Genomic Profiling of Multidrug-Resistant Swine Escherichia coli and Clonal Relationship to Human Isolates in Peru
Abstract
:1. Introduction
2. Results
2.1. Serotype and Muli-Locus Sequence Typing (MLST)
2.2. Virulence and Resistance Genes
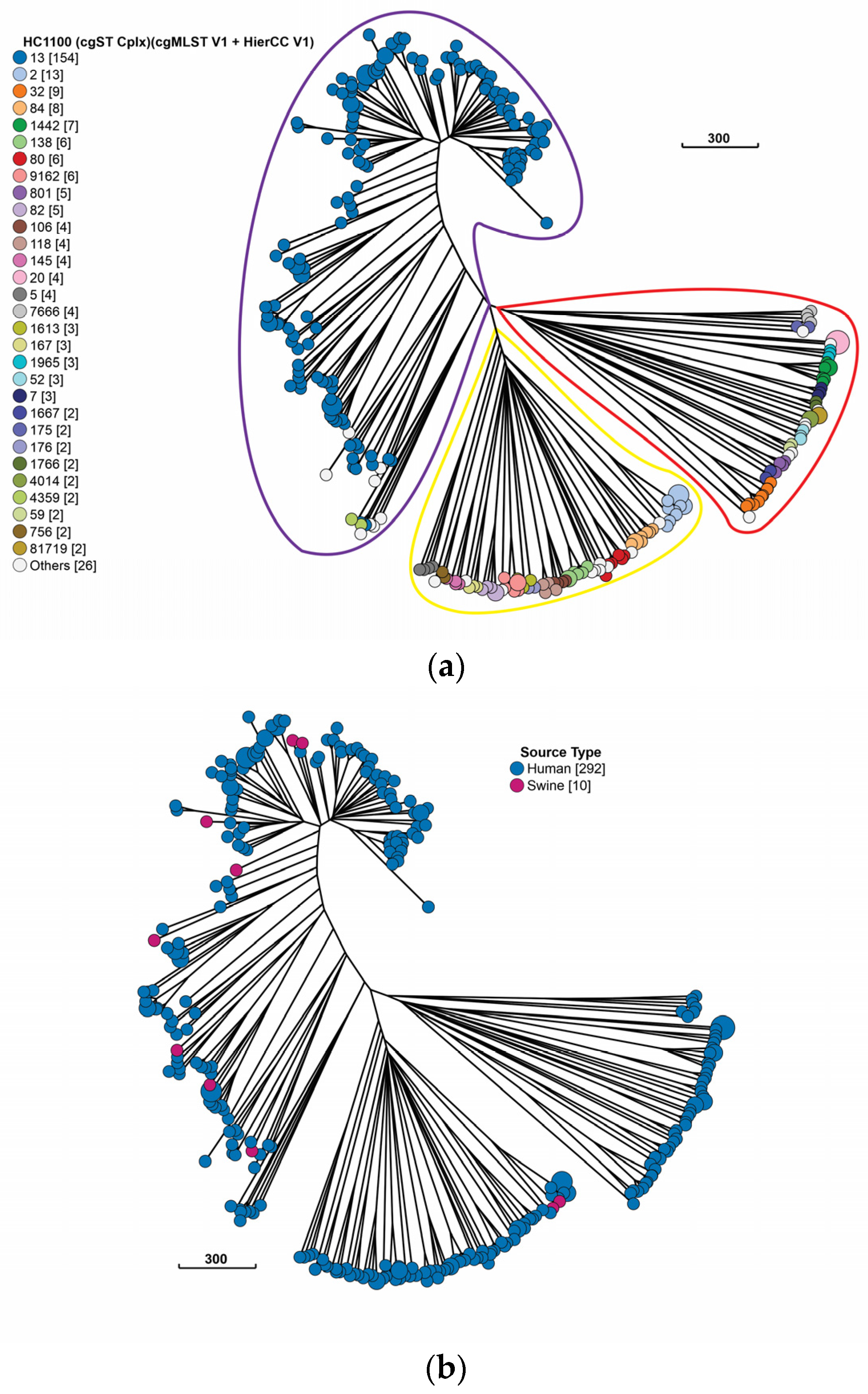
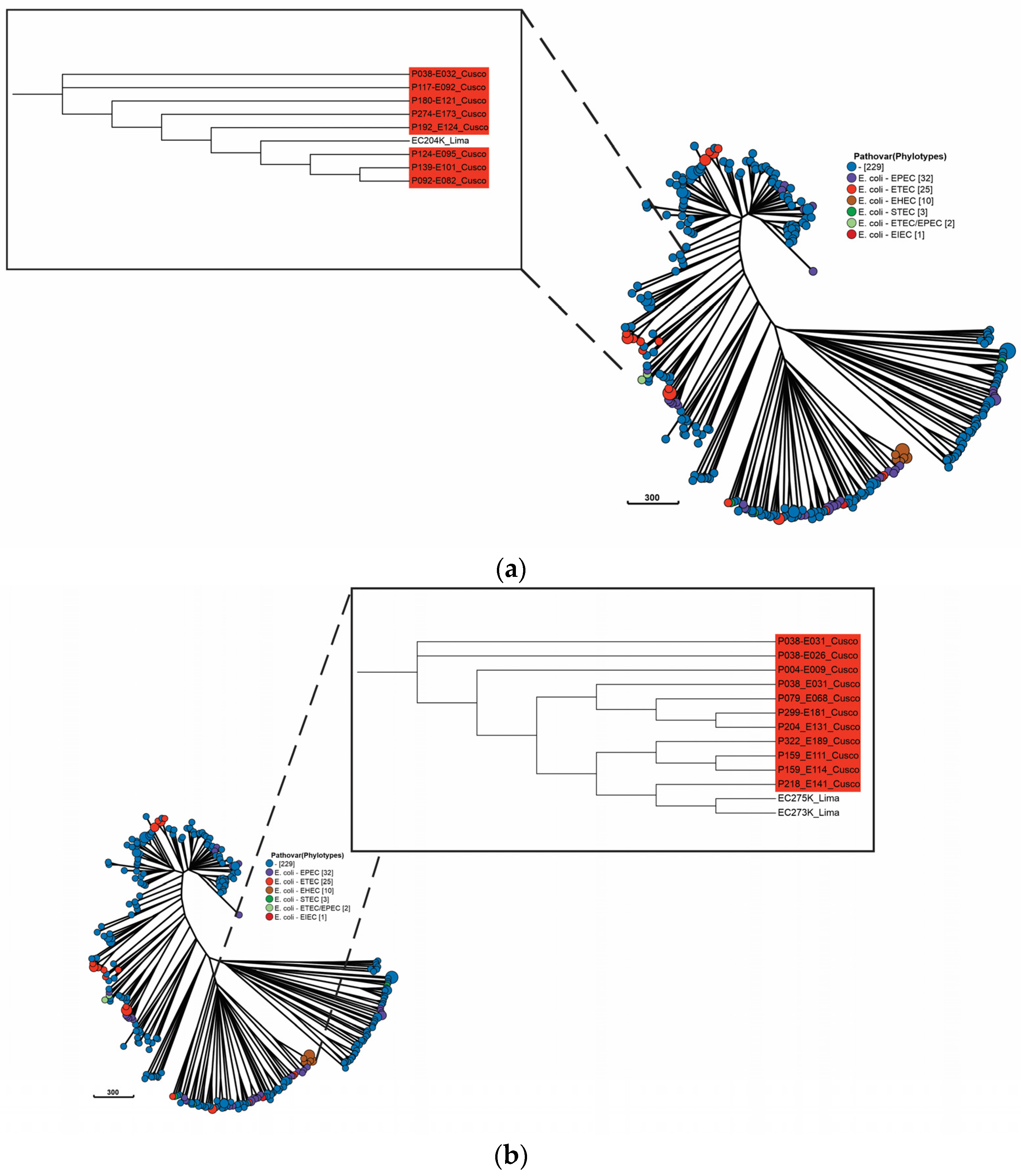
2.3. Population of Swine and Human Peruvian E. coli Isolates
3. Discussion
4. Materials and Methods
4.1. Isolation, Sequencing, and Genome Sequences
4.2. Genome Characterization
4.3. CgMLST Hierarchical Clustering Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tseng, M.; Fratamico, P.M.; Manning, S.D.; Funk, J.A. Shiga Toxin-Producing Escherichia coli in Swine: The Public Health Perspective. In Animal Health Research Reviews; Cambridge University Press: Cambridge, UK, 2014; pp. 63–75. [Google Scholar] [CrossRef]
- Losinger, W.C. Economic Impacts of the Mortality Rate for Suckling Pigs in the United States. J. Am. Vet. Med. Assoc. 2005, 227, 896–902. [Google Scholar] [CrossRef]
- Cocker, D.; Chidziwisano, K.; Mphasa, M.; Mwapasa, T.; Lewis, J.M.; Rowlingson, B.; Sammarro, M.; Bakali, W.; Salifu, C.; Zuza, A.; et al. Investigating One Health Risks for Human Colonisation with Extended Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella Pneumoniae in Malawian Households: A Longitudinal Cohort Study. Lancet Microbe 2023, 4, E534–E543. [Google Scholar] [CrossRef]
- Renzhammer, R.; Loncaric, I.; Roch, F.F.; Pinior, B.; Käsbohrer, A.; Spergser, J.; Ladinig, A.; Unterweger, C. Prevalence of Virulence Genes and Antimicrobial Resistances in e. Coli Associated with Neonatal Diarrhea, Postweaning Diarrhea, and Edema Disease in Pigs from Austria. Antibiotics 2020, 9, 208. [Google Scholar] [CrossRef]
- Kuhnke, D. Occurrence of ESBL-Producing Escherichia coli in Healthy, Living Food-Producing Animals in Europe: A Systematic Review. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 2020, 15, 1–13. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Toleman, M.A.; Walsh, T.R. Does Broad-Spectrum β -Lactam Resistance Due to NDM-1 Herald the End of the Antibiotic Era for Treatment of Infections Caused by Gram-Negative Bacteria? J. Antimicrob. Chemother. 2011, 66, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Asakura, H.; Sakata, J.; Nakamura, H.; Yamamoto, S.; Murakami, S. Phylogenetic Diversity and Antimicrobial Resistance of Campylobacter coli from Humans and Animals in Japan. Microbes Environ. 2019, 34, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Boerlin, P.; Boerlin, P. Molecular Epidemiology of Antimicrobial Resistance in Veterinary Medicine: Where Do We Go? Anim. Health Res. Rev. 2004, 5, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.; Kuhnert, P.; Boerlin, P. Antimicrobial Resistance and Resistance Gene Determinants in Clinical Escherichia coli from Different Animal Species in Switzerland. Vet. Microbiol. 2003, 91, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Junker, E.; Schroeter, A.; Malorny, B.; Lehmann, S.; Helmuth, R. Phenotypic and Genotypic Characterization of Antimicrobial Resistance in German Escherichia coli Isolates from Cattle, Swine and Poultry. J. Antimicrob. Chemother. 2003, 52, 489–492. [Google Scholar] [CrossRef]
- Instituto Nacional de Salud. Plan Multisectorial para Enfrentar la Resistencia a los Antimicrobianos 2019–2021. Available online: https://antimicrobianos.ins.gob.pe/images/contenido/plan-nacional/Decreto_Supremo_010-2019-SA-c.pdf (accessed on 10 September 2022).
- Abraham, S.; Trott, D.J.; Jordan, D.; Gordon, D.M.; Groves, M.D.; Fairbrother, J.M.; Smith, M.G.; Zhang, R.; Chapman, T.A. Phylogenetic and Molecular Insights into the Evolution of Multidrug-Resistant Porcine Enterotoxigenic Escherichia coli in Australia. Int. J. Antimicrob. Agents 2014, 44, 105–111. [Google Scholar] [CrossRef] [PubMed]
- García, V.; Gambino, M.; Pedersen, K.; Haugegaard, S.; Olsen, J.E.; Herrero-Fresno, A. F4- and F18-Positive Enterotoxigenic Escherichia coli Isolates from Diarrhea of Postweaning Pigs: Genomic Characterization. Appl. Environ. Microbiol. 2020, 86, e01913-20. [Google Scholar] [CrossRef]
- de Lagarde, M.; Vanier, G.; Arsenault, J.; Fairbrother, J.M. High Risk Clone: A Proposal of Criteria Adapted to the One Health Context with Application to Enterotoxigenic Escherichia coli in the Pig Population. Antibiotics 2021, 10, 244. [Google Scholar] [CrossRef]
- Álvarez-Suárez, M.E.; Otero, A.; García-López, M.L.; Dahbi, G.; Blanco, M.; Mora, A.; Blanco, J.; Santos, J.A. Genetic Characterization of Shiga Toxin-Producing Escherichia coli (STEC) and Atypical Enteropathogenic Escherichia coli (EPEC) Isolates from Goat’s Milk and Goat Farm Environment. Int. J. Food Microbiol. 2016, 236, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Bai, X.; Jin, Y.; Hu, B.; Wang, H.; Sun, H.; Fan, R.; Fu, S.; Xiong, Y. High Prevalence of Virulence Genes in Specific Genotypes of Atypical Enteropathogenic Escherichia coli. Front. Cell Infect. Microbiol. 2017, 7, 109. [Google Scholar] [CrossRef]
- García-Meniño, I.; García, V.; Mora, A.; Díaz-Jiménez, D.; Flament-Simon, S.C.; Alonso, M.P.; Blanco, J.E.; Blanco, M.; Blanco, J. Swine Enteric Colibacillosis in Spain: Pathogenic Potential of Mcr-1 ST10 and ST131 E. coli Isolates. Front. Microbiol. 2018, 9, 2659. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (Expec) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fernandez, A.; Villa, L.; Bibbolino, G.; Bressan, A.; Trancassini, M.; Pietropaolo, V.; Venditti, M.; Antonelli, G.; Carattoli, A. Novel Insights and Features of the NDM-5-Producing Escherichia coli Sequence Type 167 High-Risk Clone. mSphere 2020, 5, e00269-20. [Google Scholar] [CrossRef]
- Sacsaquispe-Contreras, R.; Bailón-Calderón, H. Identification of Carbapenem-Resistant Genes in Enterobacteria from Peruvian Hospitals, 2013–2017. Rev. Peru. Med. Exp. Salud Publica 2018, 35, 259–264. [Google Scholar] [CrossRef]
- Schink, A.K.; Kadlec, K.; Kaspar, H.; Mankertz, J.; Schwarz, S. Analysis of Extended-Spectrum-β-Lactamase-Producing Escherichia coli Isolates Collected in the GERM-Vet Monitoring Programme. J. Antimicrob. Chemother. 2013, 68, 1741–1749. [Google Scholar] [CrossRef]
- Fischer, J.; Rodríguez, I.; Baumann, B.; Guiral, E.; Beutin, L.; Schroeter, A.; Kaesbohrer, A.; Pfeifer, Y.; Helmuth, R.; Guerra, B. BlaCTX-M-15-Carrying Escherichia coli and Salmonella Isolates from Livestock and Food in Germany. J. Antimicrob. Chemother. 2014, 69, 2951–2958. [Google Scholar] [CrossRef]
- Tamta, S.; Vinodh, V.K.; Pruthvishree, B.S.; Karthikeyan, R.; Rupner, R.N.; Chethan, G.E.; Dubal, Z.B.; Sinha, D.K.; Singh, B.R. Faecal Carriage of Extended Spectrum Beta-Lactamase (ESBL) and New Delhi Metallo Beta-Lactamase(NDM) Producing Escherichia coli between Piglets and Pig Farmworkers. Comp. Immunol. Microbiol. Infect. Dis. 2020, 73, 101564. [Google Scholar] [CrossRef]
- Krause, G.; Zimmermann, S.; Beutin, L. Investigation of Domestic Animals and Pets as a Reservoir for Intimin- (Eae) Gene Positive Escherichia coli Types. Vet. Microbiol. 2005, 106, 87–95. [Google Scholar] [CrossRef]
- Watson, V.E.; Hazen, T.H.; Rasko, D.A.; Jacob, M.E.; Elfenbein, J.R.; Stauffer, S.H.; Gookin, J.L. Comparative Genomics of Atypical Enteropathogenic Escherichia coli from Kittens and Children Identifies Bacterial Factors Associated with Virulence in Kittens. Infect. Immun. 2021, 89, e00619-20. [Google Scholar] [CrossRef] [PubMed]
- Simner, P.J.; Bergman, Y.; Conzemius, R.; Jacobs, E.; Tekle, T.; Beisken, S.; Tamma, P.D. An NDM-Producing Escherichia coli Clinical Isolate Exhibiting Resistance to Cefiderocol and the Combination of Ceftazidime-Avibactam and Aztreonam: Another Step Towards Pan-β-Lactam Resistance. Open Forum Infect. Dis. 2023, 10, ofad276. [Google Scholar] [CrossRef]
- Carhuaricra, D.; Duran Gonzales, C.G.; Rodríguez Cueva, C.L.; Ignacion León, Y.; Silvestre Espejo, T.; Marcelo Monge, G.; Rosadio Alcántara, R.H.; Lincopan, N.; Espinoza, L.L.; Maturrano Hernández, L. Occurrence and Genomic Characterization of Mcr-1-Harboring Escherichia coli Isolates from Chicken and Pig Farms in Lima, Peru. Antibiotics 2022, 11, 1781. [Google Scholar] [CrossRef]
- Lentz, S.A.M.; Dalmolin, T.V.; Barth, A.L.; Martins, A.F. Mcr-1 Gene in Latin America: How Is It Disseminated Among Humans, Animals, and the Environment? Front. Public Health 2021, 9, 648940. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, H.M.; Nguyen, C.V.; Nguyen, T.V.; Nguyen, M.T.; Thai, H.Q.; Ho, M.H.; Thwaites, G.; Ngo, H.T.; Baker, S.; et al. Use of Colistin and Other Critical Antimicrobials on Pig and Chicken Farms in Southern Vietnam and Its Association with Resistance in Commensal Escherichia coli Bacteria. Appl. Environ. Microbiol. 2016, 82, 3727. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Malhotra-Kumar, S.; Xavier, B.B.; Das, A.J.; Lammens, C.; Butaye, P.; Goossens, H. Colistin Resistance Gene Mcr-1 Harboured on a Multidrug Resistant Plasmid. Lancet Infect. Dis. 2016, 16, 283–284. [Google Scholar] [CrossRef] [PubMed]
- Rhouma, M.; Beaudry, F.; Thériault, W.; Letellier, A. Colistin in Pig Production: Chemistry, Mechanism of Antibacterial Action, Microbial Resistance Emergence, and One Health Perspectives. Front. Microbiol. 2016, 7, 1789. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Yang, Y.; Miao, M.; Chavda, K.D.; Mediavilla, J.R.; Xie, X.; Feng, P.; Tang, Y.W.; Kreiswirth, B.N.; Chen, L.; et al. Complete Sequences of Mcr-1-Harboring Plasmids from Extended-Spectrum-β-Lactamase- and Carbapenemase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2016, 60, 4351–4354. [Google Scholar] [CrossRef] [PubMed]
- Valiakos, G.; Kapna, I. Colistin Resistant Mcr Genes Prevalence in Livestock Animals (Swine, Bovine, Poultry) from a Multinational Perspective. A Systematic Review. Vet. Sci. 2021, 8, 265. [Google Scholar] [CrossRef]
- Zhang, S.; Abbas, M.; Rehman, M.U.; Wang, M.; Jia, R.; Chen, S.; Liu, M.; Zhu, D.; Zhao, X.; Gao, Q.; et al. Updates on the Global Dissemination of Colistin-Resistant Escherichia coli: An Emerging Threat to Public Health. Sci. Total Environ. 2021, 799, 149280. [Google Scholar] [CrossRef] [PubMed]
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin Use and Colistin Resistance in Bacteria from Animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef]
- SENASA. Resolucion Directoral-No 0091-2019-MINAGRI-SENASA-DIAIA. Disponen Prohibir la Importación, Comercialización, Fabricación o Elaboración de Productos Veterinarios que Contengan el Principio Activo Colistina (Polimixina E) o Cualquiera de sus Sales. Available online: https://www.senasa.gob.pe/senasacontigo/minagri-prohiben-importacion-comercializacion-y-fabricacion-de-colistina-en-el-pais/ (accessed on 6 November 2023).
- Dawangpa, A.; Lertwatcharasarakul, P.; Boonsoongnern, A.; Ratanavanichrojn, N.; Sanguankiat, A.; Pinniam, N.; Jala, S.; Laopiem, S.; Tulayakul, P. Multidrug Resistance Problems Targeting Piglets and Environmental Health by Escherichia coli in Intensive Swine Farms. Emerg. Contam. 2022, 8, 123–133. [Google Scholar] [CrossRef]
- Peng, Z.; Hu, Z.; Li, Z.; Zhang, X.; Jia, C.; Li, T.; Dai, M.; Tan, C.; Xu, Z.; Wu, B.; et al. Antimicrobial Resistance and Population Genomics of Multidrug-Resistant Escherichia coli in Pig Farms in Mainland China. Nat. Commun. 2022, 13, 1116. [Google Scholar] [CrossRef] [PubMed]
- García-Meniño, I.; García, V.; Alonso, M.P.; Blanco, J.E.; Blanco, J.; Mora, A. Clones of Enterotoxigenic and Shiga Toxin-Producing Escherichia coli Implicated in Swine Enteric Colibacillosis in Spain and Rates of Antibiotic Resistance. Vet. Microbiol. 2021, 252, 108924. [Google Scholar] [CrossRef]
- Kolář, M.; Urbánek, K.; Látal, T. Antibiotic Selective Pressure and Development of Bacterial Resistance. Int. J. Antimicrob. Agents 2001, 17, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.E.; Pitout, J.D.D. Extended-Spectrum β-Lactamase, Carbapenemase and AmpC Producing Enterobacteriaceae in Companion Animals. Vet. Microbiol. 2014, 170, 10–18. [Google Scholar] [CrossRef]
- Snow, L.C.; Warner, R.G.; Cheney, T.; Wearing, H.; Stokes, M.; Harris, K.; Teale, C.J.; Coldham, N.G. Risk Factors Associated with Extended Spectrum Beta-Lactamase Escherichia coli (CTX-M) on Dairy Farms in North West England and North Wales. Prev. Vet. Med. 2012, 106, 225–234. [Google Scholar] [CrossRef]
- Cuong, N.V.; Padungtod, P.; Thwaites, G.; Carrique-Mas, J.J. Antimicrobial Usage in Animal Production: A Review of the Literature with a Focus on Low-and Middle-Income Countries. Antibiotics 2018, 7, 75. [Google Scholar] [CrossRef]
- Murray, M.; Salvatierra, G.; Dávila-Barclay, A.; Ayzanoa, B.; Castillo-Vilcahuaman, C.; Huang, M.; Pajuelo, M.J.; Lescano, A.G.; Cabrera, L.; Calderón, M.; et al. Market Chickens as a Source of Antibiotic-Resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 2021, 12, 327. [Google Scholar] [CrossRef] [PubMed]
- Boolchandani, M.; Blake, K.S.; Tilley, D.H.; Cabada, M.M.; Schwartz, D.J.; Patel, S.; Morales, M.L.; Meza, R.; Soto, G.; Isidean, S.D.; et al. Impact of International Travel and Diarrhea on Gut Microbiome and Resistome Dynamics. Nat. Commun. 2022, 13, 7485. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Tang, X.; Lu, P.; Wu, B.; Xu, Z.; Liu, W.; Zhang, R.; Bei, W.; Chen, H.; Tan, C. Clonal Analysis and Virulent Traits of Pathogenic Extraintestinal Escherichia coli Isolates from Swine in China. BMC Vet. Res. 2012, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Chen, X.; Zheng, S.; Han, D.; Wang, Y.; Wang, R.; Wang, B.; Chen, Y. Prevalence and Genetic Diversity of Human Diarrheagenic Escherichia coli Isolates by Multilocus Sequence Typing. Int. J. Infect. Dis. 2018, 67, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.J.; De Maere, M.Z.; Djordjevic, S.P. Australian Porcine Clonal Complex 10 (CC10) Escherichia coli Belong to Multiple Sublineages of a Highly Diverse Global CC10 Phylogeny. Microb. Genom. 2019, 5, e000225. [Google Scholar] [CrossRef]
- Sahl, J.W.; Steinsland, H.; Redman, J.C.; Angiuoli, S.V.; Nataro, J.P.; Sommerfelt, H.; Rasko, D.A. A Comparative Genomic Analysis of Diverse Clonal Types of Enterotoxigenic Escherichia coli Reveals Pathovar-Specific Conservation. Infect. Immun. 2011, 79, 950. [Google Scholar] [CrossRef]
- Hao, W.; Allen, V.G.; Jamieson, F.B.; Low, D.E.; Alexander, D.C. Phylogenetic Incongruence in E. coli O104: Understanding the Evolutionary Relationships of Emerging Pathogens in the Face of Homologous Recombination. PLoS ONE 2012, 7, e33971. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Achtman, M.; Brown, D.; Chattaway, M.; Dallman, T.; Delahay, R.; Kornschober, C.; et al. The EnteroBase User’s Guide, with Case Studies on Salmonella Transmissions, Yersinia Pestis Phylogeny, and Escherichia Core Genomic Diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Yan, S.; Jiang, Z.; Zhang, W.; Liu, Z.; Dong, X.; Li, D.; Liu, Z.; Li, C.; Liu, X.; Zhu, L. Genomes-Based MLST, CgMLST, WgMLST and SNP Analysis of Salmonella Typhimurium from Animals and Humans. Comp. Immunol. Microbiol. Infect. Dis. 2023, 96, 101973. [Google Scholar] [CrossRef]
- Alvarez, L.; Carhuaricra, D.; Palomino-Farfan, J.; Calle, S.; Maturrano, L.; Siuce, J. Draft Genome Sequences of 10 Multidrug-Resistant Escherichia coli Strains Isolated from Piglets in Peru. Microbiol. Resour. Announc. 2022, 11, e00706-22. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.; Brenner, D.J.; Krieg, N.R.; Staley, J.R. Bergey’s Manual® of Systematic Bacteriology; Springer: New York, NY, USA, 2005; Volume 2. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, 32nd ed.; CLSI Document M02-A11; Clinical and Laboratory Standards: Wayne, PA, USA, 2018. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An Integrated Platform for Visualization and Analysis of High-Throughput Sequence-Based Experimental Data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Zhou, Z.; Charlesworth, J.; Achtman, M. HierCC: A Multi-Level Clustering Scheme for Population Assignments Based on Core Genome MLST. Bioinformatics 2021, 37, 3645. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
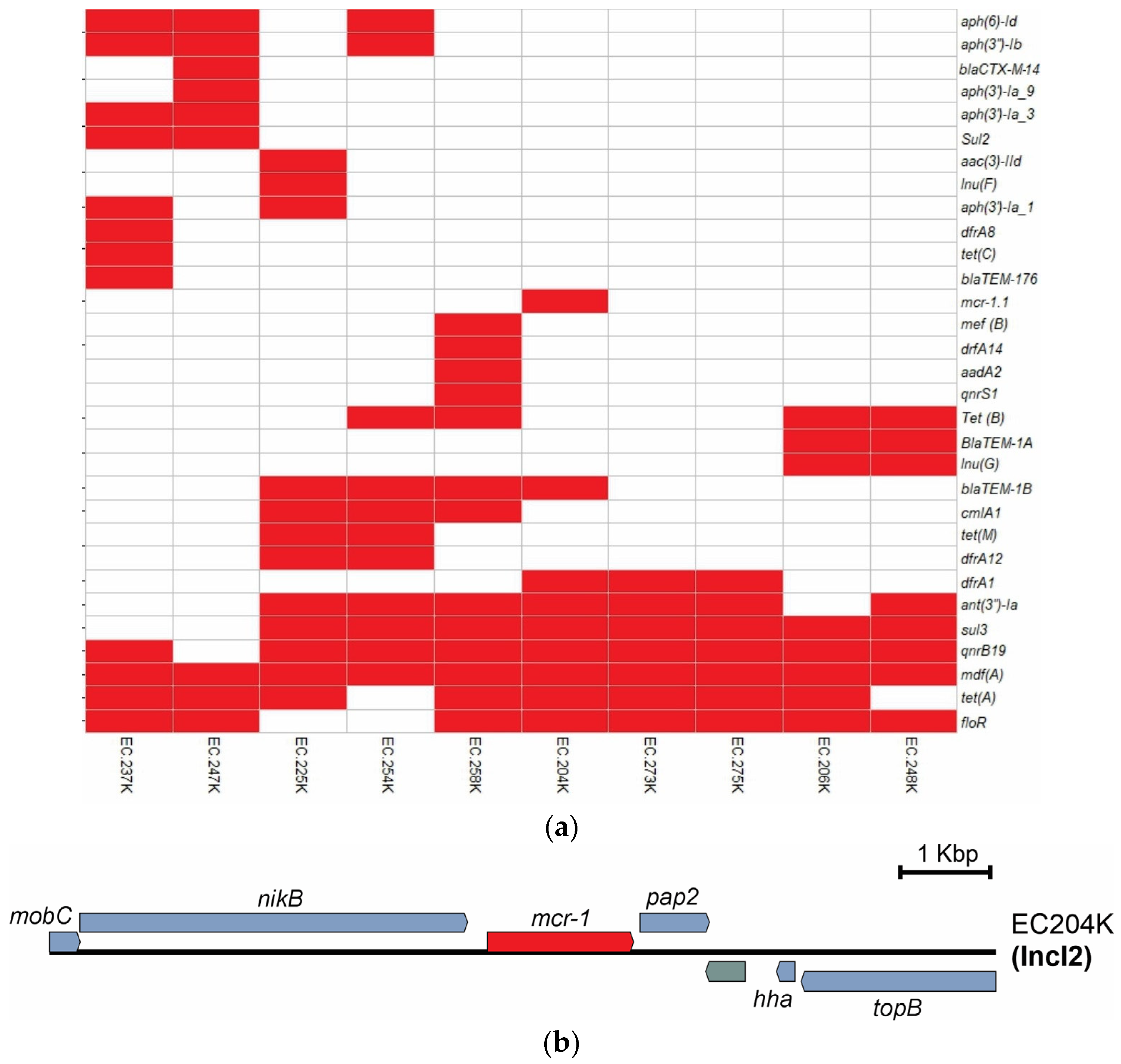
| Classes of Antibiotics | Gene | Gene No./Isolate No. |
|---|---|---|
| Tetracyclines | tet(A) | 8/10 |
| tet(B) | 4/10 | |
| tet(M) | 2/10 | |
| tet(C) | 1/10 | |
| Beta-lactam | blaTEM-1A | 2/10 |
| blaTEM-1B | 4/10 | |
| blaTEM-176 | 1/10 | |
| blaCTX-M-14 | 1/10 | |
| Amphenicol | floR | 8/10 |
| cmlA1 | 3/10 | |
| Aminoglycosides | aac(3)-IId | 1/10 |
| aadA2 | 1/10 | |
| ant(3″)-Ia | 7/10 | |
| aph(3′)-Ia_3 | 2/10 | |
| aph(3′)-Ia_1 | 2/10 | |
| aph(3′)-Ia_9 | 1/10 | |
| aph(6)-Id | 3/10 | |
| aph(3″)-Ib | 3/10 | |
| Sulfonamides | sul3 | 8/10 |
| sul2 | 2/10 | |
| Quinolones | qnrS1 | 1/10 |
| qnrB19 | 9/10 | |
| Lincomycin | lnu(F) | 1/10 |
| lnu(G) | 2/10 | |
| Trimethoprim | dfrA12 | 2/10 |
| dfrA1 | 3/10 | |
| drfA14 | 1/10 | |
| dfrA8 | 1/10 | |
| Macrolides | mef(B) | 1/10 |
| mdf(A) | 10/10 | |
| Colistin | mcr-1.1 | 1/10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez, L.; Carhuaricra, D.; Palomino-Farfan, J.; Calle, S.; Maturrano, L.; Siuce, J. Genomic Profiling of Multidrug-Resistant Swine Escherichia coli and Clonal Relationship to Human Isolates in Peru. Antibiotics 2023, 12, 1748. https://doi.org/10.3390/antibiotics12121748
Alvarez L, Carhuaricra D, Palomino-Farfan J, Calle S, Maturrano L, Siuce J. Genomic Profiling of Multidrug-Resistant Swine Escherichia coli and Clonal Relationship to Human Isolates in Peru. Antibiotics. 2023; 12(12):1748. https://doi.org/10.3390/antibiotics12121748
Chicago/Turabian StyleAlvarez, Luis, Dennis Carhuaricra, Joel Palomino-Farfan, Sonia Calle, Lenin Maturrano, and Juan Siuce. 2023. "Genomic Profiling of Multidrug-Resistant Swine Escherichia coli and Clonal Relationship to Human Isolates in Peru" Antibiotics 12, no. 12: 1748. https://doi.org/10.3390/antibiotics12121748
APA StyleAlvarez, L., Carhuaricra, D., Palomino-Farfan, J., Calle, S., Maturrano, L., & Siuce, J. (2023). Genomic Profiling of Multidrug-Resistant Swine Escherichia coli and Clonal Relationship to Human Isolates in Peru. Antibiotics, 12(12), 1748. https://doi.org/10.3390/antibiotics12121748





