Benzodioxane–Benzamides as FtsZ Inhibitors: Effects of Linker’s Functionalization on Gram-Positive Antimicrobial Activity
Abstract
:1. Introduction
2. Results
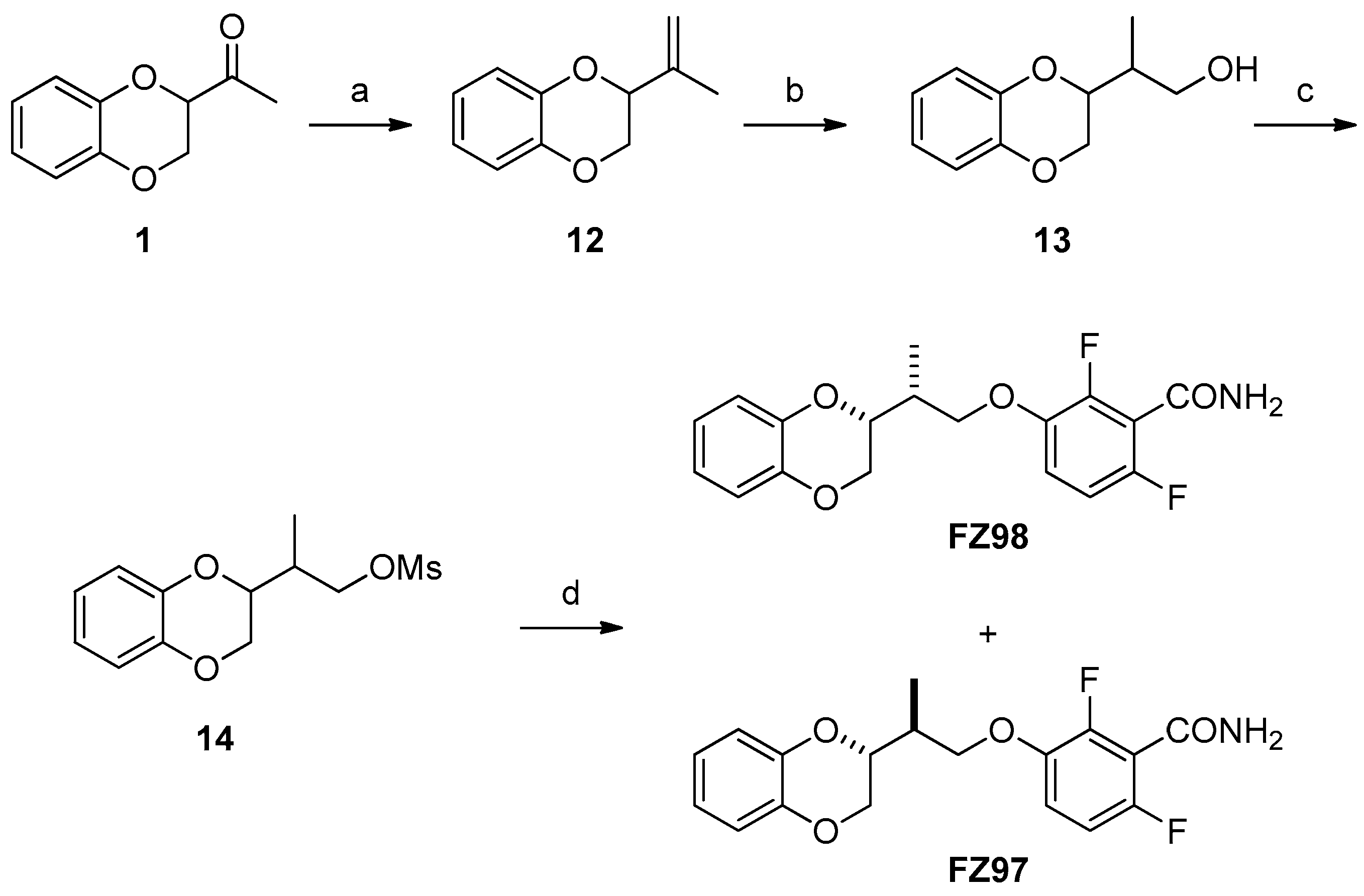
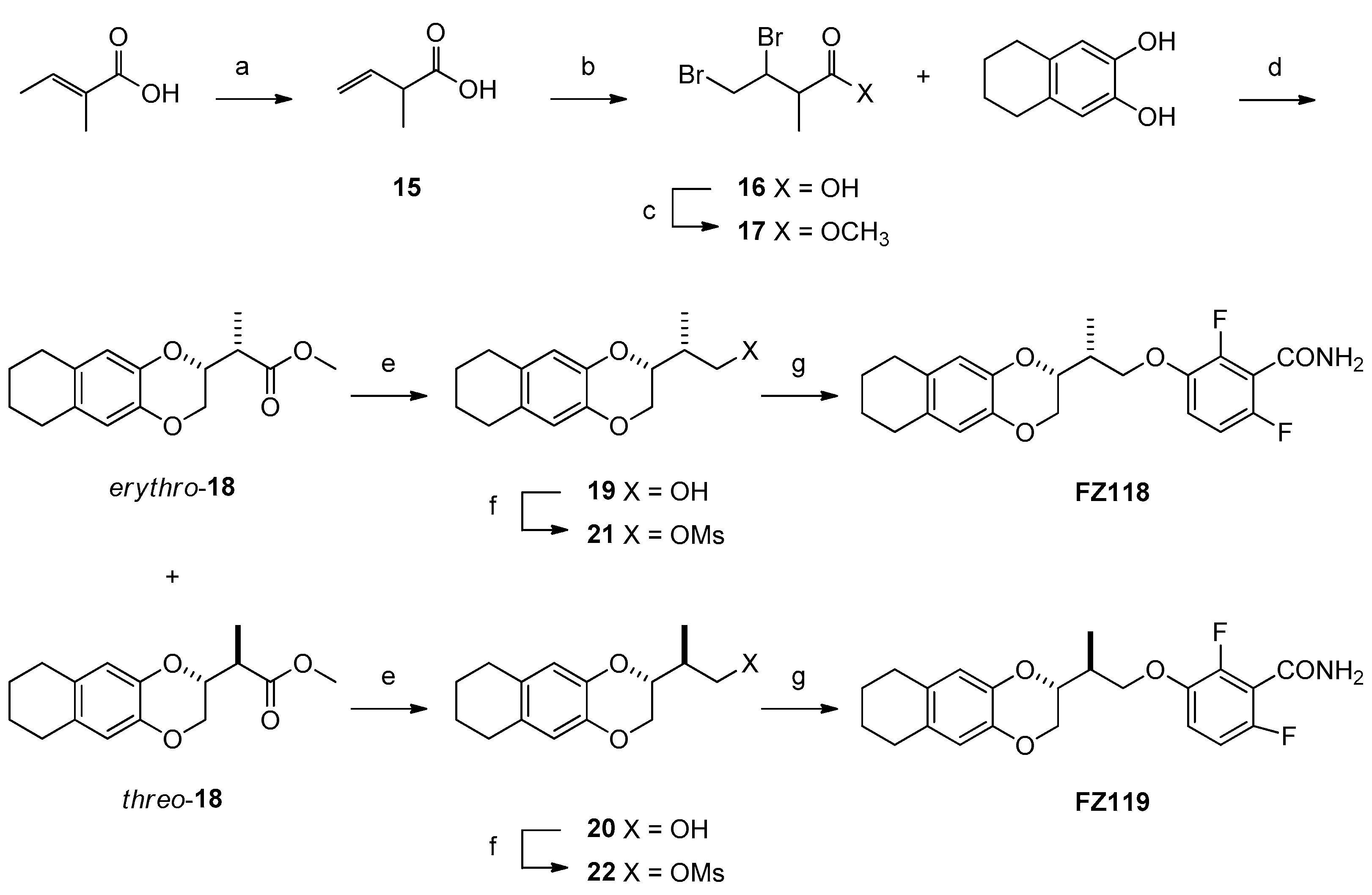
2.1. Chemistry
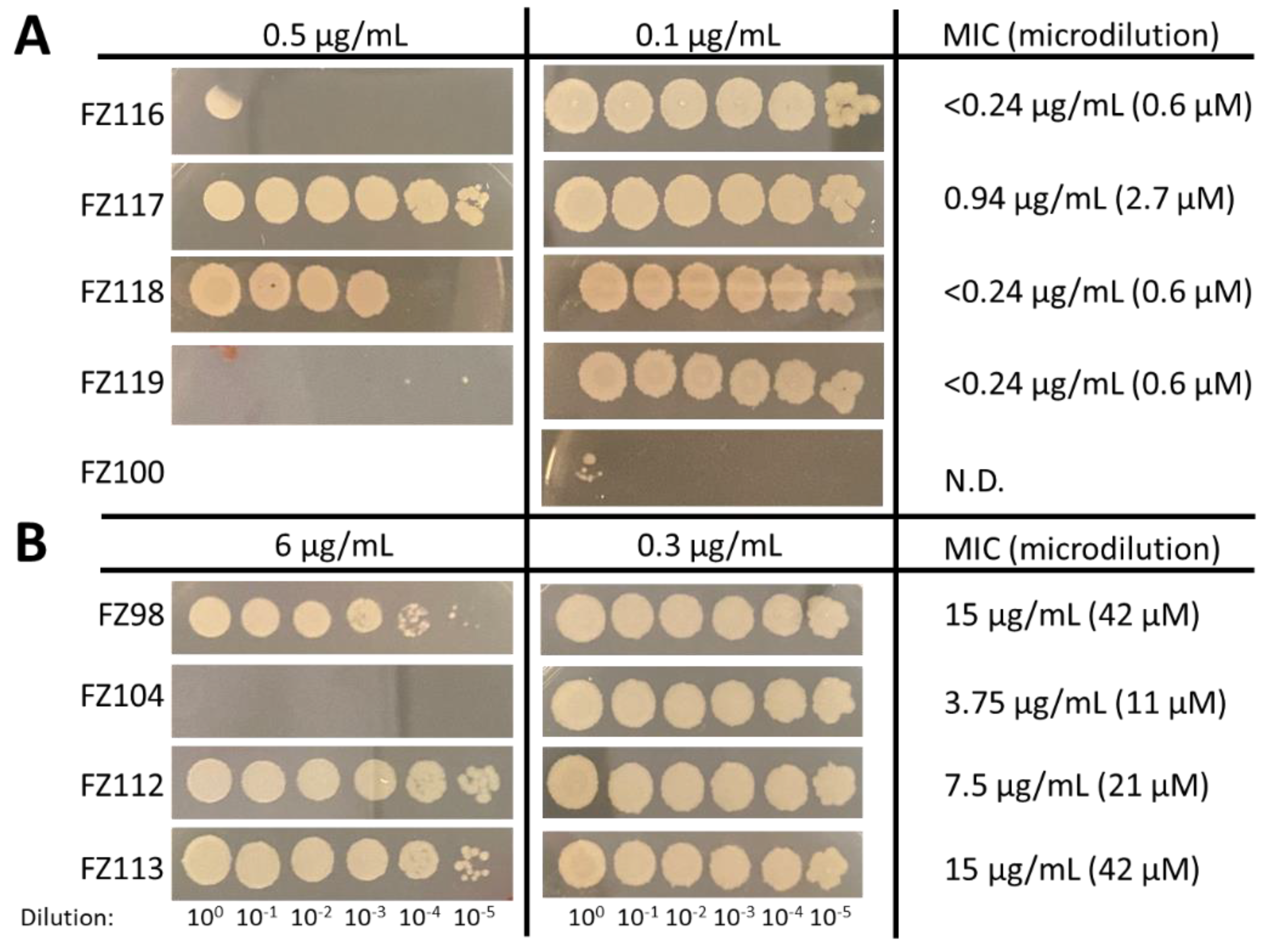
2.2. Antimicrobial Activity on Staphylococcus aureus
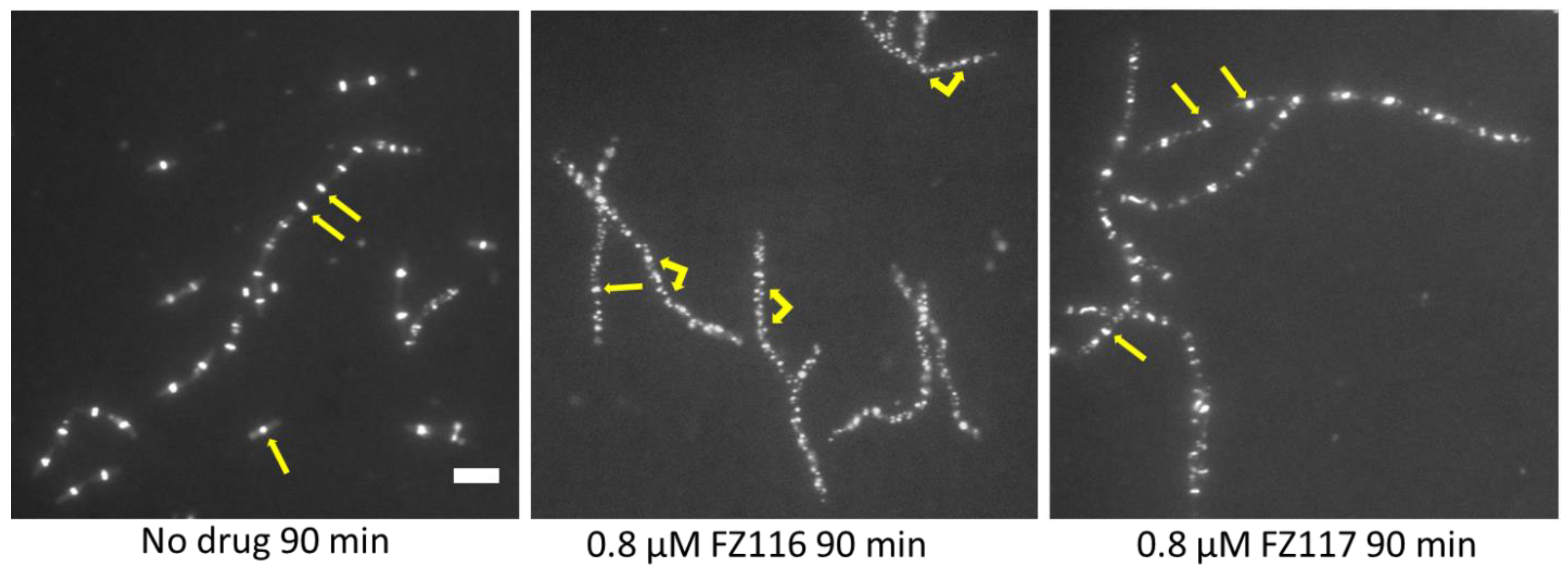
2.3. Antimicrobial Activity on Bacillus subtilis and Effects of FZ116 and FZ117 on Its Inhibition
2.4. Computational Studies
2.4.1. Physicochemical and Drug-like Profile Calculations
2.4.2. Docking Studies
2.5. Linker Effect about the Accomodation into the Binding Pocket
3. Discussion
4. Materials and Methods
4.1. Chemistry
Synthesis
4.2. Cells
4.3. Antibacterial Activity
4.3.1. MSSA and MDRSA Protocol
4.3.2. Antibacterial Activity against B. subtilis
4.4. Thiazolyl Blue Tetrazolium Bromide (MTT) Cytotoxicity Assay
4.5. Computational Studies
4.5.1. Ligand and Protein Preparation
4.5.2. Docking
4.5.3. Validations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria-A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Holmes, N.E.; Johnson, P.D.R.; Howden, B.P. Relationship between vancomycin-resistant Staphylococcus aureus, vancomycin-intermediate S. aureus, high vancomycin MIC, and outcome in serious S. aureus infections. J. Clin. Microbiol. 2012, 50, 2548–2552. [Google Scholar] [CrossRef]
- Casiraghi, A.; Suigo, L.; Valoti, E.; Straniero, V. Targeting Bacterial Cell Division: A Binding Site-Centered Approach to the Most Promising Inhibitors of the Essential Protein FtsZ. Antibiotics 2020, 9, 69. [Google Scholar] [CrossRef]
- Pradhan, P.; Margolin, W.; Beuria, T.K. Targeting the Achilles Heel of FtsZ: The Interdomain Cleft. Front. Microbiol. 2021, 12, 732796. [Google Scholar] [CrossRef]
- Cameron, T.A.; Margolin, W. Insights into the assembly and regulation of the bacterial divisome. Nat. Rev. Microbiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Haeusser, D.P.; Margolin, W. Splitsville: Structural and functional insights into the dynamic bacterial Z ring. Nat. Rev. Microbiol. 2016, 14, 305–319. [Google Scholar] [CrossRef] [PubMed]
- Haydon, D.J.; Stokes, N.R.; Ure, R.; Galbraith, G.; Bennett, J.M.; Brown, D.R.; Baker, P.J.; Barynin, V.V.; Rice, D.W.; Sedelnikova, S.E.; et al. An inhibitor of FtsZ with potent and selective anti-staphylococcal activity. Science 2008, 321, 1673–1675. [Google Scholar] [CrossRef] [PubMed]
- Chiodini, G.; Pallavicini, M.; Zanotto, C.; Bissa, M.; Radaelli, A.; Straniero, V.; Bolchi, C.; Fumagalli, L.; Ruggeri, P.; de Giuli Morghen, C.; et al. Benzodioxane-benzamides as new bacterial cell division inhibitors. Eur. J. Med. Chem. 2015, 89, 252–265. [Google Scholar] [CrossRef]
- Straniero, V.; Pallavicini, M.; Chiodini, G.; Zanotto, C.; Volontè, L.; Radaelli, A.; Bolchi, C.; Fumagalli, L.; Sanguinetti, M.; Menchinelli, G.; et al. 3-(Benzodioxan-2-ylmethoxy)-2,6-difluorobenzamides bearing hydrophobic substituents at the 7-position of the benzodioxane nucleus potently inhibit methicillin-resistant Sa and Mtb cell division. Eur. J. Med. Chem. 2016, 120, 227–243. [Google Scholar] [CrossRef]
- Straniero, V.; Zanotto, C.; Straniero, L.; Casiraghi, A.; Duga, S.; Radaelli, A.; de Giuli Morghen, C.; Valoti, E. 2,6-Difluorobenzamide Inhibitors of Bacterial Cell Division Protein FtsZ: Design, Synthesis, and Structure-Activity Relationships. ChemMedChem 2017, 12, 1303–1318. [Google Scholar] [CrossRef]
- Straniero, V.; Suigo, L.; Casiraghi, A.; Sebastián-Pérez, V.; Hrast, M.; Zanotto, C.; Zdovc, I.; de Giuli Morghen, C.; Radaelli, A.; Valoti, E. Benzamide Derivatives Targeting the Cell Division Protein FtsZ: Modifications of the Linker and the Benzodioxane Scaffold and Their Effects on Antimicrobial Activity. Antibiotics 2020, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Straniero, V.; Sebastián-Pérez, V.; Hrast, M.; Zanotto, C.; Casiraghi, A.; Suigo, L.; Zdovc, I.; Radaelli, A.; de Giuli Morghen, C.; Valoti, E. Benzodioxane-Benzamides as Antibacterial Agents: Computational and SAR Studies to Evaluate the Influence of the 7-Substitution in FtsZ Interaction. ChemMedChem 2020, 15, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Straniero, V.; Sebastián-Pérez, V.; Suigo, L.; Margolin, W.; Casiraghi, A.; Hrast, M.; Zanotto, C.; Zdovc, I.; Radaelli, A.; Valoti, E. Computational Design and Development of Benzodioxane-Benzamides as Potent Inhibitors of FtsZ by Exploring the Hydrophobic Subpocket. Antibiotics 2021, 10, 442. [Google Scholar] [CrossRef] [PubMed]
- Suigo, L.; Monterroso, B.; Sobrinos-Sanguino, M.; Alfonso, C.; Straniero, V.; Rivas, G.; Zorrilla, S.; Valoti, E.; Margolin, W. Benzodioxane-benzamides as promising inhibitors of Escherichia coli FtsZ. Int. J. Biol. Macromol. 2023, 253, 126398. [Google Scholar] [CrossRef]
- Rosado-Lugo, J.D.; Sun, Y.; Banerjee, A.; Cao, Y.; Datta, P.; Zhang, Y.; Yuan, Y.; Parhi, A.K. Evaluation of 2,6-difluoro-3-(oxazol-2-ylmethoxy)benzamide chemotypes as Gram-negative FtsZ inhibitors. J. Antibiot. 2022, 75, 385–395. [Google Scholar] [CrossRef]
- Stokes, N.R.; Baker, N.; Bennett, J.M.; Chauhan, P.K.; Collins, I.; Davies, D.T.; Gavade, M.; Kumar, D.; Lancett, P.; Macdonald, R.; et al. Design, synthesis and structure-activity relationships of substituted oxazole-benzamide antibacterial inhibitors of FtsZ. Bioorg. Med. Chem. Lett. 2014, 24, 353–359. [Google Scholar] [CrossRef] [PubMed]
- Stokes, N.R.; Baker, N.; Bennett, J.M.; Berry, J.; Collins, I.; Czaplewski, L.G.; Logan, A.; Macdonald, R.; Macleod, L.; Peasley, H.; et al. An improved small-molecule inhibitor of FtsZ with superior in vitro potency, drug-like properties, and in vivo efficacy. Antimicrob. Agents Chemother. 2013, 57, 317–325. [Google Scholar] [CrossRef]
- Straniero, V.; Casiraghi, A.; Fumagalli, L.; Valoti, E. How do reaction conditions affect the enantiopure synthesis of 2-substituted-1,4-benzodioxane derivatives? Chirality 2018, 30, 943–950. [Google Scholar] [CrossRef]
- Abushanab, E.; Vemishetti, P.; Leiby, R.W.; Singh, H.K.; Mikkilineni, A.B.; Wu, D.C.J.; Saibaba, R.; Panzica, R.P. The chemistry of L-ascorbic and D-isoascorbic acids. 1. The preparation of chiral butanetriols and -tetrols. J. Org. Chem. 1988, 53, 2598–2602. [Google Scholar] [CrossRef]
- Holý, A. Preparation and synthetic utilization of 3-(adenin-9-yl)-2-hydroxyalkanoic acids and their derivatives. Collect. Czech. Chem. Commun. 1984, 49, 2148–2166. [Google Scholar] [CrossRef]
- Mohapatra, D.K.; Reddy, D.S.; Reddy, G.S.; Yadav, J.S. Synthesis of the C-8-C-24 Fragment of Maltepolide C by Using a Tandem Di-hydroxylation/S N 2 Cyclization Sequence. Eur. J. Org. Chem. 2015, 2015, 5266–5274. [Google Scholar] [CrossRef]
- Bolchi, C.; Catalano, P.; Fumagalli, L.; Gobbi, M.; Pallavicini, M.; Pedretti, A.; Villa, L.; Vistoli, G.; Valoti, E. Structure-affinity studies for a novel series of homochiral naphtho and tetrahydronaphtho analogues of alpha 1 antagonist WB-4101. Bioorg. Med. Chem. 2004, 12, 4937–4951. [Google Scholar] [CrossRef]
- Adams, D.W.; Wu, L.J.; Czaplewski, L.G.; Errington, J. Multiple effects of benzamide antibiotics on FtsZ function. Mol. Microbiol. 2011, 80, 68–84. [Google Scholar] [CrossRef] [PubMed]
- Ralston, K.J.; Ramstadius, H.C.; Brewster, R.C.; Niblock, H.S.; Hulme, A.N. Self-Assembly of Disorazole C1 through a One-Pot Alkyne Metathesis Homodimerization Strategy. Angew. Chem. Int. Ed Engl. 2015, 54, 7086–7090. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2021-1: Glide; Schrödinger, LLC: New York, NY, USA, 2021.
- Schrödinger Release 2021-1: LigPrep; Schrödinger, LLC: New York, NY, USA, 2021.
- Schrödinger Release 2021-1: Maestro; Schrödinger, LLC: New York, NY, USA, 2021.
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Release 2021-1: Protein Preparation Wizard; Schrödinger, LLC: New York, NY, USA, 2021.
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Fujita, J.; Maeda, Y.; Mizohata, E.; Inoue, T.; Kaul, M.; Parhi, A.K.; Lavoie, E.J.; Pilch, D.S.; Matsumura, H. Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ. ACS Chem. Biol. 2017, 12, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Farid, R.; Day, T.; Friesner, R.A.; Pearlstein, R.A. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg. Med. Chem. 2006, 14, 3160–3173. [Google Scholar] [CrossRef]
- Sherman, W.; Beard, H.S.; Farid, R. Use of an induced fit receptor structure in virtual screening. Chem. Biol. Drug Des. 2006, 67, 83–84. [Google Scholar] [CrossRef] [PubMed]
- Sherman, W.; Day, T.; Jacobson, M.P.; Friesner, R.A.; Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger Software Release 2021-1: Induced Fit Docking Protocol; Glide, Schrödinger, LLC: New York, NY, USA; Prime, Schrödinger, LLC: New York, NY, USA, 2021.
- QikProp 4.4 User Manual; Schrödinger Press LLC: New York, NY, USA, 2011.
- Schrödinger Release 2021-1: QikProp; Schrödinger, LLC: New York, NY, USA, 2023.
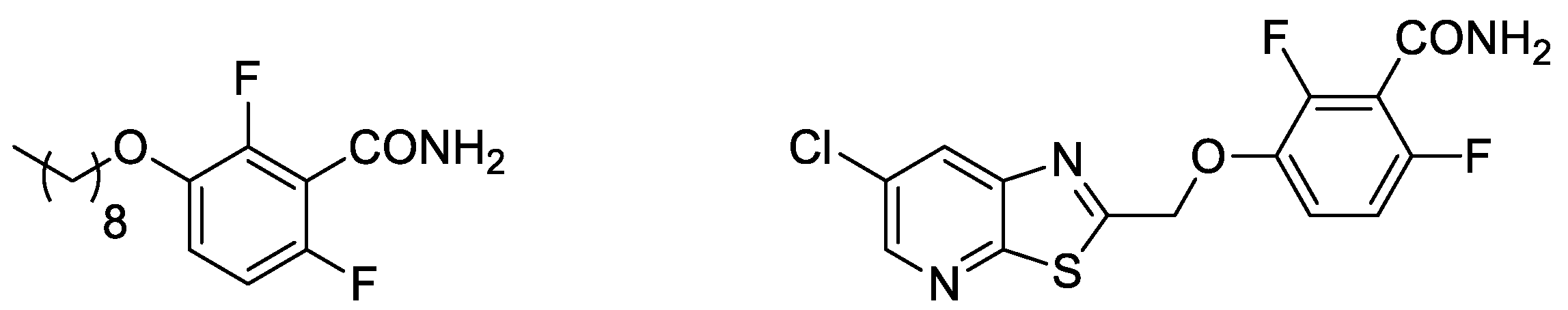






| Compound | MSSA ATCC 29213 | MDRSA 12.1 | MDRSA 11.7 | MRC-5 |
|---|---|---|---|---|
| MIC (μM) | MIC (μM) | MIC (μM) | TD90 (μM) | |
| FZ14 | 15.6 | 15.6 | 15.6 | 49.8 |
| FZ88 | 14.9 | 14.9 | 14.9 | 226 |
| FZ100 | 0.25 | 0.25 | 0.25 | 2054 |
| FZ104 (erythro) | 23.9 | 23.9 | 23.9 | - |
| FZ105 (threo) | 190.9 | 190.9 | 190.9 | - |
| FZ98 (erythro) | 91.6 | 91.6 | 91.6 | - |
| FZ97 (threo) | 366.4 | 366.4 | 366.4 | - |
| FZ118 (erythro) | 9.9 | 9.9 | 9.9 | 471.0 |
| FZ119 (threo) | 9.9 | 9.9 | 9.9 | 247.9 |
| FZ112 (erythro) | 364.4 | 364.4 | 364.4 | - |
| FZ113 (threo) | 364.4 | 364.4 | 364.4 | - |
| FZ116 (erythro) | 4.9 | 4.9 | 4.9 | 468.7 |
| FZ117 (threo) | 19.7 | 19.7 | 19.7 | 468.7 |
| Property (Recommended Values) | FZ104 S′R | FZ104 R′S | FZ105 S′S | FZ105 R′R | FZ97 R′S | FZ97 S′R | FZ98 R′R | FZ98 S′S | FZ118 R′R | FZ118 S′S | FZ119 R′S | FZ119 S′R |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #(*)stars (0–5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| #(*)rotor (0–15) | 4 | 4 | 4 | 4 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| MW (130.0–725.0) | 335.307 | 335.307 | 335.307 | 335.307 | 349.333 | 349.333 | 349.333 | 349.333 | 403.425 | 403.425 | 403.425 | 403.425 |
| dipole (1.0–12.5) | 6.780 | 6.779 | 6.854 | 6.854 | 6.912 | 7.801 | 6.889 | 6.705 | 6.798 | 7.736 | 7.333 | 7.525 |
| donorHB (0.0–6.0) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| acceptHB (2.0–20.0) | 4.75 | 4.75 | 4.75 | 4.75 | 4.75 | 4.75 | 4.75 | 4.75 | 4.750 | 4.750 | 4.750 | 4.750 |
| QplogPo/w (−2.0–6.5) | 3.196 | 3.196 | 3.157 | 3.157 | 3.603 | 3.604 | 3.595 | 3.527 | 4.480 | 4.579 | 4.486 | 4.589 |
| QPlogS (−6.5–0.5) | −4.718 | −4.718 | −4.619 | −4.619 | −5.049 | −5.056 | −5.030 | −4.852 | −6.325 | −6.551 | −6.335 | −6.589 |
| CIQPlogS (−6.5–0.5) | −4.899 | −4.899 | −4.899 | −4.899 | −5.178 | −5.178 | −5.178 | −5.178 | −6.244 | −6.244 | −6.244 | −6.244 |
| QPlogHERG (>−5) | −5.532 | −5.532 | −5.464 | −5.464 | −5.725 | −5.726 | −5.710 | −5.566 | −5.456 | −5.637 | −5.459 | −5.663 |
| QPPCaco (>500 great) | 996.278 | 996.277 | 996.339 | 996.232 | 996.100 | 996.149 | 1003.54 | 1003.22 | 996.168 | 996.423 | 1003.206 | 996.285 |
| QPPMDCK (>500 great) | 1248.09 | 1248.09 | 1210.43 | 1210.25 | 1193.20 | 1195.24 | 1198.92 | 1114.05 | 1192.965 | 1190.759 | 1199.771 | 1193.676 |
| #(*)metab (1–8) | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 4 | 4 | 4 |
| QplogKhsa (−1.5–1.5) | 0.192 | 0.192 | 0.182 | 0.182 | 0.300 | 0.301 | 0.297 | 0.281 | 0.718 | 0.754 | 0.721 | 0.757 |
| HumanOralAbsorption (1 low, 2 medium, 3 high) | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 1 | 1 | 1 | 1 |
| %HumanOralAbsorption (>80% high, <25% poor) | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 | 100.000 |
| PSA (7.0–200.0) | 75.043 | 75.043 | 74.717 | 74.715 | 74.307 | 74.198 | 74.245 | 72.669 | 74.462 | 74.271 | 74.465 | 74.212 |
| #(*)NandO (2–15) | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| RuleOfFive (max: 4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RuleOfThree (max 3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| #(*)ringatoms | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 20 | 20 | 20 | 20 |
| #(*)in34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #(*)in56 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 16 | 20 | 20 | 20 | 20 |
| #(*)noncon | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 6 | 6 | 6 | 6 |
| #(*)nonHatm | 24 | 24 | 24 | 24 | 25 | 25 | 25 | 25 | 29 | 29 | 29 | 29 |
| Property (Recommended Values) | FZ112 S′R | FZ112 R′S | FZ113 S′S | FZ113 R′R | FZ116 S′R | FZ116 R′S | FZ117 S′S | FZ117 R′R |
|---|---|---|---|---|---|---|---|---|
| #(*)stars (0–5) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #(*)rotor (0–15) | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| MW (130.0–725.0) | 351.306 | 351.306 | 351.306 | 351.306 | 405.397 | 405.397 | 405.397 | 405.397 |
| dipole (1.0–12.5) | 8.187 | 7.748 | 8.753 | 8.868 | 7.600 | 8.668 | 8.919 | 7.334 |
| donorHB (0.0–6.0) | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| acceptHB (2.0–20.0) | 6.45 | 6.45 | 6.45 | 6.45 | 6.450 | 6.450 | 6.450 | 6.450 |
| QplogPo/w (−2.0–6.5) | 2.288 | 2.378 | 2.390 | 2.202 | 3.222 | 3.313 | 3.313 | 3.264 |
| QPlogS (−6.5–0.5) | −3.997 | −4.178 | −4.214 | −3.694 | −5.414 | −5.615 | −5.392 | −5.505 |
| CIQPlogS (−6.5–0.5) | −4.533 | −4.533 | −4.533 | −4.533 | −5.559 | −5.559 | −5.559 | −5.559 |
| QPlogHERG (>−5) | −5.564 | −5.737 | −5.771 | −5.214 | −5.511 | −5.681 | −5.430 | −5.591 |
| QPPCaco (>500 great) | 424.160 | 424.001 | 423.113 | 426.638 | 424.012 | 424.958 | 513.188 | 407.964 |
| QPPMDCK (>500 great) | 495.089 | 495.497 | 494.153 | 405.951 | 495.240 | 496.276 | 609.614 | 454.895 |
| #(*)metab (1–8) | 3 | 3 | 3 | 3 | 5 | 5 | 5 | 5 |
| QplogKhsa (−1.5–1.5) | −0.157 | −0.131 | −0.127 | −0.152 | 0.251 | 0.280 | 0.267 | 0.274 |
| HumanOralAbsorption (1 low, 2 medium, 3 high) | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| %HumanOralAbsorption (>80% high, <25% poor) | 87.372 | 87.892 | 87.949 | 86.911 | 92.835 | 93.388 | 94.854 | 92.783 |
| PSA (7.0–200.0) | 97.002 | 96.240 | 96.879 | 95.380 | 96.421 | 96.223 | 94.113 | 96.186 |
| #(*)NandO (2–15) | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 |
| RuleOfFive (max: 4) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RuleOfThree (max 3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #(*)ringatoms | 16 | 16 | 16 | 16 | 20 | 20 | 20 | 20 |
| #(*)in34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| #(*)in56 | 16 | 16 | 16 | 16 | 20 | 20 | 20 | 20 |
| #(*)noncon | 2 | 2 | 2 | 2 | 6 | 6 | 6 | 6 |
| #(*)nonHatm | 25 | 25 | 25 | 25 | 29 | 29 | 29 | 29 |
| Docking Score (kcal/mol) | Ligand Interactions for a Cutoff of 4.00 (Å) |
|---|---|
| Pose 1: −12.121 | Primary amide: Val207 (-CO-) 2.24 Asn263 (-CO-) 1.82 Carbonyl: Leu209 (-NH-) 2.42 |
| Pose 2: −12.399 | Primary amide: Val207 (-CO-) 2.03 Asn263 (-CO-) 2.08 Carbonyl: Leu209 (-NH-) 2.24 |
| Substituent | Cluster 1: No Residue (%) | Cluster 2: Thr309 (%)/HB Interaction (%) |
|---|---|---|
| threo-OH | 33 | 17/75 |
| erythro-OH | 38 | 13/50 |
| threo-CH3 | 63 | 5/0 |
| erythro-CH3 | 27 | 5/0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suigo, L.; Margolin, W.; Ulzurrun, E.; Hrast Rambaher, M.; Zanotto, C.; Sebastián-Pérez, V.; Campillo, N.E.; Straniero, V.; Valoti, E. Benzodioxane–Benzamides as FtsZ Inhibitors: Effects of Linker’s Functionalization on Gram-Positive Antimicrobial Activity. Antibiotics 2023, 12, 1712. https://doi.org/10.3390/antibiotics12121712
Suigo L, Margolin W, Ulzurrun E, Hrast Rambaher M, Zanotto C, Sebastián-Pérez V, Campillo NE, Straniero V, Valoti E. Benzodioxane–Benzamides as FtsZ Inhibitors: Effects of Linker’s Functionalization on Gram-Positive Antimicrobial Activity. Antibiotics. 2023; 12(12):1712. https://doi.org/10.3390/antibiotics12121712
Chicago/Turabian StyleSuigo, Lorenzo, William Margolin, Eugenia Ulzurrun, Martina Hrast Rambaher, Carlo Zanotto, Victor Sebastián-Pérez, Nuria E. Campillo, Valentina Straniero, and Ermanno Valoti. 2023. "Benzodioxane–Benzamides as FtsZ Inhibitors: Effects of Linker’s Functionalization on Gram-Positive Antimicrobial Activity" Antibiotics 12, no. 12: 1712. https://doi.org/10.3390/antibiotics12121712
APA StyleSuigo, L., Margolin, W., Ulzurrun, E., Hrast Rambaher, M., Zanotto, C., Sebastián-Pérez, V., Campillo, N. E., Straniero, V., & Valoti, E. (2023). Benzodioxane–Benzamides as FtsZ Inhibitors: Effects of Linker’s Functionalization on Gram-Positive Antimicrobial Activity. Antibiotics, 12(12), 1712. https://doi.org/10.3390/antibiotics12121712








