Heterogeneous Phenotypic Responses of Antibiotic-Resistant Salmonella Typhimurium to Food Preservative-Related Stresses
Abstract
:1. Introduction
2. Results
2.1. Susceptibilities of S. Typhimurium Serially Exposed to Ciprofloxacin and Food Processing-Related Stresses
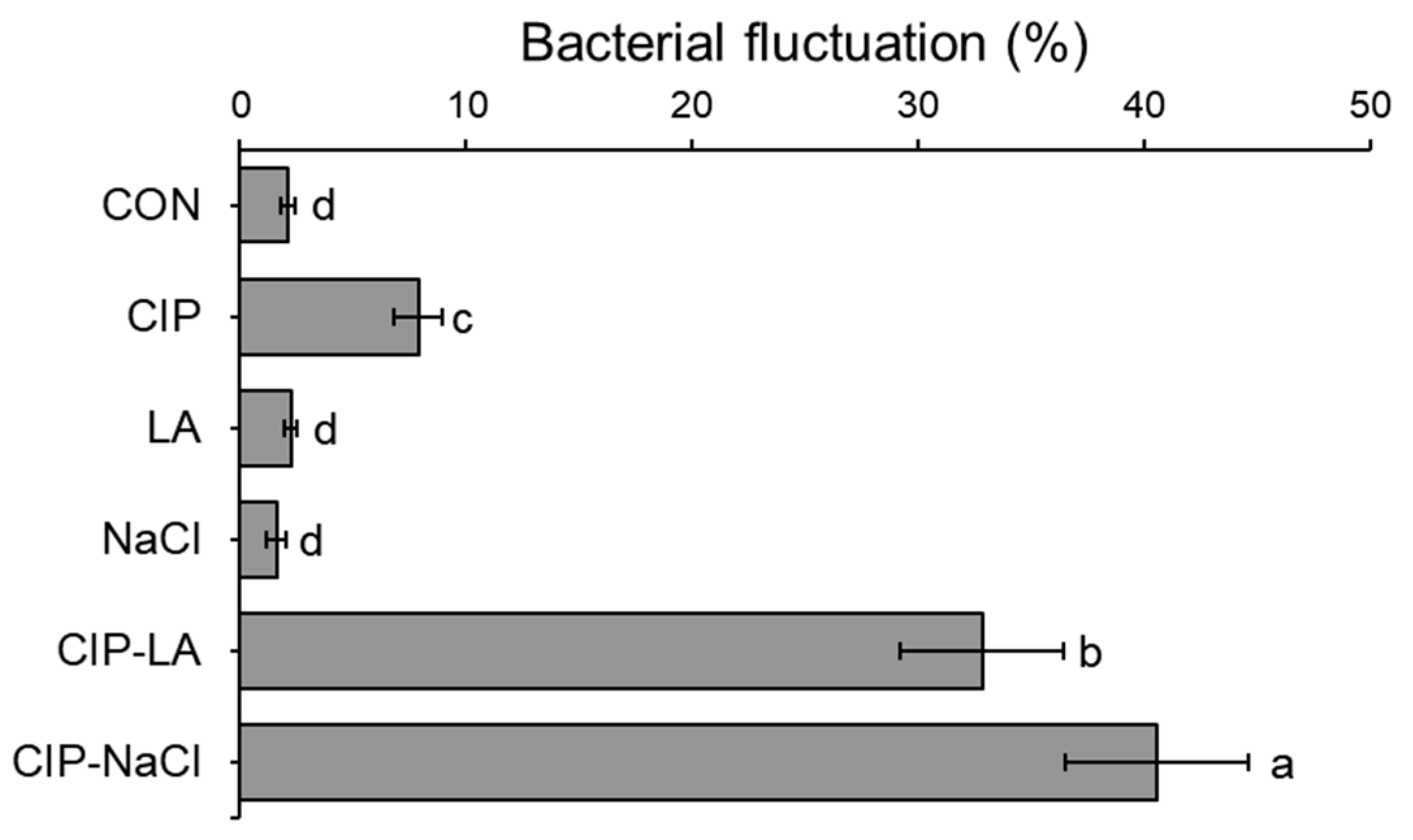
2.2. Phenotypic Heterogeneity of S. Typhimurium Serially Exposed to Ciprofloxacin and Food Processing-Related Stresses
3. Discussion
4. Materials and Methods
4.1. Bacterial Strain and Culture Condition
4.2. Antimicrobial Susceptibility Assay
4.3. Bacterial Fluctuation Assay
4.4. Agar Overlay Assay
4.5. Estimation of Persister Cells
4.6. Relative Fitness Determination
4.7. Disk Diffusion Susceptibility Test
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Pub. Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Veeraraghavan, B.; Elangovan, R.; Vivekanandan, P. Antibiotic resistance and epigenetics: More to it than meets the eye. Antimicrob. Agents Chemother. 2020, 64, e02225-02219. [Google Scholar] [CrossRef] [PubMed]
- Hossain, T.; Deter, H.S.; Peters, E.J.; Butzin, N.C. Antibiotic tolerance, persistence, and resistance of the evolved minimal cell, Mycoplasma mycoides JCVI-Syn3B. iScience 2021, 24, 102391. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Michiels, J.E.; Bergh, B.V.D.; Fauvart, M.; Michiels, J. Antibiotics: Combatting tolerance to stop resistance. MBio 2019, 10, e02095-02019. [Google Scholar] [CrossRef]
- Merker, M.; Tueffers, L.; Vallier, M.; Groth, E.E.; Sonnenkalb, L.; Unterweger, D.; Baines, J.F.; Niemann, S.; Schulenburg, H. Evolutionary approaches to combat antibiotic resistance: Opportunities and challenges for precision medicine. Front. Inmmunol. 2020, 11, 568485. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Granados, M.C.; Zenick, B.; Englander, H.E.; Mok, W.W.K. The social network: Impact of host and microbial interactions on bacterial antibiotic tolerance and persistence. Cell. Signal. 2020, 75, 109750. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Van den Bergh, B.; Michiels, J. Bacteria under antibiotic attack: Different strategies for evolutionary adaptation. PLoS Pathog. 2020, 16, e1008431. [Google Scholar] [CrossRef] [PubMed]
- Peterson, E.; Kaur, P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, C.; Grohmann, E. Horizontal gene transfer of antibiotic resistance genes in biofilms. Antibiotics 2023, 12, 328. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 2007, 5, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Horn, N.; Bhunia, A.K. Food-associated stress primes foodborne pathogens for the gastrointestinal phase of infection. Front. Microbiol. 2018, 9, 1962. [Google Scholar] [CrossRef] [PubMed]
- Zapaśnik, A.; Sokołowska, B.; Bryła, M. Role of lactic acid bacteria in food preservation and safety. Foods 2022, 11, 1283. [Google Scholar] [CrossRef] [PubMed]
- Wijnker, J.J.; Koop, G.; Lipman, L.J.A. Antimicrobial properties of salt (NaCl) used for the preservation of natural casings. Food Microbiol. 2006, 23, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef]
- Yousef, A.E.; Courtney, P.D. Basics of stress adaptation and implications in new-generation foods. Microb. Stress Adapt. Food Saf. 2003, 1, 1–30. [Google Scholar]
- Olesen, S.W.; Barnett, M.L.; MacFadden, D.R.; Brownstein, J.S.; Hernández-Díaz, S.; Lipsitch, M.; Grad, Y.H. The distribution of antibiotic use and its association with antibiotic resistance. Elife 2018, 7, e39435. [Google Scholar] [CrossRef] [PubMed]
- Drlica, K.; Malik, M.; Kerns, R.J.; Zhao, X. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 2008, 52, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef]
- Prasetyoputri, A.; Jarrad, A.M.; Cooper, M.A.; Blaskovich, M.A.T. The eagle effect and antibiotic-induced persistence: Two sides of the same coin? Trends Microbiol. 2019, 27, 339–354. [Google Scholar] [CrossRef]
- Brauner, A.; Fridman, O.; Gefen, O.; Balaban, N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016, 14, 320–330. [Google Scholar] [CrossRef]
- Fontana, R.; Boaretti, M.; Grossato, A.; Tonin, E.A.; Lleò, M.M.; Satta, G. Paradoxical response of Enterococcus faecalis to the bactericidal activity of penicillin is associated with reduced activity of one autolysin. Antimicrob. Agents Chemother. 1990, 34, 314–320. [Google Scholar] [CrossRef]
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Levin, B.R.; Perrot, V.; Walker, N. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 2000, 154, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Kint, C.I.; Verstraeten, N.; Fauvart, M.; Michiels, J. New-found fundamentals of bacterial persistence. Trends Microbiol. 2012, 20, 577–585. [Google Scholar] [CrossRef]
- Jayaraman, R. Bacterial persistence: Some new insights into an old phenomenon. J. Biosci. 2008, 33, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef]
- Rebelo, J.S.; Domingues, C.P.F.; Monteiro, F.; Nogueira, T.; Dionisio, F. Bacterial persistence is essential for susceptible cell survival in indirect resistance, mainly for lower cell densities. PLoS ONE 2021, 16, e0246500. [Google Scholar] [CrossRef]
- Wang, J.; Ray, A.J.; Hammons, S.R.; Oliver, H.F. Persistent and transient Listeria monocytogenes strains from retail deli environments vary in their ability to adhere and form biofilms and rarely have inlA premature stop codons. Foodborne Pathog. Dis. 2015, 12, 151–158. [Google Scholar] [CrossRef]
- Himeoka, Y.; Mitarai, N. When to wake up? The optimal waking-up strategies for starvation-induced persistence. PLoS Comput. Biol. 2021, 17, e1008655. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, D.; Berghoff, B.A. Type I toxin-dependent generation of superoxide affects the persister life cycle of Escherichia coli. Sci. Rep. 2019, 9, 14256. [Google Scholar] [CrossRef] [PubMed]
- Hartline, C.J.; Zhang, R.; Zhang, F. Transient antibiotic tolerance triggered by nutrient shifts from gluconeogenic carbon sources to fatty acid. Front. Microbiol. 2022, 13, 854272. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Brul, S.; Zaat, S.A.J. Bacterial persister-cells and spores in the food chain: Their potential inactivation by antimicrobial peptides (AMPs). Int. J. Mol. Sci. 2020, 21, 8967. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Bremer, H.; Dennis, P.P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus 2008, 3, 10–1128. [Google Scholar] [CrossRef]
- Li, X.; Hospital, X.F.; Hierro, E.; Fernández, M.; Sheng, L.; Wang, L. Formation of Listeria monocytogenes persister cells in the produce-processing environment. Int. J. Food Microbiol. 2023, 390, 110106. [Google Scholar] [CrossRef]
- Kaur, A.; Sharma, P.; Capalash, N. Curcumin alleviates persistence of Acinetobacter baumannii against colistin. Sci. Rep. 2018, 8, 11029. [Google Scholar] [CrossRef]
- Arvaniti, M.; Skandamis, P.N. Defining bacterial heterogeneity and dormancy with the parallel use of single-cell and population level approaches. Curr. Opin. Food Sci. 2022, 44, 100808. [Google Scholar] [CrossRef]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef]
- Jung, S.H.; Ryu, C.M.; Kim, J.S. Bacterial persistence: Fundamentals and clinical importance. J. Microbiol. 2019, 57, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Yu, P.-L.; Flint, S. Persister cell formation of Listeria monocytogenes in response to natural antimicrobial agent nisin. Food Cont. 2017, 77, 243–250. [Google Scholar] [CrossRef]
- Melnyk, A.H.; Wong, A.; Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 2015, 8, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Petersen, A.; Aarestrup, F.M.; Olsen, J.E. The in vitro fitness cost of antimicrobial resistance in Escherichia coli varies with the growth conditions. FEMS Microbiol. Lett. 2009, 299, 53–59. [Google Scholar] [CrossRef]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef]
- MacLean, R.C.; Vogwill, T. Limits to compensatory adaptation and the persistence of antibiotic resistance in pathogenic bacteria. Evol. Med. Public Health 2014, 2015, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Chellat, M.F.; Raguž, L.; Riedl, R. Targeting antibiotic resistance. Angew. Chem. Int. Ed. 2016, 55, 6600–6626. [Google Scholar] [CrossRef]
- Hughes, D.; Andersson, D.I. Evolutionary consequences of drug resistance: Shared principles across diverse targets and organisms. Nat. Rev. Genet. 2015, 16, 459–471. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug resistance in bacteria. Ann. Rev. Biochem. 2009, 78, 119–146. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Trebosc, V.; Kemmer, C.; Rosenstiel, P.; Beardmore, R.; Schulenburg, H.; Jansen, G. Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 2017, 34, 2229–2244. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Aulin, L.B.S.; Buffoni, M.; Fragkiskou, E.; Hasselt, J.G.C.V.; Rozen, D.E. Allele-specific collateral and fitness effects determine the dynamics of fluoroquinolone resistance evolution. Proc. Natl. Acad. Sci. USA 2022, 119, e2121768119. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jo, A.; Ding, T.; Lee, H.-Y.; Ahn, J. Assessment of altered binding specificity of bacteriophage for ciprofloxacin-induced antibiotic-resistant Salmonella Typhimurium . Arch. Microbiol. 2016, 198, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Dawan, J.; Ahn, J. Effectiveness of antibiotic combination treatments to control heteroresistant Salmonella Typhimurium . Microb. Drug Resist. 2021, 27, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Ruas-Madiedo, P.; Martínez, B.; Rodríguez, A.; García, P. Effective removal of staphylococcal biofilms by the endolysin LysH5. PLoS ONE 2014, 9, e107307. [Google Scholar] [CrossRef]
- Dawan, J.; Kim, S.; Ahn, J. Assessment of phenotypic heterogeneity in Salmonella Typhimurium preadapted to ciprofloxacin and tetracycline. FEMS Microbiol. Lett. 2023, 370, fnad100. [Google Scholar] [CrossRef]
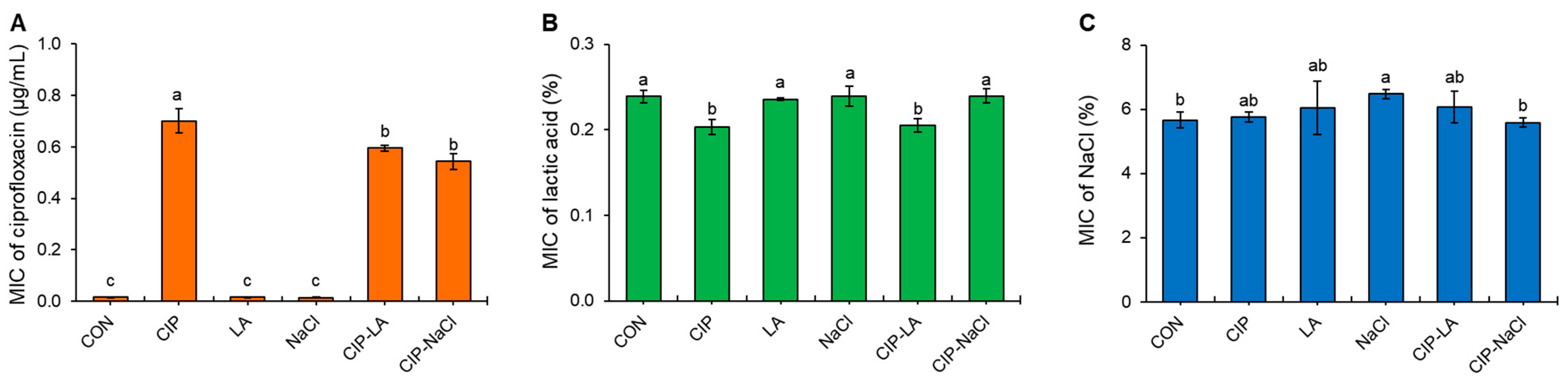



| Treatment * | MBC | EEC | MBC | EEC | MBC | EEC |
|---|---|---|---|---|---|---|
| Ciprofloxacin | Lactic Acid | NaCl | ||||
| CON | 0.03 | nd ** | 0.40 | nd | 10.0 | 10.0 |
| CIP | 2.0 | 8.0 | 0.40 | nd | 10.0 | 10.0 |
| LA | 0.03 | nd | 0.40 | nd | 10.0 | 10.0 |
| NaCl | 0.03 | nd | 0.30 | nd | 9.0 | 9.0 |
| CIP-LA | 4.0 | 8.0 | 0.40 | nd | 10.0 | 10.0 |
| CIP-NaCl | 1.0 | 16.0 | 0.30 | nd | 9.0 | 9.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, J.; Ahn, J. Heterogeneous Phenotypic Responses of Antibiotic-Resistant Salmonella Typhimurium to Food Preservative-Related Stresses. Antibiotics 2023, 12, 1702. https://doi.org/10.3390/antibiotics12121702
Yi J, Ahn J. Heterogeneous Phenotypic Responses of Antibiotic-Resistant Salmonella Typhimurium to Food Preservative-Related Stresses. Antibiotics. 2023; 12(12):1702. https://doi.org/10.3390/antibiotics12121702
Chicago/Turabian StyleYi, Jiseok, and Juhee Ahn. 2023. "Heterogeneous Phenotypic Responses of Antibiotic-Resistant Salmonella Typhimurium to Food Preservative-Related Stresses" Antibiotics 12, no. 12: 1702. https://doi.org/10.3390/antibiotics12121702
APA StyleYi, J., & Ahn, J. (2023). Heterogeneous Phenotypic Responses of Antibiotic-Resistant Salmonella Typhimurium to Food Preservative-Related Stresses. Antibiotics, 12(12), 1702. https://doi.org/10.3390/antibiotics12121702







