M.; Sgambati, N.; Miller, A.K.; Selgrade, S.E.; Miller, S.I.; Denton, M.; Conway, S.P.; et al. PmrB mutations promote polymyxin resistance of Pseudomonas aeruginosa isolated from colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2012, 56, 1019–1030. [Google Scholar] [CrossRef]
- Barrow, K.; Kwon, D.H. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 5150–5154. [Google Scholar] [CrossRef]
- Jochumsen, N.; Marvig, R.L.; Damkiær, S.; Jensen, R.L.; Paulander, W.; Molin, S.; Jelsbak, L.; Folkesson, A. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat. Commun. 2016, 7, 13002. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Imperi, F. Aminoarabinosylation of Lipid A Is Critical for the Development of Colistin Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e01820-17. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Cervoni, M.; Stefanelli, R.; Mancone, C.; Imperi, F. Effect of lipid A aminoarabinosylation on Pseudomonas aeruginosa colistin resistance and fitness. Int. J. Antimicrob. Agents 2020, 55, 105957. [Google Scholar] [CrossRef]
- Ghirga, F.; Stefanelli, R.; Cavinato, L.; Lo Sciuto, A.; Corradi, S.; Quaglio, D.; Calcaterra, A.; Casciaro, B.; Loffredo, M.R.; Cappiello, F.; et al. A novel colistin adjuvant identified by virtual screening for ArnT inhibitors. J. Antimicrob. Chemother. 2020, 75, 2564–2572. [Google Scholar] [CrossRef]
- Quaglio, D.; Mangoni, M.L.; Stefanelli, R.; Corradi, S.; Casciaro, B.; Vergine, V.; Lucantoni, F.; Cavinato, L.; Cammarone, S.; Loffredo, M.R.; et al. ent-Beyerane Diterpenes as a Key Platform for the Development of ArnT-Mediated Colistin Resistance Inhibitors. J. Org. Chem. 2020, 85, 10891–10901. [Google Scholar] [CrossRef]
- Nowicki, E.M.; O’Brien, J.P.; Brodbelt, J.S.; Trent, M.S. Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Mol. Microbiol. 2015, 97, 166–178. [Google Scholar] [CrossRef]
- Cervoni, M.; Lo Sciuto, A.; Bianchini, C.; Mancone, C.; Imperi, F. Exogenous and Endogenous Phosphoethanolamine Transferases Differently Affect Colistin Resistance and Fitness in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 778968. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Chandler, C.E.; Leung, L.M.; McElheny, C.L.; Mettus, R.T.; Shanks, R.M.Q.; Liu, J.H.; Goodlett, D.R.; Ernst, R.K.; Doi, Y. Structural Modification of Lipopolysaccharide Conferred by mcr-1 in Gram-Negative ESKAPE Pathogens. Antimicrob. Agents Chemother. 2017, 61, e00580-17. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed]
- Farr, S.B.; Arnosti, D.N.; Chamberlin, M.J.; Ames, B.N. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. USA 1989, 86, 5010–5014. [Google Scholar] [CrossRef]
- Paiardini, A.; Mantoni, F.; Giardina, G.; Paone, A.; Janson, G.; Leoni, L.; Rampioni, G.; Cutruzzolà, F.; Rinaldo, S. A novel bacterial l-arginine sensor controlling c-di-GMP levels in Pseudomonas aeruginosa. Proteins 2018, 86, 1088–1096. [Google Scholar] [CrossRef]
- Visaggio, D.; Pasqua, M.; Bonchi, C.; Kaever, V.; Visca, P.; Imperi, F. Cell aggregation promotes pyoverdine-dependent iron uptake and virulence in Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 902. [Google Scholar] [CrossRef]
- Banerjee, P.; Sahoo, P.K.; Sheenu; Adhikary, A.; Ruhal, R.; Jain, D. Molecular and structural facets of c-di-GMP signalling associated with biofilm formation in Pseudomonas aeruginosa. Mol. Asp. Med. 2021, 81, 101001. [Google Scholar] [CrossRef]
- Despotović, D.; Brandis, A.; Savidor, A.; Levin, Y.; Fumagalli, L.; Tawfik, D.S. Diadenosine tetraphosphate (Ap4A)—An E. coli alarmone or a damage metabolite? FEBS J. 2017, 284, 2194–2215. [Google Scholar] [CrossRef]
- Ferguson, F.; McLennan, A.G.; Urbaniak, M.D.; Jones, N.J.; Copeland, N.A. Re-evaluation of Diadenosine Tetraphosphate (Ap4A) From a Stress Metabolite to Bona Fide Secondary Messenger. Front. Mol. Biosci. 2020, 7, 606807. [Google Scholar] [CrossRef]
- Chung, E.S.; Lee, J.Y.; Rhee, J.Y.; Ko, K.S. Colistin resistance in Pseudomonas aeruginosa that is not linked to arnB. J. Med. Microbiol. 2017, 66, 833–841. [Google Scholar] [CrossRef]
- Sirithanakorn, C.; Cronan, J.E. Biotin, a universal and essential cofactor: Synthesis, ligation and regulation. FEMS Microbiol. Rev. 2021, 45, fuab003. [Google Scholar] [CrossRef]
- Carfrae, L.A.; MacNair, C.R.; Brown, C.M.; Tsai, C.N.; Weber, B.S.; Zlitni, S.; Rao, V.N.; Chun, J.; Junop, M.S.; Coombes, B.K.; et al. Mimicking the human environment in mice reveals that inhibiting biotin biosynthesis is effective against antibiotic-resistant pathogens. Nat. Microbiol. 2020, 5, 93–101. [Google Scholar] [CrossRef]
- Ji, X.; Zou, J.; Peng, H.; Stolle, A.S.; Xie, R.; Zhang, H.; Peng, B.; Mekalanos, J.J.; Zheng, J. Alarmone Ap4A is elevated by aminoglycoside antibiotics and enhances their bactericidal activity. Proc. Natl. Acad. Sci. USA 2019, 116, 9578–9585. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Jacobs, M.A.; Alwood, A.; Thaipisuttikul, I.; Spencer, D.; Haugen, E.; Ernst, S.; Will, O.; Kaul, R.; Raymond, C.; Levy, R.; et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 14339–14344. [Google Scholar] [CrossRef]
- Liberati, N.T.; Urbach, J.M.; Miyata, S.; Lee, D.G.; Drenkard, E.; Wu, G.; Villanueva, J.; Wei, T.; Ausubel, F.M. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 2833–2838. [Google Scholar] [CrossRef]
- Becher, A.; Schweizer, H.P. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 2000, 29, 948–950, 952. [Google Scholar] [CrossRef]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Lo Sciuto, A.; Spinnato, M.C.; Pasqua, M.; Imperi, F. Generation of Stable and Unmarked Conditional Mutants in Pseudomonas aeruginosa. Methods Mol. Biol. 2022, 2548, 21–35. [Google Scholar] [CrossRef]
- Milton, D.L.; O’Toole, R.; Horstedt, P.; Wolf-Watz, H. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1996, 178, 1310–1319. [Google Scholar] [CrossRef] [PubMed]
- Heeb, S.; Blumer, C.; Haas, D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J. Bacteriol. 2002, 184, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Rocchio, S.; Santorelli, D.; Rinaldo, S.; Franceschini, M.; Malatesta, F.; Imperi, F.; Federici, L.; Travaglini-Allocatelli, C.; Di Matteo, A. Structural and functional investigation of the Small Ribosomal Subunit Biogenesis GTPase A (RsgA) from Pseudomonas aeruginosa. FEBS J. 2019, 286, 4245–4260. [Google Scholar] [CrossRef]
- Pasqua, M.; Visaggio, D.; Lo Sciuto, A.; Genah, S.; Banin, E.; Visca, P.; Imperi, F. Ferric Uptake Regulator Fur Is Conditionally Essential in Pseudomonas aeruginosa. J. Bacteriol. 2017, 199, e00472-17. [Google Scholar] [CrossRef] [PubMed]
- Dubern, J.F.; Cigana, C.; De Simone, M.; Lazenby, J.; Juhas, M.; Schwager, S.; Bianconi, I.; Döring, G.; Eberl, L.; Williams, P.; et al. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ. Microbiol. 2015, 17, 4379–4393. [Google Scholar] [CrossRef]
- Imperi, F.; Fiscarelli, E.V.; Visaggio, D.; Leoni, L.; Visca, P. Activity and Impact on Resistance Development of Two Antivirulence Fluoropyrimidine Drugs in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2019, 9, 49. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in Gram negative bacteria. Bio/Technology 1983, 1, 784–790. [Google Scholar] [CrossRef]
- Rahme, L.G.; Stevens, E.J.; Wolfort, S.F.; Shao, J.; Tompkins, R.G.; Ausubel, F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science 1995, 268, 1899–1902. [Google Scholar] [CrossRef]

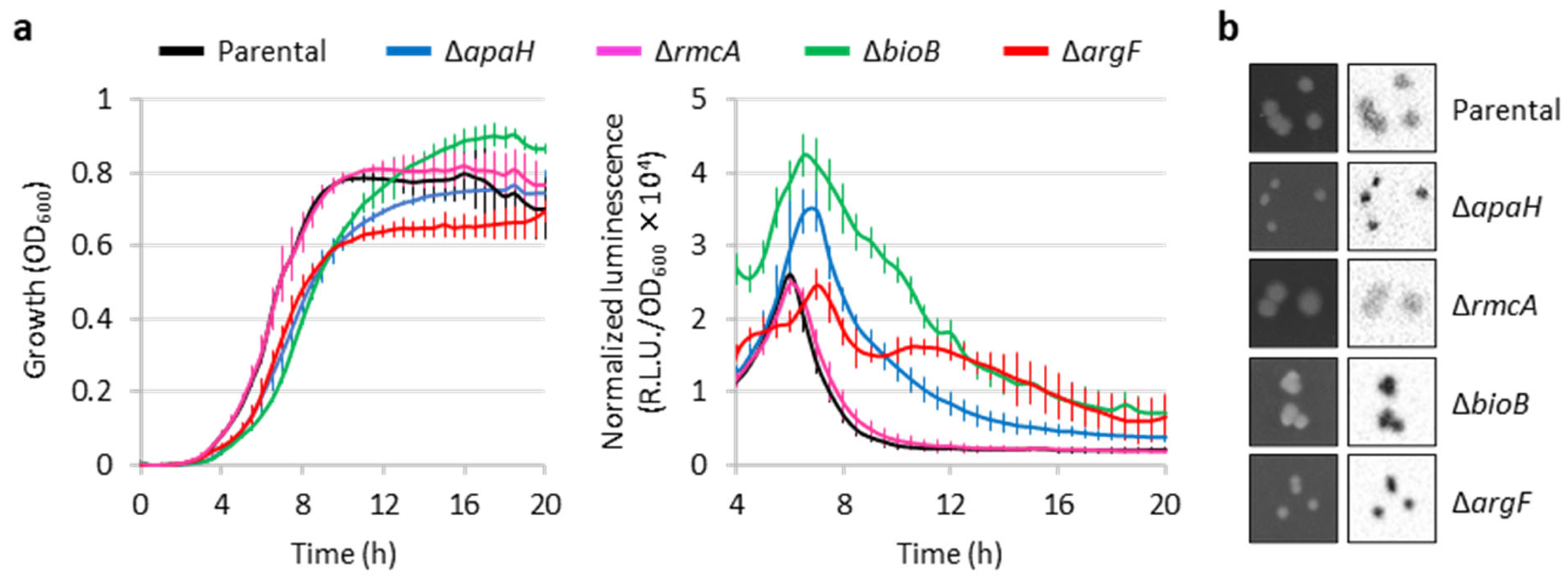

| Tn Mutant | Disrupted Gene (PA Number) | Insertion Site (Gene Length) | Gene Product | Function/Pathway |
|---|---|---|---|---|
| P34-96 | apaH (PA0590) | 95 (852 bp) | Diadenosine (Ap4A) tetraphosphatase | Ap4A degradation |
| P36-26 | rmcA (PA0575) | 2502 (3738 bp) | c-di-GMP phosphodiesterase | Redox regulator of c-di-GMP |
| P54-25 | bioB (PA0500) | 788 (1059 bp) | Biotin synthase | Biotin biosynthesis |
| P94-73 | argF (PA3537) | 379 (918 bp) | Anabolic ornithine carbamoyltransferase | Arginine biosynthesis |
| P117-04 | bioA (PA0420) | 1161 (1404 bp) | 7,8-diaminononanoate (DAPA) synthase | Biotin biosynthesis |
| Strain | Frequency of Resistant Mutants (±SD) a |
|---|---|
| PAO1 lux | 4.30 × 10−8 (±3.99 × 10−8) |
| PAO1 Parn::lux | 6.64 × 10−8 (±8.32 × 10−8) |
| PAO1 PeptA::lux | 3.72 × 10−8 (±8.45 × 10−8) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cervoni, M.; Sposato, D.; Lo Sciuto, A.; Imperi, F. Regulatory Landscape of the Pseudomonas aeruginosa Phosphoethanolamine Transferase Gene eptA in the Context of Colistin Resistance. Antibiotics 2023, 12, 200. https://doi.org/10.3390/antibiotics12020200
Cervoni M, Sposato D, Lo Sciuto A, Imperi F. Regulatory Landscape of the Pseudomonas aeruginosa Phosphoethanolamine Transferase Gene eptA in the Context of Colistin Resistance. Antibiotics. 2023; 12(2):200. https://doi.org/10.3390/antibiotics12020200
Chicago/Turabian StyleCervoni, Matteo, Davide Sposato, Alessandra Lo Sciuto, and Francesco Imperi. 2023. "Regulatory Landscape of the Pseudomonas aeruginosa Phosphoethanolamine Transferase Gene eptA in the Context of Colistin Resistance" Antibiotics 12, no. 2: 200. https://doi.org/10.3390/antibiotics12020200
APA StyleCervoni, M., Sposato, D., Lo Sciuto, A., & Imperi, F. (2023). Regulatory Landscape of the Pseudomonas aeruginosa Phosphoethanolamine Transferase Gene eptA in the Context of Colistin Resistance. Antibiotics, 12(2), 200. https://doi.org/10.3390/antibiotics12020200






