Analysis of Resistance Gene Diversity in the Intestinal Microbiome of Broilers from Two Types of Broiler Farms in Hebei Province, China
Abstract
:1. Introduction
2. Results
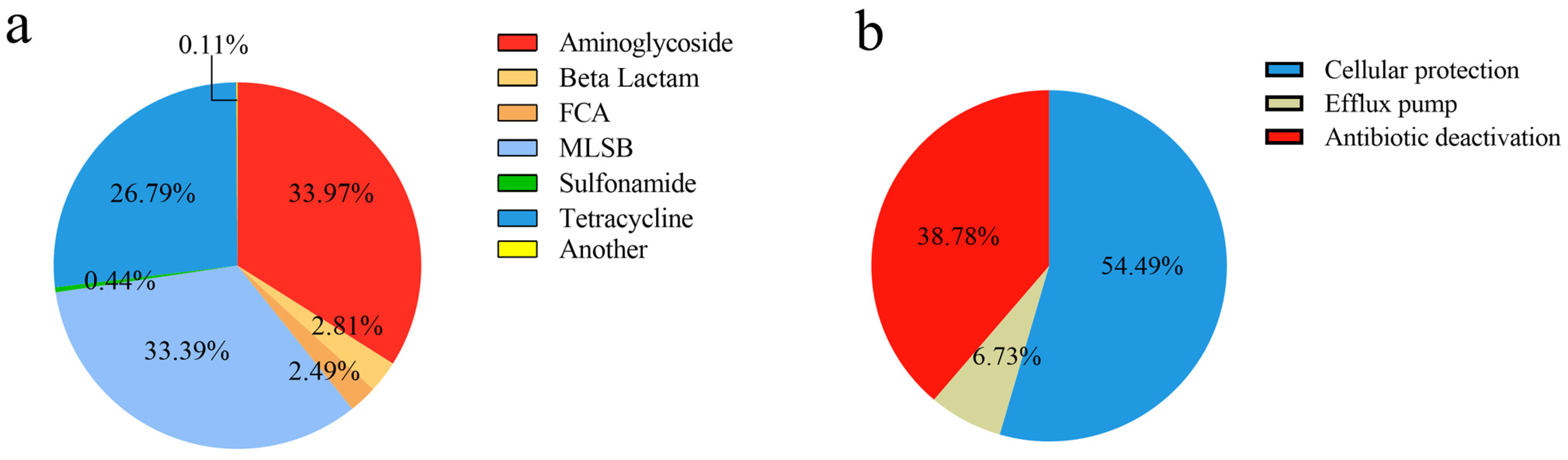
2.1. The Diversity of ARGs and MGEs
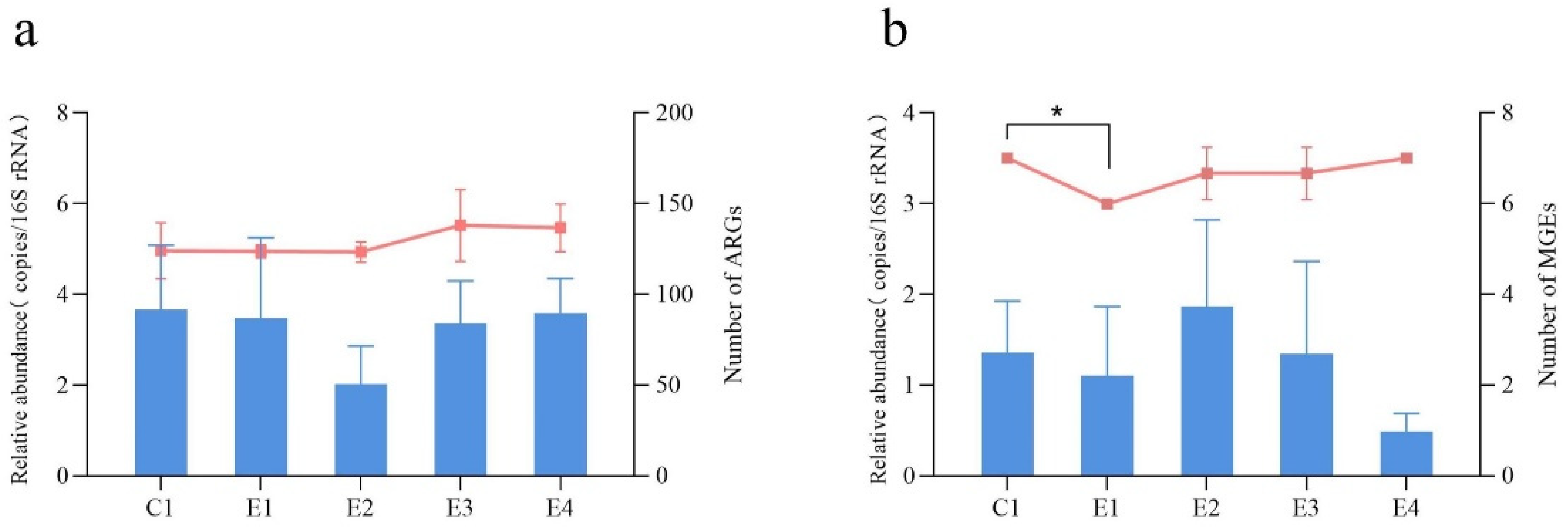
2.2. Changes in the Abundance of ARGs and MGEs between the SFs and the NSF
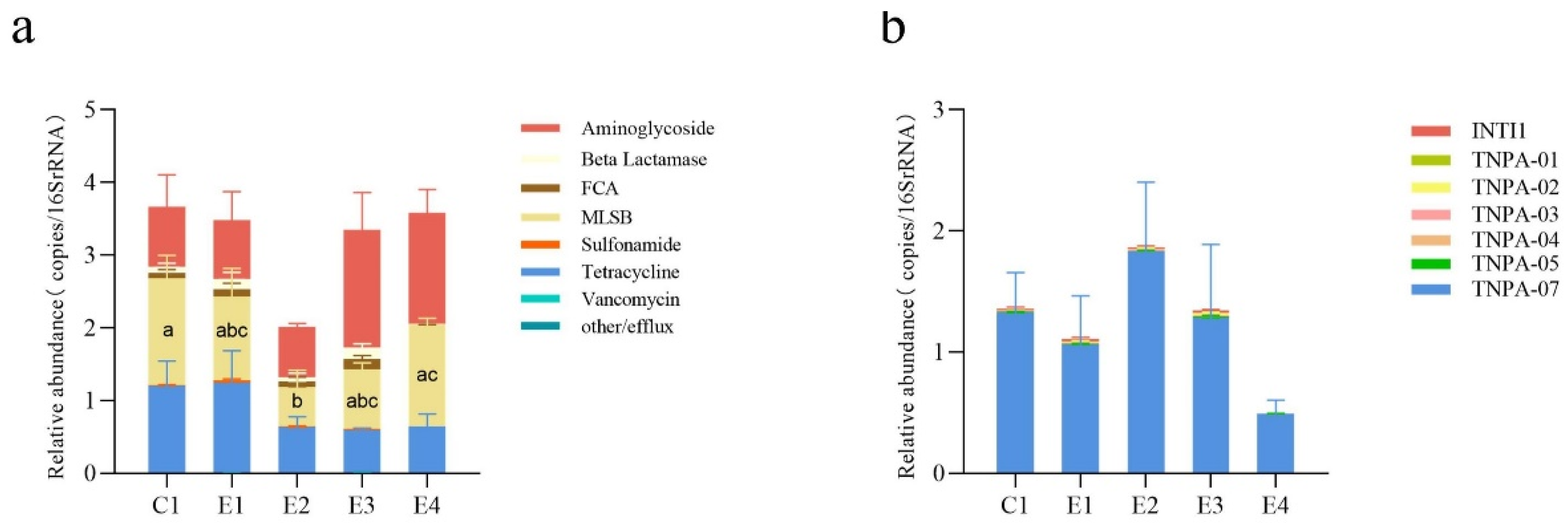
2.3. Co-occurrence of ARGs and MGEs
3. Materials and Methods
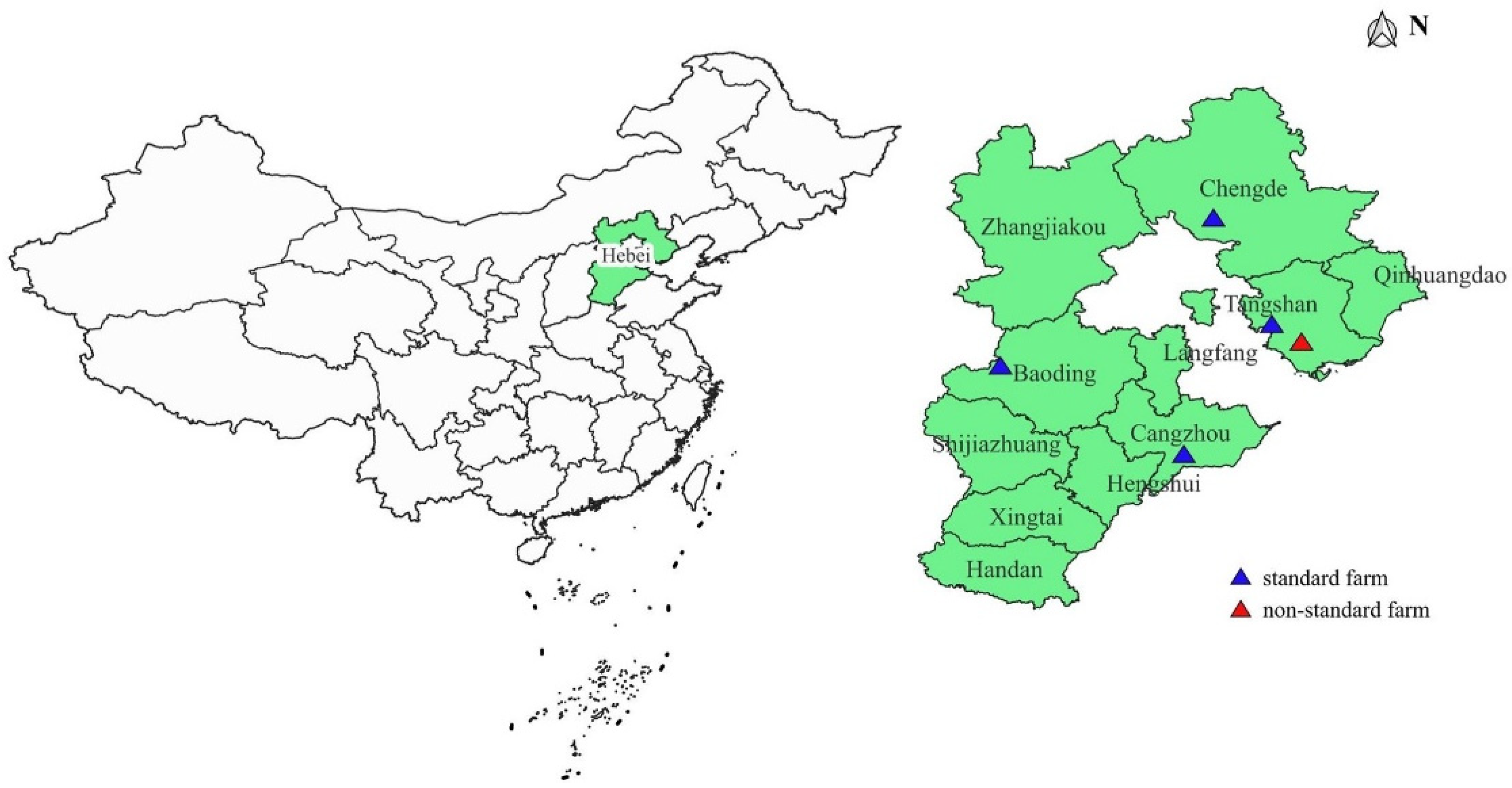
3.1. Sample Source
3.2. DNA Extraction and The Real-Time qPCR
3.3. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, H.; He, L.Y.; Wu, D.L.; Gao, F.Z.; Zhang, M.; Zou, H.Y.; Yao, M.S.; Ying, G.G. Spread of airborne antibiotic resistance from animal farms to the environment: Dispersal pattern and exposure risk. Environ. Int. 2022, 158, 106927. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef] [PubMed]
- Ying, G.G.; He, L.Y.; Ying, A.J.; Zhang, Q.Q.; Liu, Y.S.; Zhao, J.L. China Must Reduce Its Antibiotic Use. Environ. Sci. Technol. 2017, 51, 1072–1073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.Q.; Ying, G.G.; Pan, C.G.; Liu, Y.S.; Zhao, J.L. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef]
- Plata, G.; Baxter, N.T.; Susanti, D.; Volland-Munson, A.; Gangaiah, D.; Nagireddy, A.; Mane, S.P.; Balakuntla, J.; Hawkins, T.B.; Kumar Mahajan, A. Growth promotion and antibiotic induced metabolic shifts in the chicken gut microbiome. Commun. Biol. 2022, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.S.; Rama Rao, S.V.; Hegde, N.; Williams, N.J.; Chatterjee, R.N.; Raju, M.; Reddy, G.N.; Kumar, V.; Phani Kumar, P.S.; Mallick, S.; et al. Effects of Dietary Antimicrobial Growth Promoters on Performance Parameters and Abundance and Diversity of Broiler Chicken Gut Microbiome and Selection of Antibiotic Resistance Genes. Front. Microbiol. 2022, 13, 905050. [Google Scholar] [CrossRef] [PubMed]
- Castanon, J.I. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLoS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef]
- Diallo, O.O.; Baron, S.A.; Abat, C.; Colson, P.; Chaudet, H.; Rolain, J.M. Antibiotic resistance surveillance systems: A review. J. Glob. Antimicrob. Resist. 2020, 23, 430–438. [Google Scholar] [CrossRef]
- Hu, Y.J.; Cowling, B.J. Reducing antibiotic use in livestock, China. Bull. World Health Organ. 2020, 98, 360–361. [Google Scholar] [CrossRef]
- O’Brien, T.F. Emergence, spread, and environmental effect of antimicrobial resistance: How use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 2002, 34 (Suppl. S3), S78–S84. [Google Scholar] [CrossRef]
- Rolain, J.M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 2013, 4, 173. [Google Scholar] [CrossRef]
- Wang, C.; Li, P.; Yan, Q.; Chen, L.; Li, T.; Zhang, W.; Li, H.; Chen, C.; Han, X.; Zhang, S.; et al. Characterization of the Pig Gut Microbiome and Antibiotic Resistome in Industrialized Feedlots in China. mSystems 2019, 4, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Y.; Cao, J.; Bi, Y.; Lv, N.; Liu, F.; Liang, S.; Shi, Y.; Jiao, X.; Gao, G.F.; et al. Antibiotic resistance gene reservoir in live poultry markets. J. Infect. 2019, 78, 445–453. [Google Scholar] [CrossRef]
- Rui, Y.; Qiu, G. Analysis of Gut Microbial Communities and Resistance Genes in Pigs and Chickens in Central China. Animals 2022, 12, 3404. [Google Scholar] [CrossRef] [PubMed]
- Zyoud, S.H.; Shakhshir, M.; Abushanab, A.S.; Koni, A.; Taha, A.A.; Abushamma, F.; Sabateen, A.; Al-Jabi, S.W. Global trends in research related to the links between microbiota and antibiotics: A visualization study. Sci. Rep. 2023, 13, 6890. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Wolters, B.; Widyasari-Mehta, A.; Kreuzig, R.; Smalla, K. Contaminations of organic fertilizers with antibiotic residues, resistance genes, and mobile genetic elements mirroring antibiotic use in livestock? Appl. Microbiol. Biotechnol. 2016, 100, 9343–9353. [Google Scholar] [CrossRef]
- Peng, S.; Feng, Y.; Wang, Y.; Guo, X.; Chu, H.; Lin, X. Prevalence of antibiotic resistance genes in soils after continually applied with different manure for 30 years. J. Hazard. Mater. 2017, 340, 16–25. [Google Scholar] [CrossRef]
- Wang, Y.; Lyu, N.; Liu, F.; Liu, W.J.; Bi, Y.; Zhang, Z.; Ma, S.; Cao, J.; Song, X.; Wang, A.; et al. More diversified antibiotic resistance genes in chickens and workers of the live poultry markets. Environ. Int. 2021, 153, 106534. [Google Scholar] [CrossRef]
- Mazhar, S.H.; Li, X.; Rashid, A.; Su, J.; Xu, J.; Brejnrod, A.D.; Su, J.Q.; Wu, Y.; Zhu, Y.G.; Zhou, S.G.; et al. Co-selection of antibiotic resistance genes, and mobile genetic elements in the presence of heavy metals in poultry farm environments. Sci. Total Environ. 2021, 755, 142702. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; Liu, H.; Xin, Y.; Wang, G.; Dai, X.; Yao, J. Composting of oxytetracycline fermentation residue in combination with hydrothermal pretreatment for reducing antibiotic resistance genes enrichment. Bioresour. Technol. 2020, 318, 124271. [Google Scholar] [CrossRef]
- Patel, T.; Marmulak, T.; Gehring, R.; Pitesky, M.; Clapham, M.O.; Tell, L.A. Drug residues in poultry meat: A literature review of commonly used veterinary antibacterials and anthelmintics used in poultry. J. Vet. Pharmacol. Ther. 2018, 41, 761–789. [Google Scholar] [CrossRef]
- Xu, J.; Sangthong, R.; McNeil, E.; Tang, R.; Chongsuvivatwong, V. Antibiotic use in chicken farms in northwestern China. Antimicrob. Resist. Infect. Control 2020, 9, 10. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Tian, G.M.; Jin, R.C. The occurrence, maintenance, and proliferation of antibiotic resistance genes (ARGs) in the environment: Influencing factors, mechanisms, and elimination strategies. Appl. Microbiol. Biotechnol. 2018, 102, 8261–8274. [Google Scholar] [CrossRef] [PubMed]
- Tong, P.; Ji, X.; Chen, L.; Liu, J.; Xu, L.; Zhu, L.; Zhou, W.; Liu, G.; Wang, S.; Guo, X. Metagenome analysis of antibiotic resistance genes in fecal microbiota of chickens. Agri. Gene 2017, 5, 1–6. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Liu, F.; Cao, J.; Lv, N.; Zhu, B.; Zhang, G.; Gao, G.F. Integrated metagenomic and metatranscriptomic profiling reveals differentially expressed resistomes in human, chicken, and pig gut microbiomes. Environ. Int. 2020, 138, 105649. [Google Scholar] [CrossRef]
- Koorakula, R.; Schiavinato, M.; Ghanbari, M.; Wegl, G.; Grabner, N.; Koestelbauer, A.; Klose, V.; Dohm, J.C.; Domig, K.J. Metatranscriptomic Analysis of the Chicken Gut Resistome Response to In-Feed Antibiotics and Natural Feed Additives. Front. Microbiol. 2022, 13, 833790. [Google Scholar] [CrossRef]
- Qian, X.; Gu, J.; Sun, W.; Wang, X.J.; Su, J.Q.; Stedfeld, R. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 2018, 344, 716–722. [Google Scholar] [CrossRef]
- Xie, W.Y.; Yang, X.P.; Li, Q.; Wu, L.H.; Shen, Q.R.; Zhao, F.J. Changes in antibiotic concentrations and antibiotic resistome during commercial composting of animal manures. Environ. Pollut. 2016, 219, 182–190. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, Y.; Cai, Y.; Xing, S.; Mi, J.; Liao, X. The relationship between culturable doxycycline-resistant bacterial communities and antibiotic resistance gene hosts in pig farm wastewater treatment plants. Ecotoxicol. Environ. Saf. 2020, 206, 111164. [Google Scholar] [CrossRef]
- Chen, T.; Zhang, S.; Zhu, R.; Zhao, M.; Zhang, Y.; Wang, Y.; Liao, X.; Wu, Y.; Mi, J. Distribution and driving factors of antibiotic resistance genes in treated wastewater from different types of livestock farms. Sci. Total Environ. 2022, 849, 157837. [Google Scholar] [CrossRef]
- Wooten, K.J.; Mayer, G.D.; Smith, P.N. Persistence of elevated concentrations of PM, affiliated pharmaceuticals, and tetracycline resistance genes downwind of feedyards. Environ. Pollut. 2019, 247, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Qiao, M.; Zhang, B.; Cheng, W.D.; Zhu, Y.G. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ. Sci. Technol. 2010, 44, 6933–6939. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; He, M.; Zhang, J.; Liu, J.; Su, J.; Dai, J. Effects of the coexistence of antibiotics and heavy metals on the fate of antibiotic resistance genes in chicken manure and surrounding soils. Ecotoxicol. Environ. Saf. 2023, 263, 115367. [Google Scholar] [CrossRef] [PubMed]
- Rampelli, S.; Schnorr, S.L.; Consolandi, C.; Turroni, S.; Severgnini, M.; Peano, C.; Brigidi, P.; Crittenden, A.N.; Henry, A.G.; Candela, M. Metagenome Sequencing of the Hadza Hunter-Gatherer Gut Microbiota. Curr. Biol. CB 2015, 25, 1682–1693. [Google Scholar] [CrossRef]
- Zeng, J.; Pan, Y.; Yang, J.; Hou, M.; Zeng, Z.; Xiong, W. Metagenomic insights into the distribution of antibiotic resistome between the gut-associated environments and the pristine environments. Environ. Int. 2019, 126, 346–354. [Google Scholar] [CrossRef]
- Gupta, C.L.; Blum, S.E.; Kattusamy, K.; Daniel, T.; Druyan, S.; Shapira, R.; Krifucks, O.; Zhu, Y.G.; Zhou, X.Y.; Su, J.Q.; et al. Longitudinal study on the effects of growth-promoting and therapeutic antibiotics on the dynamics of chicken cloacal and litter microbiomes and resistomes. Microbiome 2021, 9, 178. [Google Scholar] [CrossRef]
- Joyce, A.; McCarthy, C.G.P.; Murphy, S.; Walsh, F. Antibiotic resistomes of healthy pig faecal metagenomes. Microb. Genom. 2019, 5, 5. [Google Scholar] [CrossRef]
- Ma, X.; Yang, Z.; Xu, T.; Qian, M.; Jiang, X.; Zhan, X.; Han, X. Chlortetracycline alters microbiota of gut or faeces in pigs and leads to accumulation and migration of antibiotic resistance genes. Sci. Total Environ. 2021, 796, 148976. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, Y.; Zhang, L.; Wu, Z.; Huang, Y.; Yan, H.; Zhong, J.; Wang, L.J.; Abdullah, H.M.; Wang, H.H. Antibiotic Administration Routes and Oral Exposure to Antibiotic Resistant Bacteria as Key Drivers for Gut Microbiota Disruption and Resistome in Poultry. Front. Microbiol. 2020, 11, 1319. [Google Scholar] [CrossRef]
- Zhou, Y.; Fu, H.; Yang, H.; Wu, J.; Chen, Z.; Jiang, H.; Liu, M.; Liu, Q.; Huang, L.; Gao, J.; et al. Extensive metagenomic analysis of the porcine gut resistome to identify indicators reflecting antimicrobial resistance. Microbiome 2022, 10, 39. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Hallowell, H.A.; Koutouvalis, K.; Suez, J. Good microbes, bad genes? The dissemination of antimicrobial resistance in the human microbiome. Gut Microbes 2022, 14, 2055944. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chai, B. Fate of Antibiotic Resistance Genes and Changes in Bacterial Community with Increasing Breeding Scale of Layer Manure. Front. Microbiol. 2022, 13, 857046. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Zhang, H.; Song, D.; Chen, H.; Lin, X.; Wang, Y.; Ji, L. Distribution of antibiotic, heavy metals and antibiotic resistance genes in livestock and poultry feces from different scale of farms in Ningxia, China. J. Hazard. Mater. 2022, 440, 129719. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tong, C.; Xiao, D.; Xie, L.; Zhao, R.; Huo, Z.; Tang, Z.; Hao, J.; Zeng, Z.; Xiong, W. Metagenomic Insights into Chicken Gut Antibiotic Resistomes and Microbiomes. Microbiol. Spectr. 2022, 10, e0190721. [Google Scholar] [CrossRef]
- Yue, Z.; Zhang, J.; Zhou, Z.; Ding, C.; Wan, L.; Liu, J.; Chen, L.; Wang, X. Pollution characteristics of livestock faeces and the key driver of the spread of antibiotic resistance genes. J. Hazard. Mater. 2021, 409, 124957. [Google Scholar] [CrossRef] [PubMed]
- Stokes, H.W.; Gillings, M.R. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011, 35, 790–819. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.Y.; Zhu, D.; Giles, M.; Daniell, T.; Neilson, R.; Yang, X.R. Does reduced usage of antibiotics in livestock production mitigate the spread of antibiotic resistance in soil, earthworm guts, and the phyllosphere? Environ. Int. 2020, 136, 105359. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Xia, X.; Wang, Z.; Fu, Y.; Du, X.D.; Gao, B.; Zhou, Y.; He, J.; Wang, Y.; Shen, J.; Jiang, H.; et al. Association of colistin residues and manure treatment with the abundance of mcr-1 gene in swine feedlots. Environ. Int. 2019, 127, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Wang, Y.; Sun, Y.; Ma, L.; Zeng, Q.; Jiang, X.; Li, A.; Zeng, Z.; Zhang, T. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome 2018, 6, 34. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, C.; Wei, Y.; Wang, X.; Gao, J.; Cui, H.; Zhang, C.; Liu, J. Analysis of Resistance Gene Diversity in the Intestinal Microbiome of Broilers from Two Types of Broiler Farms in Hebei Province, China. Antibiotics 2023, 12, 1664. https://doi.org/10.3390/antibiotics12121664
Liang C, Wei Y, Wang X, Gao J, Cui H, Zhang C, Liu J. Analysis of Resistance Gene Diversity in the Intestinal Microbiome of Broilers from Two Types of Broiler Farms in Hebei Province, China. Antibiotics. 2023; 12(12):1664. https://doi.org/10.3390/antibiotics12121664
Chicago/Turabian StyleLiang, Chuncai, Yujie Wei, Xiaolan Wang, Jinduo Gao, Huan Cui, Cheng Zhang, and Juxiang Liu. 2023. "Analysis of Resistance Gene Diversity in the Intestinal Microbiome of Broilers from Two Types of Broiler Farms in Hebei Province, China" Antibiotics 12, no. 12: 1664. https://doi.org/10.3390/antibiotics12121664
APA StyleLiang, C., Wei, Y., Wang, X., Gao, J., Cui, H., Zhang, C., & Liu, J. (2023). Analysis of Resistance Gene Diversity in the Intestinal Microbiome of Broilers from Two Types of Broiler Farms in Hebei Province, China. Antibiotics, 12(12), 1664. https://doi.org/10.3390/antibiotics12121664





