Antibiotic Resistance: Do We Need Only Cutting-Edge Methods, or Can New Visions Such as One Health Be More Useful for Learning from Nature?
Abstract
:1. Introduction
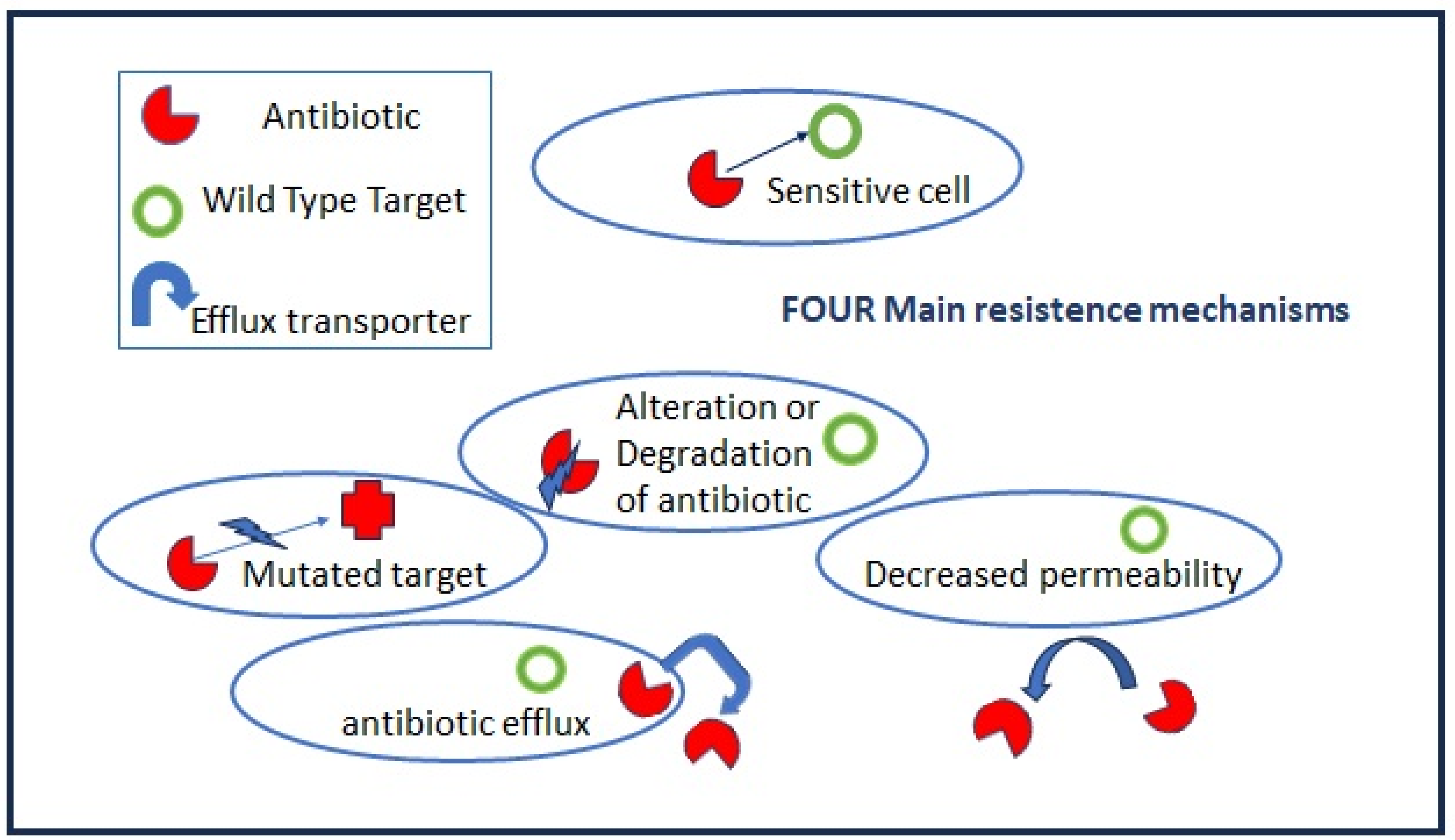
2. Antibiotics’ Primary Targets and Resistance Mechanisms
2.1. Lessons from Nature: From Intrinsic to Acquired Resistance
2.1.1. Intrinsic Resistance
2.1.2. Acquired Resistance
- (a)
- Mutations in target genes located at chromosomal or extrachromosomal elements which are vertically transmitted in the same bacteria species. The mutation rate increases when bacteria are actively multiplying as, for example, during the acute phase of host infection.
- (b)
- Horizontal gene transfers occur through mobile elements that can be transmitted both intraspecies and among different bacteria genera, i.e., the vancomycin-resistant gene (vanA) from Enterococcus to S. aureus. Plasmids, prophages, pathogenicity islands, restriction and modification systems, transposons, and insertion sequences are able to move within the host genome as well as jump across genomes. Mobile elements can change their insertion location and copy number and produce frequent gene gain and loss, modifying and co-evolving with chromosomal genomes. The genetic modifications induced by mobile elements can deeply affect bacterial fitness, contributing to their adaptation to new environments and, ultimately, producing evolutionarily distinct species over time. Once the acquisition of resistance determinants is established in few strains, antibiotic misuse and pressure drive the positive selection of resistant over sensitive strains.
2.1.3. Acquired Resistance through Bacterial Cooperation
3. Lessons from Nature: Biofilms
- (1)
- The extracellular polysaccharide matrix (EPS), which is produced upon biofilm organization, may slow down or impair antibiotic penetration.
- (2)
- In biofilm microenvironments, metabolic byproducts, waste, and nutrients accumulate. Additionally, oxygen may be greatly reduced, creating an anaerobic environment. For example, low oxygen levels reduce the bactericidal effects of the antibiotics tobramycin and ciprofloxacin, while pH changes can negatively impact aminoglycoside’s action.
- (3)
- Strict cellular contact and communication and the presence of a large amount of extracellular DNA in the biofilm EPS facilitate horizontal gene transfer from resistant to sensitive bacteria.
- (4)
- The presence of different metabolic stages of bacteria in the community create an environment in which antibiotics that are active on dividing cells are ineffective toward more quiescent cells.
- (5)
- The resistance of bacterial “Persister” cells: small subpopulations of bacteria that enter a “spore-like” state in which they are resistant to extreme conditions, like chemical treatment or antibiotic activity. These persisters exist in a dormant state without performing any genetic changes and do not divide in the presence of antibiotics. But once the organisms are released from the biofilm or begin dividing again, they return to their pre-persister susceptibility profile.
4. Lessons from Nature: The Power of the Host Defense—AMPs and MoABs
4.1. Host First-Line Defense: Antimicrobial Peptides
Bacteriocins and Relative Resistance
4.2. Antibodies Linked to Antibiotics
5. Lesson from Nature: “Living Killers for Pathogens”—Phages and Predatory Bacteria
5.1. Bacteriophages and Relative Resistance
5.2. Predatory Bacteria
5.3. Main Comparisons of the “Two Living Antibiotic Classes”: Phages versus Predatory Bacteria
- Host/prey threshold: both phages and predatory bacteria cannot destroy the entire population of their host. The amplification process inside the host is self-limited, and it stops in the absence of the prey. However, this can also be an advantage for therapy since a reduction in pathogen load will reduce or eliminate disease symptomatology.
- Host range: Individual phages have high species-specificity ranges, and a rapid acquisition of bacterial resistance to phages often occurs in contrast to the broad bacterial prey range and the lack of simple resistance mechanisms.
- DNA transferance: Phages can transfer DNA through generalized and specialized transduction, with a potential risk for uncontrolled mutations which does not apply to predatory bacteria.
6. Nature’s Lessons Need Receptive Students
6.1. Education
6.2. Social and Political
7. Future Directions
Conclusions: The Beneficial Circle from Nature to Humans and Back to Nature
Funding
Acknowledgments
Conflicts of Interest
References
- Adriaenssens, N.; Coenen, S.; Versporten, A.; Muller, A.; Minalu, G.; Faes, C.; Vankerckhoven, V.; Aerts, M.; Hens, N.; Molenberghs, G.; et al. European Surveillance of Antimicrobial Consumption (ESAC): Outpatient antibiotic use in Europe (1997–2009). J. Antimicrob. Chemother. 2011, 66, vi3–vi12. [Google Scholar] [CrossRef]
- Kim, D.-W.; Cha, C.J. Antibiotic resistome from the One-Health perspective: Understanding and controlling antimicrobial resistance transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Youle, M.; Rohwer, F.; Stacy, A.; Whiteley, M.; Steel, B.C.; Delalez, N.J.; Nord, A.L.; Berry, R.M.; Armitage, J.P.; Kamoun, S.; et al. The Microbial Olympics. Nat. Rev. Microbiol. 2012, 10, 583–588. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; World Health Organization: Geneva, Switzerland, 2017; pp. 1–7. [Google Scholar]
- Krawczyk-Balska, A.; Markiewicz, Z. The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J. Appl. Microbiol. 2016, 120, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012, 3, 421–569. [Google Scholar] [CrossRef] [PubMed]
- Strateva, T.; Yordanov, D. Pseudomonas aeruginosa—A phenomenon of bacterial resistance. J. Med. Microbiol. 2009, 58 Pt 9, 1133–1148. [Google Scholar] [CrossRef]
- Nguyen, M.C.P.; Woerther, P.L.; Bouvet, M.; Andremont, A.; Leclercq, R.; Canu, A. Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 2009, 15, 1648–1650. [Google Scholar] [CrossRef]
- Bernardini, A.; Cuesta, T.; Tomás, A.; Bengoechea, J.A.; Martínez, J.L.; Sánchez, M.B. The intrinsic resistome of Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2019, 53, 29–33. [Google Scholar] [CrossRef]
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Yurtsev, E.A.; Conwill, A.; Gore, J. Oscillatory dynamics in a bacterial cross-protection mutualism. Proc. Natl. Acad. Sci. USA 2016, 113, 6236–6241. [Google Scholar] [CrossRef] [PubMed]
- Dawan, J.; Ahn, J. Assessment of cooperative antibiotic resistance of Salmonella Typhimurium within heterogeneous population. Microb. Pathog. 2021, 157, 104973. [Google Scholar] [CrossRef]
- Williams, P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 2007, 153 Pt 12, 3923–3938. [Google Scholar] [CrossRef] [PubMed]
- Bassler, B.L.; Greenberg, E.P.; Stevens, A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1997, 179, 4043–4045. [Google Scholar] [CrossRef]
- Kim, C.S.; Gatsios, A.; Cuesta, S.; Lam, Y.C.; Wei, Z.; Chen, H.; Russell, R.M.; Shine, E.E.; Wang, R.; Wyche, T.P.; et al. Characterization of autoinducer-3 structure and biosynthesis in E. coli. ACS Cent. Sci. 2020, 6, 197–206. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory Mechanisms and Promising Applications of Quorum Sensing-Inhibiting Agents in Control of Bacterial Biofilm Formation. Front. Microbiol. 2020, 11, 589640. [Google Scholar] [CrossRef]
- D’Angelo, F.; Baldelli, V.; Halliday, N.; Pantalone, P.; Polticelli, F.; Fiscarelli, E.; Williams, P.; Visca, P.; Leoni, L.; Rampioni, G. Identification of FDA-Approved Drugs as Antivirulence Agents Targeting the pqs Quorum-Sensing System of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2018, 62, e01296-18. [Google Scholar] [CrossRef]
- Ahmed, S.A.K.S.; Rudden, M.; Smyth, T.J.; Dooley, J.S.G.; Marchant, R.; Banat, I.M. Natural quorum sensing inhibitors effectively downregulate gene expression of Pseudomonas aeruginosa virulence factors. Appl. Microbiol. Biotechnol. 2019, 103, 3521–3535. [Google Scholar] [CrossRef]
- Borges, A.; Simões, M. Quorum Sensing Inhibition by Marine Bacteria. Mar. Drugs 2019, 17, 427. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Melchior, M.B.; Vaarkamp, H.; Fink-Gremmels, J. Biofilms: A role in recurrent mastitis infections? Vet. J. 2006, 171, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Clutterbuck, A.L.; Woods, E.J.; Knottenbelt, D.C.; Clegg, P.D.; Cochrane, C.A.; Percival, S.L. Biofilms and their relevance to veterinary medicine. Vet. Microbiol. 2007, 121, 1–17. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef]
- Punginelli, D.; Catania, V.; Vazzana, M.; Mauro, M.; Spinello, A.; Barone, G.; Barberi, G.; Fiorica, C.; Vitale, M.; Cunsolo, V.; et al. A Novel Peptide with Antifungal Activity from Red Swamp Crayfish Procambarus clarkii. Antibiotics 2022, 11, 1792. [Google Scholar] [CrossRef]
- Falanga, A.; Lombardi, L.; Franci, G.; Vitiello, M.; Iovene, M.R.; Morelli, G.; Galdiero, M.; Galdiero, S. Marine antimicrobial peptides: Nature provides templates for the design of novel compounds against pathogenic bacteria. Int. J. Mol. Sci. 2016, 17, 785. [Google Scholar] [CrossRef]
- Schillaci, D.; Cusimano, M.G.; Spinello, A.; Barone, G.; Russo, D.; Vitale, M.; Parrinello, D.; Arizza, V. Paracentrin 1, a synthetic antimicrobial peptide from the sea-urchin Paracentrotus lividus, interferes with staphylococcal and Pseudomonas aeruginosa biofilm formation. AMB Express 2014, 4, 78. [Google Scholar] [CrossRef]
- Cusimano, M.G.; Spinello, A.; Barone, G.; Schillaci, D.; Cascioferro, S.; Magistrato, A.; Parrino, B.; Arizza, V.; Vitale, M. A Synthetic Derivative of Antimicrobial Peptide Holothuroidin 2 from Mediterranean Sea Cucumber (Holothuria tubulosa) in the Control of Listeria monocytogenes. Mar. Drugs 2019, 17, 159. [Google Scholar] [CrossRef] [PubMed]
- Oh, R.; Lee, M.J.; Kim, Y.-O.; Nam, B.-H.; Kong, H.J.; Kim, J.-W.; Park, J.-Y.; Seo, J.-K.; Kim, D.-G. Myticusin-beta, antimicrobial peptide from the marine bivalve, Mytilus coruscus. Fish Shellfish Immunol. 2020, 99, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.K.; Davies, B.W. Microcins reveal natural mechanisms of bacterial manipulation to inform therapeutic development. Microbiology 2022, 168, 001175. [Google Scholar] [CrossRef] [PubMed]
- de Freire Bastos, M.D.C.; Coelho, M.L.V.; Da Silva Santos, O.C. Resistance to bacteriocins produced by Gram-positive bacteria. Microbiology 2015, 161 Pt 4, 683–700. [Google Scholar] [CrossRef] [PubMed]
- Gravesen, A.; Axelsen, A.M.J.; da Silva, J.M.; Hansen, T.B.; Knøchel, S. Frequency of bacteriocin resistance development and associated fitness costs in Listeria monocytogenes. Appl. Environ. Microbiol. 2002, 68, 756–764. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Pircalabioru, G.G.; Popa, L.I.; Marutescu, L.; Gheorghe, I.; Popa, M.; Barbu, I.C.; Cristescu, R.; Chifiriuc, M.-C. Bacteriocins in the Era of Antibiotic Resistance: Rising to the Challenge. Pharmaceutics 2021, 13, 196. [Google Scholar] [CrossRef]
- Kung, P.; Goldstein, G.; Reinherz, E.L.; Schlossman, S.F. Monoclonal antibodies defining distinctive human T cell surface antigens. Science 1979, 206, 347–349. [Google Scholar] [CrossRef]
- Nagy, E.; Nagy, G.; Power, C.A.; Badarau, A.; Szijártó, V. Anti-bacterial monoclonal antibodies. Adv. Exp. Med. Biol. 2017, 1053, 119–153. [Google Scholar]
- Yamada, T. Therapeutic monoclonal antibodies. Keio J. Med. 2011, 60, 37–46. [Google Scholar] [CrossRef]
- Di Giandomenico, A.; Sellman, B.R. Antibacterial monoclonal antibodies: The next generation? Curr. Opin. Microbiol. 2015, 27, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef] [PubMed]
- Alomari, M.M.M.; Dec, M.; Urban-Chmiel, R. Bacteriophages as an Alternative Method for Control of Zoonotic and Foodborne Pathogens. Viruses 2021, 13, 2348. [Google Scholar] [CrossRef] [PubMed]
- Wójcicki, M.; Świder, O.; Gientka, I.; Błażejak, S.; Średnicka, P.; Shymialevich, D.; Cieślak, H.; Wardaszka, A.; Emanowicz, P.; Sokołowska, B.; et al. Effectiveness of a Phage Cocktail as a Potential Biocontrol Agent against Saprophytic Bacteria in Ready-To-Eat Plant-Based Food. Viruses 2023, 15, 172. [Google Scholar] [CrossRef]
- Bumunang, E.W.; Zaheer, R.; Niu, D.; Narvaez-Bravo, C.; Alexander, T.; McAllister, T.A.; Stanford, K. Bacteriophages for the Targeted Control of Foodborne Pathogens. Foods 2023, 12, 2734. [Google Scholar] [CrossRef]
- Schofield, D.A.; Westwater, C. Phage-mediated bioluminescent detection of Bacillus anthracis. J. Appl. Microbiol. 2009, 107, 1468–1478. [Google Scholar] [CrossRef] [PubMed]
- Wisuthiphaet, N.; Yang, X.; Young, G.M.; Nitin, N. Quantitative Imaging of Bacteriophage Amplification for Rapid Detection of Bacteria in Model Foods. Front. Microbiol. 2022, 13, 853048. Available online: https://www.frontiersin.org/articles/10.3389/fmicb.2022.853048 (accessed on 28 October 2023). [CrossRef]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef]
- Van Belleghem, J.D.; Clement, F.; Merabishvili, M.; Lavigne, R.; Vaneechoutte, M. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci. Rep. 2017, 7, 8004. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Torres-Barceló, C. Phage Therapy Faces Evolutionary Challenges. Viruses 2018, 10, 323. [Google Scholar] [CrossRef] [PubMed]
- Chabas, H.; van Houte, S.; Høyland-Kroghsbo, N.M.; Buckling, A.; Westra, E.R. Immigration of susceptible hosts triggers the evolution of alternative parasite defence strategies. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160721. [Google Scholar] [CrossRef] [PubMed]
- Torres-Barceló, C.; Turner, P.E.; Buckling, A. Mitigation of evolved bacterial resistance to phage therapy. Curr. Opin. Virol. 2022, 53, 101201. [Google Scholar] [CrossRef]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage treatment of human infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Uyttebroek, S.; Chen, B.; Onsea, J.; Ruythooren, F.; Debaveye, Y.; Devolder, D.; Spriet, I.; Depypere, M.; Wagemans, J.; Lavigne, R.; et al. Safety and efficacy of phage therapy in difficult-to-treat infections: A systematic review. Lancet Infect. Dis. 2022, 22, e208–e220. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Dedrick, R.M.; Schooley, R.T. Phage Therapy for Antibiotic-Resistant Bacterial Infections. Annu. Rev. Med. 2022, 73, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Johnke, J.; Fraune, S.; Bosch, T.C.G.; Hentschel, U.; Schulenburg, H. Bdellovibrio and Like Organisms Are Predictors of Microbiome Diversity in Distinct Host Groups. Microb. Ecol. 2020, 79, 252–257. [Google Scholar] [CrossRef]
- Kadouri, D.; O’Toole, G.A. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 2005, 71, 4044–4051. [Google Scholar] [CrossRef]
- Atterbury, R.J.; Tyson, J. Predatory bacteria as living antibiotics—Where are we now? Microbiology 2021, 167, 001025. [Google Scholar] [CrossRef]
- Bratanis, E.; Andersson, T.; Lood, R.; Bukowska-Faniband, E. Biotechnological Potential of Bdellovibrio and Like Organisms and Their Secreted Enzymes. Front. Microbiol. 2020, 11, 662. [Google Scholar] [CrossRef]
- Pulcini, C.; Gyssens, I.C. How to educate prescribers in antimicrobial stewardship practices. Virulence 2013, 4, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Inglis, T.J.J.; Urosevic, N. Where Sepsis and Antimicrobial Resistance Countermeasures Converge. Front. Public Health 2017, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, F.; Tarrant, C.; Hamilton, V.; Kiernan, F.M.; Jenkins, D.; Krockow, E.M. Sepsis and antimicrobial stewardship: Two sides of the same coin. BMJ Qual. Saf. 2019, 28, 758–761. [Google Scholar] [CrossRef] [PubMed]
- Bertollo, L.G.; Lutkemeyer, D.S.; Levin, A.S. Are antimicrobial stewardship programs effective strategies for preventing anti-biotic resistance? A systematic review. Am. J. Infect. Control 2018, 46, 824–836. [Google Scholar] [CrossRef]
- Morgan, D.J.; Okeke, I.N.; Laxminarayan, R.; Perencevich, E.N.; Weisenberg, S. Non-prescription antimicrobial use world-wide: A systematic review. Lancet. Infect. Dis. 2011, 11, 692–701. [Google Scholar] [CrossRef]
- Lee, C.-R.; Lee, J.H.; Kang, L.-W.; Jeong, B.C.; Lee, S.H. Educational effectiveness, target, and content for prudent antibiotic use. BioMed Res. Int. 2015, 2015, 214021. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2018, 17, e05598. [Google Scholar]
- World Health Organization (WHO) Climate Change and Health. 2021. Available online: www.who.int/news-room/fact-sheets/detail/climate-change-and-health (accessed on 6 November 2023).
- Lio, R.M.S.; Favara, G.; Maugeri, A.; Barchitta, M.; Agodi, A. How Antimicrobial Resistance Is Linked to Climate Change: An Overview of Two Intertwined Global Challenges. Int. J. Environ. Res. Public Health 2023, 20, 1681. [Google Scholar] [CrossRef]
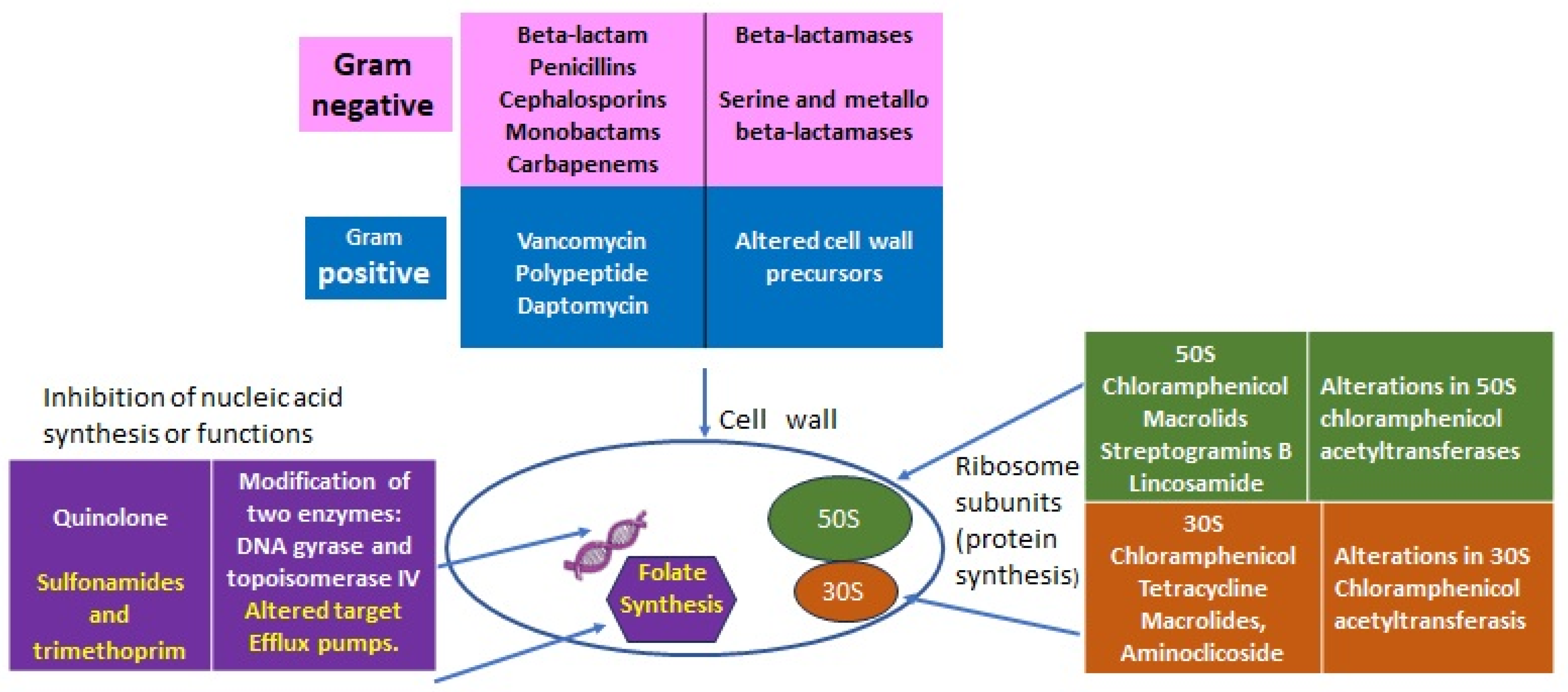
| Intrinsic Resistance (IR) | Determinants | |
|---|---|---|
| All Gram-negative diderm bacteria | Glycopeptides, lipopeptides, and antibiotic targeting the bacteria peptidoglycan wall | EPS (extra-polymeric substance) avoids the permeability of antibiotics. |
| P. aeruginosa | Sulfonamides, ampicillin, 1st- and 2nd-generation cephalosporins, chloramphenicol, and tetracycline | Constitutive expression of Amp C beta-lactamase and efflux pumps. Low permeability of the outer membrane [7]. |
| Enterococcus spp. | Aminoglycosides, cephalosporins, and lincosamides | Low cell wall permeability, aminoglycoside-modifying enzyme (AME), ribosome-modifying methyltransferase, altered cell wall, and ABC-efflux pump [6]. |
| L. monocytogenes | Cephalosporins | Penicillin-binding proteins, multidrug resistance transporters, cell envelope proteins, etc. [4]. |
| E. coli | Macrolides | Macrolides modifying genes such as mphA; efflux pump [8]. |
| K. pneumonia | Ampicillin | SHV beta-lactamase, the fosfomycin resistance gene fosA, and the nalidixic acid efflux pump OqxAB [9]. |
| A. baumanii | Cephalsporins, ampicillin, glycopeptides, and carbapenems | Class C (AmpC) and Class D beta-lactamases located in chromosome [10]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vitale, M. Antibiotic Resistance: Do We Need Only Cutting-Edge Methods, or Can New Visions Such as One Health Be More Useful for Learning from Nature? Antibiotics 2023, 12, 1694. https://doi.org/10.3390/antibiotics12121694
Vitale M. Antibiotic Resistance: Do We Need Only Cutting-Edge Methods, or Can New Visions Such as One Health Be More Useful for Learning from Nature? Antibiotics. 2023; 12(12):1694. https://doi.org/10.3390/antibiotics12121694
Chicago/Turabian StyleVitale, Maria. 2023. "Antibiotic Resistance: Do We Need Only Cutting-Edge Methods, or Can New Visions Such as One Health Be More Useful for Learning from Nature?" Antibiotics 12, no. 12: 1694. https://doi.org/10.3390/antibiotics12121694
APA StyleVitale, M. (2023). Antibiotic Resistance: Do We Need Only Cutting-Edge Methods, or Can New Visions Such as One Health Be More Useful for Learning from Nature? Antibiotics, 12(12), 1694. https://doi.org/10.3390/antibiotics12121694






