Genome-Wide Analysis of Escherichia coli Isolated from Dairy Animals Identifies Virulence Factors and Genes Enriched in Multidrug-Resistant Strains
Abstract
:1. Introduction
2. Materials and Methods
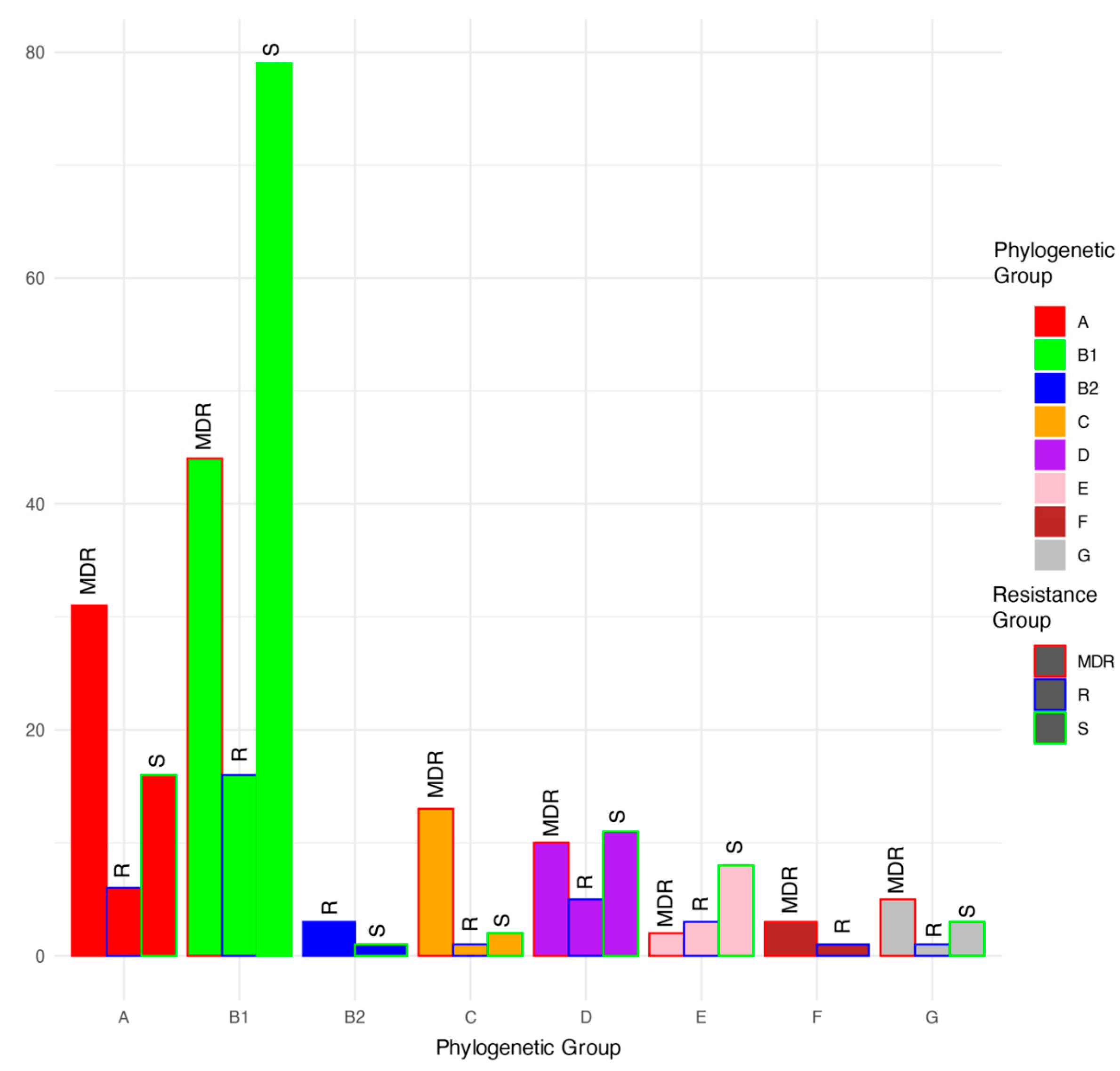
3. Results
3.1. Antimicrobial Resistance Genes
3.2. Biocide and Metal Resistance Genes
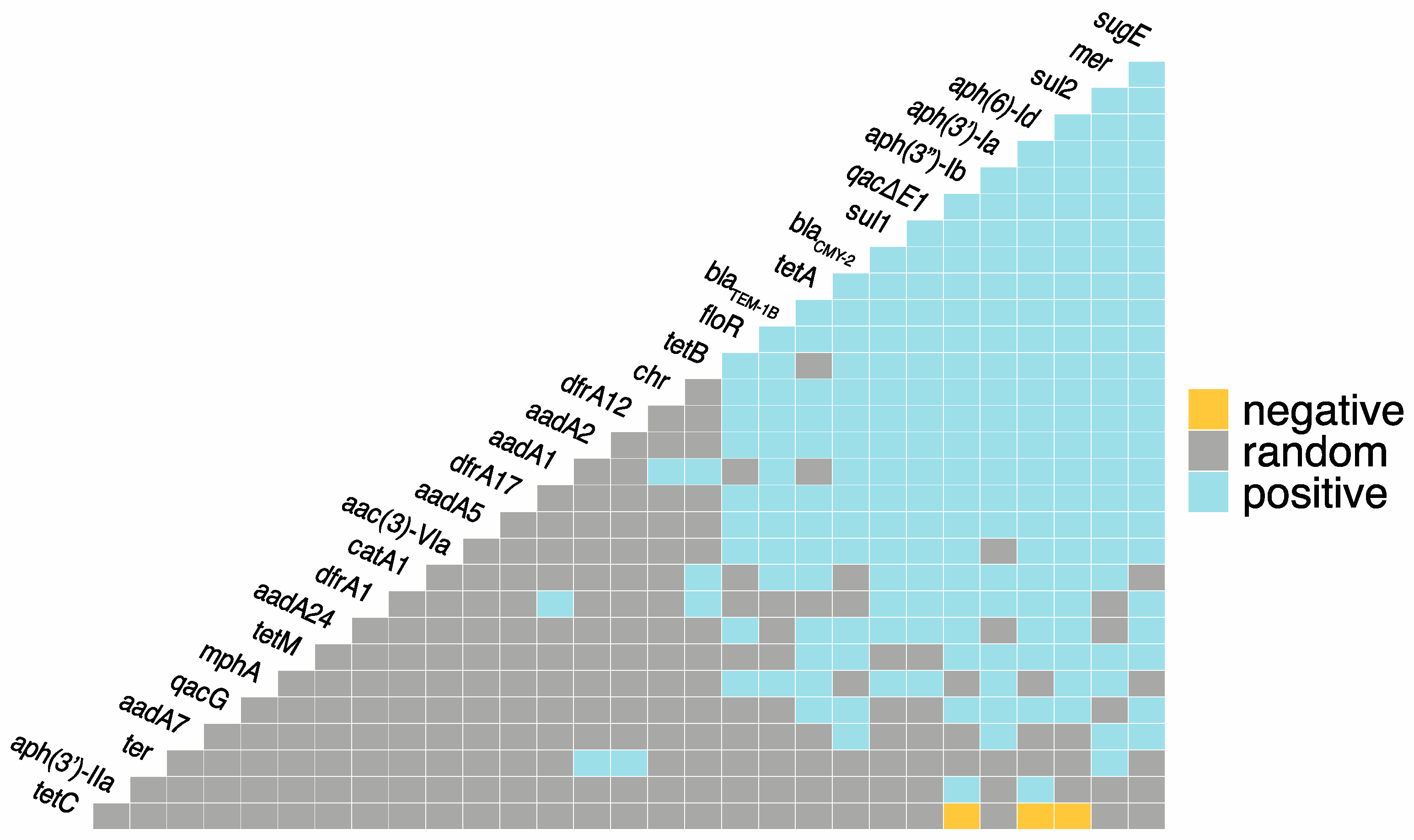
3.3. Co-Occurrence between ARGs, MRGs, and BRGs
3.4. Plasmid Replicon Diversity
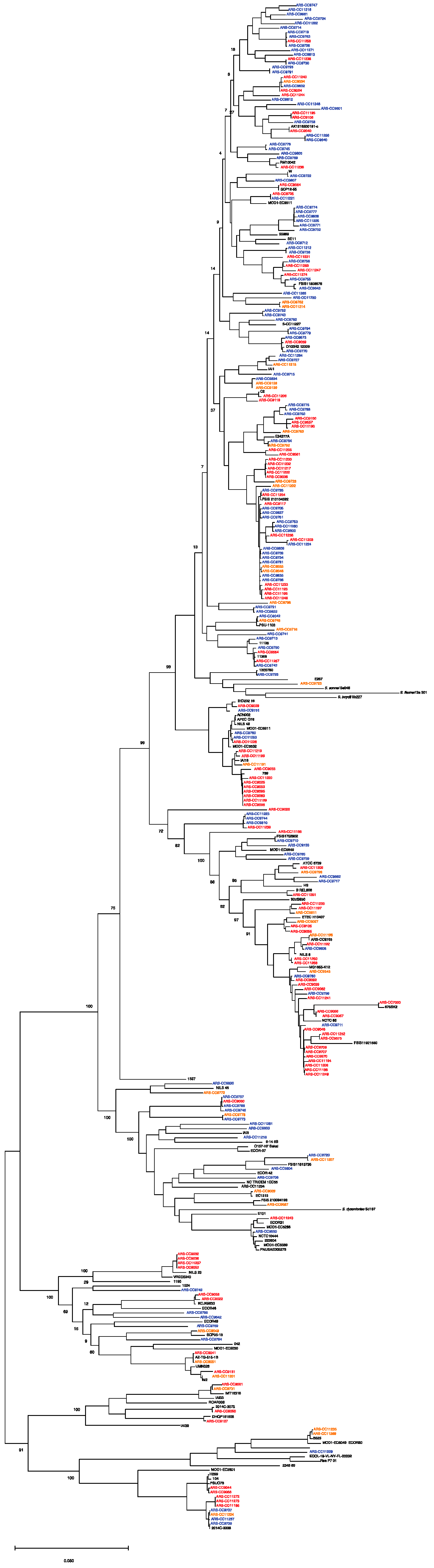
3.5. Genomic Diversity
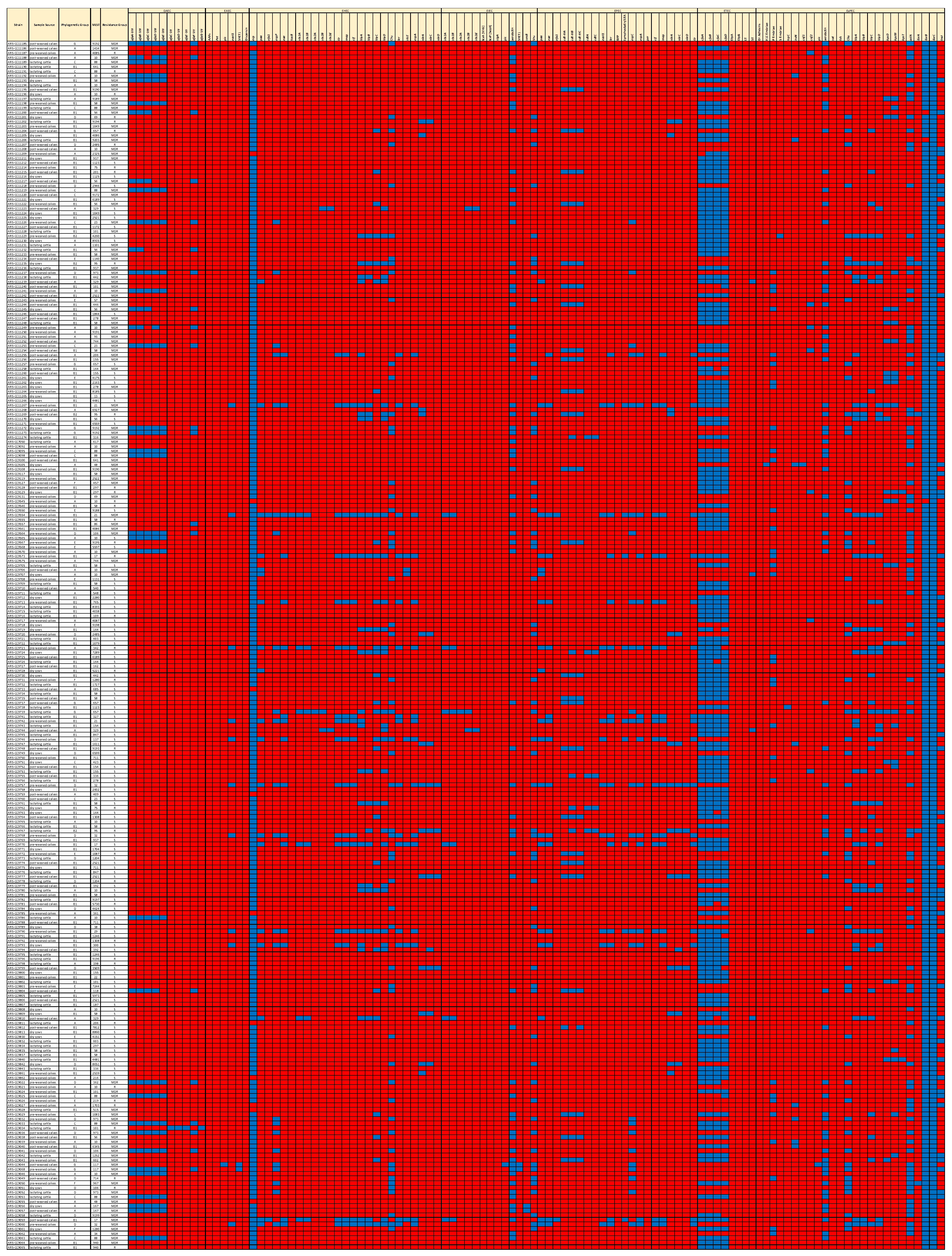
3.6. Virulence and Fitness Factors
3.7. Accessory Genes Associated with the MDR and Susceptible Genotypes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, H.; Pradhan, A.K.; Karns, J.S.; Hovingh, E.; Wolfgang, D.R.; Vinyard, B.T.; Kim, S.W.; Salaheen, S.; Haley, B.J.; Van Kessel, J.A.S. Age-Associated Distribution of Antimicrobial-Resistant Salmonella enterica and Escherichia coli Isolated from Dairy Herds in Pennsylvania, 2013–2015. Foodborne Pathog. Dis. 2019, 16, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Taft, D.H.; Maldonado-Gomez, M.X.; Johnson, D.; Treiber, M.L.; Lemay, D.G.; DePeters, E.J.; Mills, D.A. The fecal resistome of dairy cattle is associated with diet during nursing. Nat. Commun. 2019, 10, 4406. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Jay-Russell, M.; Lemay, D.G.; Mills, D.A. Reservoirs of antimicrobial resistance genes in retail raw milk. Microbiome 2020, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Gyles, C.L. Shiga toxin-producing Escherichia coli: An overview. J. Anim. Sci. 2007, 85, E45–E62. [Google Scholar] [CrossRef] [PubMed]
- Salaheen, S.; Kim, S.W.; Springer, H.R.; Hovingh, E.P.; Van Kessel, J.A.S.; Haley, B.J. Genomic diversity of antimicrobial-resistant and Shiga toxin gene-harboring non-O157 Escherichia coli from dairy calves. J. Glob. Antimicrob. Resist. 2023, 33, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Call, D.R.; Davis, M.A.; Sawant, A.A. Antimicrobial resistance in beef and dairy cattle production. Anim. Health Res. Rev. 2008, 9, 159–167. [Google Scholar] [CrossRef]
- Springer, H.R.; Denagamage, T.N.; Fenton, G.D.; Haley, B.J.; Van Kessel, J.A.S.; Hovingh, E.P. Antimicrobial Resistance in Fecal Escherichia coli and Salmonella enterica from Dairy Calves: A Systematic Review. Foodborne Pathog. Dis. 2019, 16, 23–34. [Google Scholar] [CrossRef]
- Durso, L.M.; Harhay, G.P.; Bono, J.L.; Smith, T.P. Virulence-associated and antibiotic resistance genes of microbial populations in cattle feces analyzed using a metagenomic approach. J. Microbiol. Methods 2011, 84, 278–282. [Google Scholar] [CrossRef]
- Wichmann, F.; Udikovic-Kolic, N.; Andrew, S.; Handelsman, J. Diverse Antibiotic Resistance Genes in Dairy Cow Manure. mBio 2014, 5, e01017. [Google Scholar] [CrossRef]
- Haley, B.J.; Van Kessel, J.A.S. The resistome of the bovine gastrointestinal tract. Curr. Opin. Biotechnol. 2022, 73, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Salaheen, S.; Kim, S.W.; Springer, H.R.; Hovingh, E.P.; Van Kessel, J.A.S.; Haley, B.J. Characterization of Antimicrobial Resistance Genes and Virulence Factors in the Genomes of Escherichia coli ST69 Isolates from Preweaned Dairy Calves and Their Phylogenetic Relationship with Poultry and Human Clinical Strains. Microb. Drug Resist. 2023, 29, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Schmidt, J.W. FunctionalblaKPC-2Sequences Are Present in U.S. Beef Cattle Feces Regardless of Antibiotic Use. Foodborne Pathog. Dis. 2018, 15, 444–448. [Google Scholar] [CrossRef]
- Noyes, N.R.; Yang, X.; Linke, L.M.; Magnuson, R.J.; Cook, S.R.; Zaheer, R.; Yang, H.; Woerner, D.R.; Geornaras, I.; McArt, J.A.; et al. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci. Rep. 2016, 6, 24645. [Google Scholar] [CrossRef] [PubMed]
- Rovira, P.; McAllister, T.; Lakin, S.M.; Cook, S.R.; Doster, E.; Noyes, N.R.; Weinroth, M.D.; Yang, X.; Parker, J.K.; Boucher, C.; et al. Characterization of the Microbial Resistome in Conventional and “Raised Without Antibiotics” Beef and Dairy Production Systems. Front. Microbiol. 2019, 10, 1980. [Google Scholar] [CrossRef]
- Haley, B.J.; Kim, S.-W.; Salaheen, S.; Hovingh, E.; Van Kessel, J.A.S. Differences in the Microbial Community and Resistome Structures of Feces from Preweaned Calves and Lactating Dairy Cows in Commercial Dairy Herds. Foodborne Pathog. Dis. 2020, 17, 494–503. [Google Scholar] [CrossRef]
- Gaire, T.N.; Scott, H.M.; Sellers, L.; Nagaraja, T.G.; Volkova, V.V. Age Dependence of Antimicrobial Resistance Among Fecal Bacteria in Animals: A Scoping Review. Front. Veter. Sci. 2021, 7, 622495. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontéen, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.; Lund, O.; Voldby Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Achtman, M.; the Agama Study Group; Brown, D.; Chattaway, M.; Dallman, T.; Delahay, R.; et al. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Abricate, Github. 2020. Available online: https://github.com/tseemann/abricate (accessed on 28 March 2023).
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Food Safety and Inspection Service, Federal Register. Vol. 77, No. 105 Thursday, 31 May 2012. Available online: https://www.govinfo.gov/content/pkg/FR-2012-05-31/pdf/FR-2012-05-31.pdf (accessed on 2 April 2023).
- Spurbeck, R.R.; Dinh, P.C., Jr.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Kim, K.S.; Johnson, J.R.; Mobley, H.L.T. Escherichia coli Isolates That Carry vat, fyuA, chuA, and yfcV Efficiently Colonize the Urinary Tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef]
- Mellata, M.; Ahmed, L.N.; Price, L.B.; Graham, J.P.; Gazal, L.E.S.; Puño-Sarmiento, J.J.; Medeiros, L.P.; Cyoia, P.S.; da Silveira, W.D.; Nakazato, G.; et al. Human and Avian Extraintestinal Pathogenic Escherichia coli: Infections, Zoonotic Risks, and Antibiotic Resistance Trends. Foodborne Pathog. Dis. 2013, 10, 916–932. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [PubMed]
- Springer, H.R.; Van Kessel, J.S.; Haley, B.J.; Kim, S.W.; Vinyard, B.; Hovingh, E. Impact of Weaning Age and Antibiotic Residues in the Diet on Dynamics of Antimicrobial-Resistant Fecal Escherichia coli in Dairy Calves; United States Department of Agriculture: Beltsville, MD, USA, 2023; to be submitted. [Google Scholar]
- Li, L.-G.; Xia, Y.; Zhang, T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 2017, 11, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.C.; Oladeinde, A.; Kieran, T.J.; Finger, J.W.; Bayona-Vásquez, N.J.; Cartee, J.C.; Beasley, J.C.; Seaman, J.C.; McArthur, J.V.; Rhodes, O.E.; et al. Co-occurrence of antibiotic, biocide, and heavy metal resistance genes in bacteria from metal and radionuclide contaminated soils at the Savannah River Site. Microb. Biotechnol. 2020, 13, 1179–1200. [Google Scholar] [CrossRef]
- Medardus, J.J.; Molla, B.Z.; Nicol, M.; Morrow, W.M.; Rajala-Schultz, P.J.; Kazwala, R.; Gebreyes, W.A. In-Feed Use of Heavy Metal Micronutrients in U.S. Swine Production Systems and Its Role in Persistence of Multidrug-Resistant Salmonellae. Appl. Environ. Microbiol. 2014, 80, 2317–2325. [Google Scholar] [CrossRef]
- Bearson, B.L.; Trachsel, J.M.; Shippy, D.C.; Sivasankaran, S.K.; Kerr, B.J.; Loving, C.L.; Brunelle, B.W.; Curry, S.M.; Gabler, N.K.; Bearson, S.M.D. The Role of Salmonella Genomic Island 4 in Metal Tolerance of Salmonella enterica Serovar I 4,[5],12:i:-Pork Outbreak Isolate USDA15WA-1. Genes 2020, 11, 1291. [Google Scholar] [CrossRef]
- Salaheen, S.; Cao, H.; Sonnier, J.L.; Kim, S.W.; Del Collo, L.P.; Hovingh, E.; Karns, J.S.; Haley, B.J.; Van Kessel, J.A.S. Diversity of Extended-Spectrum Cephalosporin-Resistant Escherichia coli in Feces from Calves and Cows on Pennsylvania Dairy Farms. Foodborne Pathog. Dis. 2019, 16, 368–370. [Google Scholar] [CrossRef]
- Dale, A.P.; Woodford, N. Extra-intestinal pathogenic Escherichia coli (ExPEC): Disease, carriage and clones. J. Infect. 2015, 71, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Roos, V.; Ulett, G.C.; Schembri, M.A.; Klemm, P. The Asymptomatic Bacteriuria Escherichia coli Strain 83972 Outcompetes Uropathogenic E. coli Strains in Human Urine. Infect. Immun. 2006, 74, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Hagan, E.C.; Mobley, H.L.T. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 2009, 71, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Skyberg, J.A.; Johnson, T.J.; Nolan, L.K. Mutational and transcriptional analyses of an avian pathogenic Escherichia coli ColV plasmid. BMC Microbiol. 2008, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Weiss, W.; Pinos-Rodríguez, J.; Socha, M. Effects of feeding supplemental organic iron to late gestation and early lactation dairy cows. J. Dairy Sci. 2010, 93, 2153–2160. [Google Scholar] [CrossRef]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Chu, B.C.; Garcia-Herrero, A.; Johanson, T.H.; Krewulak, K.D.; Lau, C.K.; Peacock, R.S.; Slavinskaya, Z.; Vogel, H.J. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. BioMetals 2010, 23, 601–611. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- Garénaux, A.; Caza, M.; Dozois, C.M. The Ins and Outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Veter. Microbiol. 2011, 153, 89–98. [Google Scholar] [CrossRef]
- Haley, B.J.; Kim, S.W.; Salaheen, S.; Hovingh, E.; Van Kessel, J.A.S. Virulome and genome analyses identify associations between antimicrobial resistance genes and virulence factors in highly drug-resistant Escherichia coli isolated from veal calves. PLoS ONE 2022, 17, e0265445. [Google Scholar] [CrossRef]
- Kröger, C.; Fuchs, T.M. Characterization of the myo -Inositol Utilization Island of Salmonella enterica serovar Typhimurium. J. Bacteriol. 2009, 191, 545–554. [Google Scholar] [CrossRef] [PubMed]

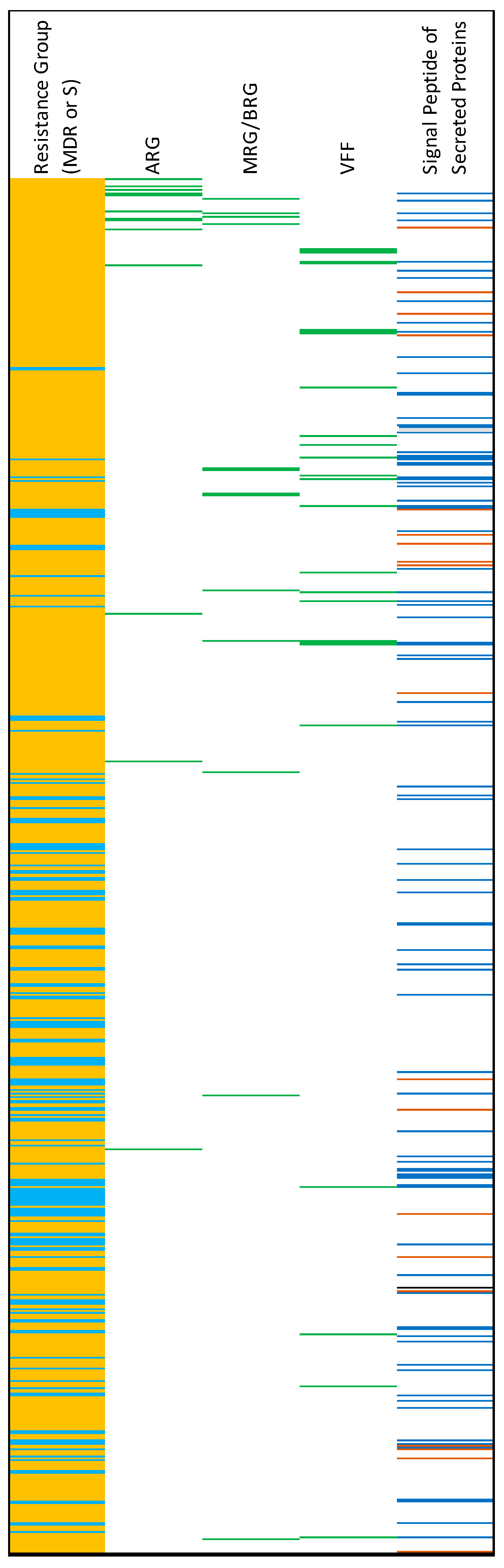
| Isolate ID | Sample Source | Phylogenetic Group | MLST | Antimicrobial Resistance Genes (ARGs) | Resistance Conferring Point Mutations | Metal Resistance Genes (MRGs) | BRGs | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ami | β-Lac | Flu | Fos | MLS | Phe | Sul | Tet | Tri | gyrA | parC | parE | ampC | As | Cu | Hg | Ag | Te | Chromate | sugE1 | QAC | ||||
| ARS-CC11185 | Postweaned calves | G | 9192 | aadA1, aph(3′)-Ia, aadA2, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | floR | sul1, sul2 | tetB, tetA | dfrA1, dfra12 | ampC promoter n.-42C>T | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1 | ||||||||||
| ARS-CC11186 | Post-weaned calves | A | 1434 | aph(3′)-Ia, aadA5, aph(6)-Id, aph(3″)-Ib | blaCMY-2, blaTEM-1B | floR | sul1, sul2 | tetA | dfrA17 | parC p.A56T | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silB, silC, silP, silR | chrA | sugE1 | qacE∆1 | |||||||||
| ARS-CC11187 | Pre-weaned calves | A | 4085 | aadA1 | sul1 | ampC promoter n.-42C>T | merA, merT, merR, merP, merC, merD, merE | qacE∆1 | ||||||||||||||||
| ARS-CC11188 | post-weaned calves | A | 10 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaTEM-1B, blaCMY-2 | sul2 | tetB | sugE1 | ||||||||||||||||
| ARS-CC11189 | lactating cattle | C | 88 | aph(6)-Id, aph(3′)-Ia, aph(3″)-Ib | sul2 | tetB | ampC promoter n.-42C>T | |||||||||||||||||
| ARS-CC11190 | lactating cattle | B1 | 641 | aph(3′)-Ia | blaCMY-2 | tetA | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silB, silC, silP, silR | ||||||||||||||||
| ARS-CC11191 | lactating cattle | C | 88 | blaCMY-2 | sugE1 | |||||||||||||||||||
| ARS-CC11192 | pre-weaned calves | A | 10 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | sul2 | tetB | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silC, silP, silR | |||||||||||||||
| ARS-CC11193 | dry cows | B1 | 58 | aph(3′)-Ia, aph(6)-Id, aadA24, aac(3)-Via, aph(3″)-Ib | blaCMY-2, blaTEM-1B | floR | sul1, sul2 | tetB, tetA | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1 | |||||||||||||
| ARS-CC11194 | lactating cattle | A | 10 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | mphA | floR | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacG | |||||||||||
| ARS-CC11195 | post-weaned calves | B1 | 9190 | aph(3″)-Ib, aph(6)-Id, aadA2 | blaCMY-2 | floR | sul1, sul2 | tetA | dfrA12 | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1 | |||||||||||
| ARS-CC11196 | dry cows | A | 10 | blaCTX-M1 | ||||||||||||||||||||
| ARS-CC11197 | lactating cattle | A | 9189 | aph(3″)-Ib, aph(6)-Id, aadA2 | blaCMY-2 | floR | sul1, sul2 | tetA | dfrA12 | parC p.A56T | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1 | ||||||||||
| ARS-CC11198 | pre-weaned calves | B1 | 58 | aph(6)-Id aph(3″)-Ib | blaCMY-2, blaTEM-1B | sul2 | dfrA5 | merA, merT, merR, merP, merC, merD, merE | sugE1 | |||||||||||||||
| ARS-CC11199 | lactating cattle | C | 88 | aac(3)-Via, aadA24, aph(6)-Id, aph(3″)-Ib | blaCMY-2, blaTEM-1B | floR | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | sugE1 | ||||||||||||||
| ARS-CC11200 | post-weaned calves | B1 | 56 | aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, aadA2 | blaCMY-2 | mphA | floR | sul1, sul2 | tetA, tetB, tetM | dfrA12 | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1, qacG | ||||||||||
| ARS-CC11201 | dry cows | D | 69 | blaCMY-2 | arsA, arsB, arsC, arsD | sugE1 | ||||||||||||||||||
| ARS-CC11202 | lactating cattle | B1 | 9194 | blaCMY-2 | arsA, arsB, arsC, arsD | sugE1 | ||||||||||||||||||
| ARS-CC11203 | pre-weaned calves | B1 | 1049 | blaCTX-M1 | sul2 | tetA | ||||||||||||||||||
| ARS-CC11204 | post-weaned calves | G | 657 | blaCMY-2 | sugE1 | |||||||||||||||||||
| ARS-CC11205 | dry cows | B1 | 4086 | aph(6)-Id, aph(3″)-Ib | blaCMY-2 | floR | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | sugE1 | ||||||||||||||
| ARS-CC11206 | lactating cattle | B1 | 9203 | aph(3″)-Ib, aph(6)-Id, aadA1, aac(3)-Via, aadA5 | floR | sul1, sul2 | tetA | dfrA17 | chrA | qacE∆1 | ||||||||||||||
| ARS-CC11207 | post-weaned calves | D | 2485 | blaCMY-2 | tetA | sugE1 | ||||||||||||||||||
| ARS-CC11208 | post-weaned calves | A | 10 | aac(3)-Via, aph(3″)-Ib, aph(6)-Id | blaCMY-2 | floR | sul2 | tetA | merT | sugE1 | ||||||||||||||
| ARS-CC11209 | pre-weaned calves | A | 2325 | aph(6)-Id, aadA1, aph(3′)-Ia, aph(3″)-Ib | blaCMY-2, blaTEM-1B | sul1 | tetB | merA, merT, merR, merP, merC, merD | sugE1 | qacE∆1 | ||||||||||||||
| ARS-CC11211 | dry cows | B1 | 937 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11212 | post-weaned calves | B1 | 1123 | |||||||||||||||||||||
| ARS-CC11214 | pre-weaned calves | B1 | 75 | fosA7.5 | ||||||||||||||||||||
| ARS-CC11215 | post-weaned calves | B1 | 201 | aph(3′)-Ia | ||||||||||||||||||||
| ARS-CC11216 | dry cows | B1 | 1125 | |||||||||||||||||||||
| ARS-CC11217 | post-weaned calves | B1 | 56 | aph(6)-Id, aph(3″)-Ib | blaTEM-1B | sul2 | tetB | |||||||||||||||||
| ARS-CC11218 | pre-weaned calves | D | 2946 | |||||||||||||||||||||
| ARS-CC11219 | pre-weaned calves | C | 88 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ampC promoter n.-42C>T | |||||||||||||||||
| ARS-CC11220 | post-weaned calves | C | 9172 | aph(3″)-Ib, aph(6)-Id, aph(3′)-Ia | floR | sul2 | tetB | ampC promoter n.-32T>A | ||||||||||||||||
| ARS-CC11221 | dry cows | B1 | 6189 | |||||||||||||||||||||
| ARS-CC11222 | pre-weaned calves | B1 | 56 | aadA2, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id | blaTEM-1B | sul1, sul2 | tetA | dfrA12 | merA, merT, merR, merP, merC, merD, merE | terY2, terY1, terW, terZ, terA, terB, terC, terD, terE, terF | qacE∆1 | |||||||||||||
| ARS-CC11223 | post-weaned calves | A | 329 | |||||||||||||||||||||
| ARS-CC11224 | dry cows | B1 | 1049 | |||||||||||||||||||||
| ARS-CC11225 | dry cows | B1 | 2521 | |||||||||||||||||||||
| ARS-CC11226 | pre-weaned calves | C | 23 | aadA1, aph(3′)-Iia, aph(6)-Ic, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | dfrA1 | ampC promoter n.-42C>T | terW, terZ, terA, terB, terC, terD, terE, terF | |||||||||||||||
| ARS-CC11227 | post-weaned calves | B1 | 1172 | |||||||||||||||||||||
| ARS-CC11228 | lactating cattle | B1 | 101 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11229 | pre-weaned calves | B2 | 4260 | |||||||||||||||||||||
| ARS-CC11230 | dry cows | A | 8935 | |||||||||||||||||||||
| ARS-CC11231 | lactating cattle | A | 1101 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11232 | lactating cattle | B1 | 56 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11233 | pre-weaned calves | B1 | 58 | aac(3)-Iid, aadA5 | blaTEM-1B | sul1, sul2 | tetB | dfrA17 | chrA | qacE∆1 | ||||||||||||||
| ARS-CC11234 | post-weaned calves | E | 1140 | aph(6)-Id, aph(3″)-Ib | blaCMY-2 | floR | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | sugE1 | ||||||||||||||
| ARS-CC11235 | dry cows | B2 | 95 | tetC | ||||||||||||||||||||
| ARS-CC11236 | lactating cattle | B1 | 937 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11237 | pre-weaned calves | D | 973 | aadA7, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | sul1, sul2 | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1 | |||||||||||||||
| ARS-CC11238 | lactating cattle | B1 | 442 | aph(3″)-Ib,aph(6)-Id | floR | sul2 | tetA | |||||||||||||||||
| ARS-CC11239 | post-weaned calves | A | 329 | aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id | sul2 | tetB | terW, terZ, terA, terB, terC, terD, terE, terF | |||||||||||||||||
| ARS-CC11240 | post-weaned calves | B1 | 101 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11241 | pre-weaned calves | A | 10 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | sul2 | tetB | sugE1 | ||||||||||||||||
| ARS-CC11242 | post-weaned calves | B1 | 2522 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11243 | pre-weaned calves | E | 57 | aph(3′)-Ia, aadA5, aph(6)-Id, aac(3)-Iia, aadA1, rmtE, aph(3″)-Ib | blaCMY-2 | mphB | catA1 | sul2, sul1 | tetM, tetA | dfrA1, dfrA17 | gyrA p.S83L | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1 | |||||||||
| ARS-CC11244 | post-weaned calves | B1 | 446 | aph(3′)-Ia, aadA2, aph(6)-Id, aph(3″)-Ib | blaTEM-1A | sul1 | tetA | merA, merC, merD, merE | qacE∆1 | |||||||||||||||
| ARS-CC11245 | dry cows | B1 | 56 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11246 | post-weaned calves | B1 | 1844 | |||||||||||||||||||||
| ARS-CC11247 | post-weaned calves | B1 | 278 | aph(3″)-Ib, aph(6)-Id | floR | sul2 | tetA | |||||||||||||||||
| ARS-CC11248 | lactating cattle | B1 | 58 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11249 | pre-weaned calves | A | 10 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2, blaTEM-1B | sul2 | tetB | sugE1 | ||||||||||||||||
| ARS-CC11250 | pre-weaned calves | A | 9191 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | tetB | sugE1 | |||||||||||||||||
| ARS-CC11251 | pre-weaned calves | A | 93 | aph(3″)-Ib, aph(6)-Id, aadA1 | sul1,sul2 | tetB | dfrA1 | gyrA p.S83L | merA, merT, merR, merP, merC, merD, merE | qacE∆1 | ||||||||||||||
| ARS-CC11252 | post-weaned calves | A | 744 | aph(6)-Id, aph(3″)-Ib | blaTEM-1B | mphA | catA1 | sul2 | tetB | dfrA17 | gyrA p.S83L, gyrA p.D87N | parC p.A56T, parC p.S80I, parC p.E84K | merA, merT, merR, merP, merC, merD, merE | |||||||||||
| ARS-CC11253 | pre-weaned calves | C | 23 | aph(3″)-Ib, aph(6)-Id, aph(3′)-Ia, aadA1 | sul2 | tetB | dfrA1 | ampC promoter n.-42C>T | terW, terZ, terA, terB, terC, terD, terE, terF | |||||||||||||||
| ARS-CC11254 | post-weaned calves | B1 | 58 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11255 | post-weaned calves | A | 206 | aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, aadA2 | blaTEM-1B | catA1, floR | sul1, sul2 | tetA | dfrA12 | parC p.A56T | merA, merT, merR, merP, merC, merD, merE | terY2, terY1, terW, terZ, terA, terB, terC, terD, terE, terF | qacE∆1 | |||||||||||
| ARS-CC11256 | post-weaned calves | B1 | 155 | aac(3)-Via, aadA24 | floR | sul1 | tetA | qacE∆1 | ||||||||||||||||
| ARS-CC11257 | pre-weaned calves | G | 657 | |||||||||||||||||||||
| ARS-CC11258 | lactating cattle | B1 | 164 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC11260 | post-weaned calves | B1 | 155 | |||||||||||||||||||||
| ARS-CC11261 | dry cows | E | 4175 | |||||||||||||||||||||
| ARS-CC11262 | dry cows | B1 | 2163 | |||||||||||||||||||||
| ARS-CC11263 | dry cows | B1 | 278 | aph(6)-Id, aph(3″)-Ib | blaTEM-1A | floR | sul2 | tetA | ||||||||||||||||
| ARS-CC11264 | pre-weaned calves | B1 | 8185 | |||||||||||||||||||||
| ARS-CC11265 | dry cows | B1 | 13 | |||||||||||||||||||||
| ARS-CC11266 | dry cows | B1 | 4481 | |||||||||||||||||||||
| ARS-CC11267 | pre-weaned calves | B1 | 21 | aac(3)-Via, aph(3″)-Ib, aph(6)-Id | blaCMY-2 | floR | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | terW, terZ, terA, terB, terC, terD, terE, terF | sugE1 | |||||||||||||
| ARS-CC11268 | post-weaned calves | A | 6927 | aadA24, aac(3)-Via, aph(3″)-Ib, aph(6)-Id | blaCMY-2 | sul1, sul2 | tetA | merA, merT, merR, merP, merC, merE | sugE1 | qacE∆1 | ||||||||||||||
| ARS-CC11269 | post-weaned calves | B2 | 95 | tetC | ||||||||||||||||||||
| ARS-CC11270 | dry cows | B1 | 56 | |||||||||||||||||||||
| ARS-CC11271 | pre-weaned calves | B1 | 6559 | |||||||||||||||||||||
| ARS-CC11272 | dry cows | G | 9192 | aph(6)-Id, aph(3′)-Ia, aph(3″)-Ib, aadA1 | sul2 | tetB | dfrA1 | ampC promoter n.-42C>T | ||||||||||||||||
| ARS-CC11273 | lactating cattle | G | 9192 | aph(6)-Id, aph(3′)-Ia, aph(3″)-Ib, aadA1 | sul2 | tetB | dfrA1 | ampC promoter n.-42C>T | ||||||||||||||||
| ARS-CC11274 | lactating cattle | B1 | 316 | aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC7050 | lactating cattle | A | 617 | aadA5, aac(6′)-Ib-cr | blaCTX-M-15 | aac(6′)-Ib-cr | mphA | catB3 | sul1, sul2 | tetA | dfrA17 | gyrA p.S83L, gyrA p.D87N | parC p.S80I | parE p.S458A | chrA | qacE∆1 | ||||||||
| ARS-CC9092 | pre-weaned calves | A | 10 | aadA1, aph(3′)-Iia, aph(6)-Ic, aph(6)-Id, aph(3″)-Ib | blaTEM-1B, blaCMY-2 | tetB | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silB, silC, silP, silR | sugE1 | |||||||||||||||
| ARS-CC9095 | pre-weaned calves | C | 88 | aph(6)-Id, aph(3′)-Ia, aph(3″)-Ib | sul2 | tetB | ampC promoter n.-42C>T | |||||||||||||||||
| ARS-CC9098 | post-weaned calves | C | 88 | aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id | sul2 | tetB | ampC promoter n.-42C>T | |||||||||||||||||
| ARS-CC9100 | post-weaned calves | B1 | 641 | aac(3)-Via, aadA24, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, aadA5 | blaTEM-1B, blaCMY-2 | floR | sul1, sul2 | tetM, tetA | dfrA17 | sugE1 | qacE∆1 | |||||||||||||
| ARS-CC9105 | dry cows | A | 48 | aadA5, aac(3)-Via, aph(3′)-Ia, rmtE, aph(6)-Id, aadA24, aph(3″)-Ib | blaTEM-1B, blaCMY-2 | floR | sul1, sul2 | tetM, tetA | dfrA17 | sugE1 | qacE∆1, qacG | |||||||||||||
| ARS-CC9108 | pre-weaned calves | B1 | 9190 | aph(3″)-Ib, aph(6)-Id, aadA2 | blaCMY-2 | floR | sul1, sul2 | tetA | dfrA12 | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1 | |||||||||||
| ARS-CC9117 | dry cows | B1 | 58 | aph(3″)-Ib, aph(6)-Id | blaCMY-2 | floR | sul2 | tetA | ||||||||||||||||
| ARS-CC9119 | pre-weaned calves | B1 | 2522 | aph(3″)-Ib, aac(6′)-Iia, aph(6)-Id | blaCMY-2, blaTEM-1B | floR | sul1, sul2 | tetA, tetB | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1 | ||||||||||||
| ARS-CC9127 | post-weaned calves | F | 457 | aph(3″)-Ib, aph(6)-Id | blaCMY-2 | floR | sul2 | tetA | sugE1 | |||||||||||||||
| ARS-CC9128 | post-weaned calves | B1 | 297 | blaCMY-2 | sugE1 | |||||||||||||||||||
| ARS-CC9129 | dry cows | B1 | 297 | blaCMY-2 | sugE1 | |||||||||||||||||||
| ARS-CC9131 | pre-weaned calves | D | 69 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | floR | sul2 | tetB | arsA, arsB, arsC, arsD | |||||||||||||||
| ARS-CC9545 | pre-weaned calves | A | 10 | blaTEM-1D | tetA | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silB, silC, silP, silR | |||||||||||||||||
| ARS-CC9546 | pre-weaned calves | B1 | 58 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | tetB | |||||||||||||||||||
| ARS-CC9550 | pre-weaned calves | E | 9188 | |||||||||||||||||||||
| ARS-CC9554 | pre-weaned calves | B1 | 21 | aac(3)-Iid, aadA2, aph(6)-Id, aph(3″)-Ib | blaTEM-1B | mphA | sul1, sul2 | dfrA12 | ampC promoter n.-42C>T | merA, merT, merR, merP, merC, merD, merE | terW, terZ, terA, terB, terC, terD, terE, terF | chrA | qacE∆1 | |||||||||||
| ARS-CC9555 | pre-weaned calves | B1 | 58 | blaCMY-2 | sugE1 | |||||||||||||||||||
| ARS-CC9557 | pre-weaned calves | B1 | 86 | aac(3)-Via, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id | blaCMY-2, blaTEM-1B | floR | sul1, sul2 | tetA, tetM | gyrA p.S83L | parC p.S80I | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1, qacG | |||||||||||
| ARS-CC9561 | pre-weaned calves | B1 | 4086 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ||||||||||||||||||
| ARS-CC9564 | pre-weaned calves | D | 106 | aadA7, aph(3′)-Ia | blaTEM-1B | sul1 | tetA | parC p.S57T | arsA, arsB, arsC, arsD | merA, merT, merR, merP, merD, merE | qacE∆1 | |||||||||||||
| ARS-CC9565 | pre-weaned calves | A | 10 | |||||||||||||||||||||
| ARS-CC9567 | pre-weaned calves | E | 9195 | tetA | ||||||||||||||||||||
| ARS-CC9568 | pre-weaned calves | E | 5597 | |||||||||||||||||||||
| ARS-CC9570 | pre-weaned calves | A | 10 | aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, aph(4)-Ia, aac(3)-IV, aadA1 | blaCMY-2 | floR | sul2 | tetB, tet31, tetA | dfrA1 | sugE1 | ||||||||||||||
| ARS-CC9573 | pre-weaned calves | B1 | 17 | blaCTX-M-14 | terW, terZ, terA, terB, terC, terD, terE, terF | |||||||||||||||||||
| ARS-CC9575 | pre-weaned calves | A | 744 | aph(3′)-Ia, aph(3′)-Iia, aadA5, aph(6)-Id, aph(3″)-Ib | blaTEM-214 | mphA | floR, catA1 | sul1, sul2 | tetB, tetA | dfrA17 | gyrA p.S83L, gyrA p.D87N | parC p.A56T, parC p.S80I | merA, merT, merR, merP, merD, merE | chrA | qacE∆1 | |||||||||
| ARS-CC9705 | lactating cattle | B1 | 58 | |||||||||||||||||||||
| ARS-CC9706 | post-weaned calves | A | 10 | aadA13 | sul1 | tetB | merA, merT, merR, merP, merC, merD, merE | qacE∆1 | ||||||||||||||||
| ARS-CC9707 | dry cows | A | 10 | aadA13 | sul1 | tetB | merA, merT, merR, merP. merC, merD,merE | qacE∆1 | ||||||||||||||||
| ARS-CC9708 | pre-weaned calves | E | 1131 | |||||||||||||||||||||
| ARS-CC9709 | lactating cattle | B1 | 58 | |||||||||||||||||||||
| ARS-CC9710 | post-weaned calves | A | 540 | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR,pcoS | silA, silB, silC, silP, silR | |||||||||||||||||||
| ARS-CC9711 | lactating cattle | A | 548 | |||||||||||||||||||||
| ARS-CC9712 | dry cows | B1 | 2280 | |||||||||||||||||||||
| ARS-CC9713 | pre-weaned calves | B1 | 765 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9714 | lactating cattle | B1 | 8393 | |||||||||||||||||||||
| ARS-CC9715 | lactating cattle | B1 | 4038 | |||||||||||||||||||||
| ARS-CC9716 | lactating cattle | B1 | 109 | aadA1 | sul1 | merA, merT, merR, merP, merC, merD, merE | qacE∆1 | |||||||||||||||||
| ARS-CC9717 | pre-weaned calves | A | 4087 | |||||||||||||||||||||
| ARS-CC9718 | dry cows | E | 9198 | |||||||||||||||||||||
| ARS-CC9719 | dry cows | B1 | 164 | |||||||||||||||||||||
| ARS-CC9720 | pre-weaned calves | D | 2485 | |||||||||||||||||||||
| ARS-CC9721 | lactating cattle | B1 | 603 | terY1, terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9722 | lactating cattle | B1 | 1079 | |||||||||||||||||||||
| ARS-CC9723 | pre-weaned calves | A | 342 | fosA7.5 | terW, terZ, terA, terB, terC, terD, terE, terF | |||||||||||||||||||
| ARS-CC9724 | dry cows | B1 | 7289 | |||||||||||||||||||||
| ARS-CC9725 | post-weaned calves | B1 | 6189 | |||||||||||||||||||||
| ARS-CC9726 | lactating cattle | B1 | 164 | |||||||||||||||||||||
| ARS-CC9727 | post-weaned calves | B1 | 162 | |||||||||||||||||||||
| ARS-CC9728 | dry cows | B1 | 5221 | fosA7.5 | ||||||||||||||||||||
| ARS-CC9730 | dry cows | B1 | 442 | |||||||||||||||||||||
| ARS-CC9731 | pre-weaned calves | F | 1280 | tetB | ||||||||||||||||||||
| ARS-CC9732 | lactating cattle | B1 | 1727 | |||||||||||||||||||||
| ARS-CC9733 | post-weaned calves | A | 685 | |||||||||||||||||||||
| ARS-CC9734 | lactating cattle | B1 | 58 | |||||||||||||||||||||
| ARS-CC9735 | post-weaned calves | B1 | 58 | |||||||||||||||||||||
| ARS-CC9737 | post-weaned calves | G | 657 | |||||||||||||||||||||
| ARS-CC9738 | lactating cattle | B1 | 1123 | |||||||||||||||||||||
| ARS-CC9739 | lactating cattle | G | 657 | |||||||||||||||||||||
| ARS-CC9741 | lactating cattle | B1 | 327 | |||||||||||||||||||||
| ARS-CC9742 | pre-weaned calves | B1 | 21 | |||||||||||||||||||||
| ARS-CC9743 | lactating cattle | B1 | 154 | |||||||||||||||||||||
| ARS-CC9744 | post-weaned calves | A | 329 | |||||||||||||||||||||
| ARS-CC9745 | lactating cattle | B1 | 847 | |||||||||||||||||||||
| ARS-CC9746 | pre-weaned calves | D | 137 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9747 | lactating cattle | B1 | 1611 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9748 | post-weaned calves | B1 | 9193 | aph(3′)-Ia | ||||||||||||||||||||
| ARS-CC9749 | dry cows | D | 6599 | |||||||||||||||||||||
| ARS-CC9750 | pre-weaned calves | B1 | 711 | |||||||||||||||||||||
| ARS-CC9751 | dry cows | C | 423 | |||||||||||||||||||||
| ARS-CC9752 | post-weaned calves | B1 | 154 | |||||||||||||||||||||
| ARS-CC9753 | lactating cattle | B1 | 155 | |||||||||||||||||||||
| ARS-CC9755 | post-weaned calves | B1 | 336 | |||||||||||||||||||||
| ARS-CC9756 | lactating cattle | B1 | 278 | |||||||||||||||||||||
| ARS-CC9757 | pre-weaned calves | D | 32 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9758 | dry cows | B1 | 2602 | |||||||||||||||||||||
| ARS-CC9759 | post-weaned calves | A | 409 | arsA, arsB, arsC, arsD, arsR | ||||||||||||||||||||
| ARS-CC9760 | post-weaned calves | C | 23 | |||||||||||||||||||||
| ARS-CC9761 | lactating cattle | B1 | 58 | |||||||||||||||||||||
| ARS-CC9762 | dry cows | B1 | 75 | fosA7.5 | ||||||||||||||||||||
| ARS-CC9763 | dry cows | B1 | 164 | |||||||||||||||||||||
| ARS-CC9764 | post-weaned calves | B1 | 1308 | |||||||||||||||||||||
| ARS-CC9765 | lactating cattle | A | 10 | |||||||||||||||||||||
| ARS-CC9766 | lactating cattle | B1 | 58 | |||||||||||||||||||||
| ARS-CC9767 | lactating cattle | B2 | 95 | |||||||||||||||||||||
| ARS-CC9768 | pre-weaned calves | D | 32 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9769 | lactating cattle | B1 | 937 | |||||||||||||||||||||
| ARS-CC9770 | pre-weaned calves | B1 | 17 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9771 | dry cows | B1 | 1704 | |||||||||||||||||||||
| ARS-CC9772 | pre-weaned calves | E | 1087 | tetC | ||||||||||||||||||||
| ARS-CC9773 | lactating cattle | D | 1204 | |||||||||||||||||||||
| ARS-CC9774 | post-weaned calves | B1 | 2521 | |||||||||||||||||||||
| ARS-CC9775 | dry cows | B1 | 711 | |||||||||||||||||||||
| ARS-CC9776 | lactating cattle | B1 | 847 | |||||||||||||||||||||
| ARS-CC9777 | post-weaned calves | B1 | 2521 | |||||||||||||||||||||
| ARS-CC9778 | lactating cattle | D | 1204 | tetC | ||||||||||||||||||||
| ARS-CC9779 | post-weaned calves | B1 | 392 | |||||||||||||||||||||
| ARS-CC9780 | lactating cattle | A | 10 | |||||||||||||||||||||
| ARS-CC9781 | pre-weaned calves | B1 | 58 | |||||||||||||||||||||
| ARS-CC9782 | lactating cattle | B1 | 9197 | |||||||||||||||||||||
| ARS-CC9783 | post-weaned calves | B1 | 5730 | tetA | ||||||||||||||||||||
| ARS-CC9784 | dry cows | D | 4624 | |||||||||||||||||||||
| ARS-CC9785 | pre-weaned calves | A | 361 | |||||||||||||||||||||
| ARS-CC9786 | lactating cattle | A | 10 | |||||||||||||||||||||
| ARS-CC9788 | post-weaned calves | B1 | 711 | |||||||||||||||||||||
| ARS-CC9789 | dry cows | D | 38 | |||||||||||||||||||||
| ARS-CC9790 | pre-weaned calves | B1 | 29 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9791 | lactating cattle | B1 | 1246 | |||||||||||||||||||||
| ARS-CC9792 | pre-weaned calves | B1 | 1308 | tetC | ||||||||||||||||||||
| ARS-CC9793 | dry cows | B1 | 300 | terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9794 | post-weaned calves | B1 | 392 | |||||||||||||||||||||
| ARS-CC9795 | lactating cattle | B1 | 1246 | |||||||||||||||||||||
| ARS-CC9796 | lactating cattle | B1 | 9196 | fosA7.5 | tetC | |||||||||||||||||||
| ARS-CC9798 | lactating cattle | A | 398 | tetC | arsB, arsD, arsC, arsD, asrR | pcoA, pcoB, pcoC, pcoD, pcoR, pcoS | ||||||||||||||||||
| ARS-CC9799 | post-weaned calves | D | 3509 | terY2, tertY1, terW, terZ, terA, terB, terC, terD, terE, terF | ||||||||||||||||||||
| ARS-CC9800 | dry cows | B1 | 155 | |||||||||||||||||||||
| ARS-CC9801 | pre-weaned calves | B1 | 22 | |||||||||||||||||||||
| ARS-CC9802 | lactating cattle | B1 | 101 | |||||||||||||||||||||
| ARS-CC9803 | pre-weaned calves | E | 7244 | |||||||||||||||||||||
| ARS-CC9804 | post-weaned calves | E | 118 | |||||||||||||||||||||
| ARS-CC9805 | lactating cattle | B1 | 5973 | |||||||||||||||||||||
| ARS-CC9806 | post-weaned calves | B1 | 2521 | |||||||||||||||||||||
| ARS-CC9807 | lactating cattle | B1 | 187 | |||||||||||||||||||||
| ARS-CC9808 | dry cows | A | 10 | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silB, silC, silP, silR | |||||||||||||||||||
| ARS-CC9809 | dry cows | B1 | 58 | |||||||||||||||||||||
| ARS-CC9810 | post-weaned calves | A | 329 | |||||||||||||||||||||
| ARS-CC9811 | lactating cattle | A | 206 | parC p.A56T | ||||||||||||||||||||
| ARS-CC9812 | post-weaned calves | B1 | 7812 | |||||||||||||||||||||
| ARS-CC9813 | dry cows | B1 | 8860 | |||||||||||||||||||||
| ARS-CC9830 | dry cows | E | 4151 | |||||||||||||||||||||
| ARS-CC9832 | lactating cattle | B1 | 603 | |||||||||||||||||||||
| ARS-CC9834 | lactating cattle | B1 | 297 | |||||||||||||||||||||
| ARS-CC9835 | lactating cattle | B1 | 58 | |||||||||||||||||||||
| ARS-CC9837 | lactating cattle | B1 | 58 | |||||||||||||||||||||
| ARS-CC9840 | lactating cattle | B1 | 4481 | |||||||||||||||||||||
| ARS-CC9842 | dry cows | D | 8651 | |||||||||||||||||||||
| ARS-CC9843 | lactating cattle | B1 | 336 | |||||||||||||||||||||
| ARS-CC9861 | pre-weaned calves | B1 | 2539 | |||||||||||||||||||||
| ARS-CC9862 | pre-weaned calves | A | 216 | arsA, arsB, arsC, arsD, arsR | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | merA, merT, merR, merP, merC, merD, merE | silA, silB, silC, silP, silR | |||||||||||||||||
| ARS-CC9022 | pre-weaned calves | D | 362 | aac(3)-Via, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id | blaCMY-2, blaTEM-1B | floR | sul1, sul2 | tetA, tetB | merR, merP, merD, merE | sugE1 | qacE∆1, qacG | |||||||||||||
| ARS-CC9023 | pre-weaned calves | A | 10 | aph(6)-Id, aadA1, aph(3′)-Ia, aph(3″)-Ib | blaCMY-2, blaTEM-1B | tetB | sugE1 | |||||||||||||||||
| ARS-CC9024 | pre-weaned calves | B1 | 101 | aac(3)-Via, aph(3″)-Ib, aph(6)-Id | blaCMY-2 | floR | sul2 | tetA, tetB | sugE1 | |||||||||||||||
| ARS-CC9025 | pre-weaned calves | C | 88 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ampC promoter n.-42C>T | |||||||||||||||||
| ARS-CC9026 | pre-weaned calves | E | 219 | blaCMY-2 | tetA | sugE1 | ||||||||||||||||||
| ARS-CC9027 | pre-weaned calves | A | 1703 | blaCMY-2 | sugE1 | |||||||||||||||||||
| ARS-CC9028 | lactating cattle | B1 | 515 | aph(6)-Id, aph(3′)-Ia, aph(3″)-Ib | blaTEM-1B, blaCMY-2 | tetB | arsA, arsB, arsC, arsD | sugE1 | ||||||||||||||||
| ARS-CC9029 | pre-weaned calves | C | 1083 | aadA1, aph(3′)-Ia | blaTEM-1A | catA1 | sul1 | tetA | merA, merT, merR, merC, merD, merE | qacE∆1 | ||||||||||||||
| ARS-CC9032 | pre-weaned calves | D | 973 | aadA7, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib, aadA1, aadA7 | blaCMY-2, blaTEM-1B | catA1 | sul1, sul2 | tetB | dfrA1 | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1 | ||||||||||||
| ARS-CC9033 | lactating cattle | C | 88 | aph(6)-Id, aph(3″)-Ib | blaCMY-2 | sul2 | dfrA8 | |||||||||||||||||
| ARS-CC9034 | lactating cattle | B1 | 101 | blaTEM-1B | tetA | |||||||||||||||||||
| ARS-CC9036 | post-weaned calves | D | 973 | aadA7, aadA7, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | sul1, sul2 | tetB | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1 | ||||||||||||||
| ARS-CC9038 | post-weaned calves | B1 | 56 | aph(3′)-Ia, aadA2, aph(6)-Id, aph(3″)-Ib | blaTEM-1B | sul1, sul2 | tetA | dfrA12 | merA, merT, merR, merP, merC, merD, merE | terY2, terY1, terW, terZ, terA, terB, terC, terD, terE, terF | qacE∆1 | |||||||||||||
| ARS-CC9039 | pre-weaned calves | A | 10 | aadA2, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id | blaCMY-2, blaTEM-1B | mphA | floR | sul1, sul2 | tetA, tetB, tetM | dfrA12 | merT, merR, merP, merC, merD, merE | chrA | sugE1 | qcG2 | ||||||||||
| ARS-CC9040 | post-weaned calves | B1 | 6345 | aadA2, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2 | floR | sul1, sul2 | tetA, tetB | dfrA12 | merA, merT, merR, merP, merC, merD, merE | chrA | sugE1 | qacE∆1 | |||||||||||
| ARS-CC9041 | pre-weaned calves | D | 106 | aadA7, aph(3′)-Ia | blaTEM-1B, blaCTX-M-1 | mphA | sul1 | tetA | parC p.S57T | arsA, arsB, arsC, arsD | merA, merT, merR, merP, merC, merD, merE | qacE∆1 | ||||||||||||
| ARS-CC9042 | lactating cattle | B1 | 1252 | blaCTX-M-1 | sul2 | tetA | ||||||||||||||||||
| ARS-CC9043 | pre-weaned calves | B1 | 602 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaTEM-1B | tetB | ||||||||||||||||||
| ARS-CC9044 | post-weaned calves | G | 117 | aadA1, aadA5, aph(3′)-Ia | blaCTX-M-14, blaTEM-1B | mphA | catA1 | sul1, sul2, | tetA | dfrA17 | merA, merT, merR, merP, merC, merD, merE | chrA | qacE∆1 | |||||||||||
| ARS-CC9068 | pre-weaned calves | G | 117 | aph(3′)-Ia, aadA1, aadA2 | blaCMY-2, blaCTX-M-14 | sul1, sul2 | tetA | dfrA12 | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1 | |||||||||||||
| ARS-CC9046 | pre-weaned calves | A | 10 | aac(3)-Via, aadA1, aph(6)-Id, aph(3″)-Ib, aph(3′)-Ia | floR | sul1, sul2 | tetB, tetA | ampC promoter n.-42C>T | qacE∆1 | |||||||||||||||
| ARS-CC9049 | post-weaned calves | D | 714 | blaCMY-2 | sugE1 | |||||||||||||||||||
| ARS-CC9050 | pre-weaned calves | F | 967 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2, blaTEM-1B | sul2 | tetB | sugE1 | ||||||||||||||||
| ARS-CC9051 | dry cows | D | 106 | blaCMY-2 | parC p.S57T | arsA, arsB, arsC, arsD | sugE1 | |||||||||||||||||
| ARS-CC9052 | lactating cattle | D | 973 | aph(3″)-Ib, aph(6)-Id, aadA1, aadA7 | blaCMY-2 | catA1 | sul2, sul1 | tetB | dfrA1 | merA, merT, merR, merP, merC, merD, merE | terY2, terY1, terW, terZ, terA, terB, terC, terD, terE, terF | sugE1 | qacE∆1 | |||||||||||
| ARS-CC9053 | lactating cattle | C | 88 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ampC promoter n.-42C>T | |||||||||||||||||
| ARS-CC9055 | post-weaned calves | A | 48 | aadA5, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2, blaTEM-1B | sul2 | tetB, tetA | dfrA17 | sugE1 | |||||||||||||||
| ARS-CC9056 | dry cows | A | 167 | aph(3′)-Ia, aph(6)-Id, aadA5, aph(3″)-Ib | blaCMY-2, blaTEM-1B | floR | sul2 | tetB, tetA | dfrA17 | ampC promoter n.-42C>T | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1 | |||||||||||
| ARS-CC9057 | post-weaned calves | A | 167 | aph(6)-Id, aadA5, aph(3′)-Ia, aph(3″)-Ib | blaCMY-2, blaTEM-1B | floR | sul2 | tetB, tetA | dfrA17 | ampC promoter n.-42C>T | merA, merT, merR, merP, merC, merD, merE | sugE1 | qacE∆1 | |||||||||||
| ARS-CC9058 | lactating cattle | D | 9199 | aph(6)-Id, aph(3″)-Ib | blaCMY-2 | floR | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | sugE1 | ||||||||||||||
| ARS-CC9059 | post-weaned calves | B1 | 17 | aph(6)-Id, aph(3″)-Ib | blaCMY-2 | floR | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | terW, terZ, terA, terB, terC, terD, terE, terF | sugE1 | |||||||||||||
| ARS-CC9060 | pre-weaned calves | D | 32 | aph(6)-Id, aph(3″)-Ib | blaCMY-2 | sul2 | tetA | merA, merT, merR, merP, merC, merD, merE | terW, terZ, terA, terB, terC, terD, terE, terF | sugE1 | ||||||||||||||
| ARS-CC9061 | dry cows | F | 1280 | aph(3″)-Ib, aph(6)-Id | blaCMY-2 | floR | sul2 | tetB, tetA | merA, merT, merR, merP, merC, merD, merE | sugE1 | ||||||||||||||
| ARS-CC9062 | pre-weaned calves | A | 34 | aadA1, aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | blaCMY-2, blaTEM-1B | sul2 | tetB | dfrA1 | sugE1 | |||||||||||||||
| ARS-CC9063 | lactating cattle | C | 88 | aph(3′)-Ia, aph(6)-Id, aph(3″)-Ib | sul2 | tetB | ampC promoter n.-42C>T | |||||||||||||||||
| ARS-CC9064 | pre-weaned calves | B1 | 940 | aadA24, aph(3″)-Ib, aph(6)-Id | blaCTX-M-14, blaOXA-1 | catA1 | sul2 | tetB | dfrA1 | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silB, silC, silP, silR | |||||||||||||
| ARS-CC9065 | lactating cattle | B1 | 940 | aadA1, aph(3″)-Ib, aph(6)-Id | blaCTX-M-14, blaOXA-1 | catA1 | sul2 | tetB | dfrA1 | pcoA, pcoB, pcoC, pcoD, pcoE, pcoR, pcoS | silA, silB, silC, silP, silR | |||||||||||||
| Isolate ID | Sample Source | Phylogenetic Group | MLST | Resistance Group | Plasmid Replicons |
|---|---|---|---|---|---|
| ARS-CC11185 | postweaned calves | G | 9192 | MDR | ColRNAI, IncA/C2, IncFIA, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11186 | postweaned calves | A | 1434 | MDR | IncA/C2, IncX1 |
| ARS-CC11187 | preweaned calves | A | 4085 | MDR | ColRNAI, IncB/O/K/Z, IncFIB(AP001918), IncQ1 |
| ARS-CC11188 | postweaned calves | A | 10 | MDR | IncFIA, IncFIB(AP001918), IncFII, IncI1_Alpha |
| ARS-CC11189 | lactating cattle | C | 88 | MDR | Col(MG828), IncFIA, IncFIB(AP001918) |
| ARS-CC11190 | lactating cattle | B1 | 641 | MDR | Col(MG828), Col440I, ColRNAI, IncFIB(AP001918), IncFII(pRSB107)_pRSB107, IncI1_Alpha, IncX1 |
| ARS-CC11191 | lactating cattle | C | 88 | R | Col156, IncI1_Alpha |
| ARS-CC11192 | preweaned calves | A | 10 | MDR | Col(MG828), Col(MP18), Col156, Col440I, ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC11193 | dry cows | B1 | 58 | MDR | IncA/C2, IncFIB(AP001918) |
| ARS-CC11194 | lactating cattle | A | 10 | MDR | IncA/C2 |
| ARS-CC11195 | postweaned calves | B1 | 9190 | MDR | Col440I, ColRNAI, IncA/C2, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11196 | dry cows | A | 10 | R | IncI1_Alpha, IncN |
| ARS-CC11197 | lactating cattle | A | 9189 | MDR | IncA/C2 |
| ARS-CC11198 | preweaned calves | B1 | 58 | MDR | IncFIB(AP001918), IncFII |
| ARS-CC11199 | lactating cattle | C | 88 | MDR | Col(MG828), Col8282, ColRNAI, IncA/C2, IncFIB(AP001918), IncFII, IncI1_Alpha |
| ARS-CC11200 | postweaned calves | B1 | 56 | MDR | IncA/C2 |
| ARS-CC11201 | dry cows | D | 69 | R | IncFII |
| ARS-CC11202 | lactating cattle | B1 | 9194 | R | IncFII |
| ARS-CC11203 | preweaned calves | B1 | 1049 | MDR | IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11204 | postweaned calves | G | 657 | R | ColRNAI, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11205 | dry cows | B1 | 4086 | MDR | ColRNAI, IncA/C2, IncFIA, IncFIB(AP001918) |
| ARS-CC11206 | lactating cattle | B1 | 9203 | MDR | ColRNAI, IncFII, p0111 |
| ARS-CC11207 | postweaned calves | D | 2485 | R | IncFIA, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11208 | postweaned calves | A | 10 | MDR | IncA/C2 |
| ARS-CC11209 | preweaned calves | A | 2325 | MDR | Col440II, ColRNAI, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11211 | dry cows | B1 | 937 | MDR | ColRNAI, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC11212 | postweaned calves | B1 | 1123 | S | IncFIB(pB171)_pB171 |
| ARS-CC11214 | preweaned calves | B1 | 75 | R | ColRNAI, IncFIA, IncFIB(AP001918), IncFIC(FII), p0111 |
| ARS-CC11215 | postweaned calves | B1 | 201 | R | IncFIB(AP001918), IncI1_Alpha, IncX1 |
| ARS-CC11216 | dry cows | B1 | 1125 | S | IncFIB(AP001918), IncFIC(FII) |
| ARS-CC11217 | postweaned calves | B1 | 56 | MDR | ColRNAI, IncY |
| ARS-CC11218 | preweaned calves | D | 2946 | S | Col440I, IncFIB(AP001918) |
| ARS-CC11219 | preweaned calves | C | 88 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC11220 | postweaned calves | C | 9172 | MDR | IncFII(pCoo)_pCoo |
| ARS-CC11221 | dry cows | B1 | 6189 | S | IncFIA, IncFIB(AP001918), IncFIC(FII), IncY |
| ARS-CC11222 | preweaned calves | B1 | 56 | MDR | IncFIB(AP001918), IncHI2A, IncHI2, RepA_pKPC-CAV1321 |
| ARS-CC11223 | postweaned calves | A | 329 | S | ColRNAI, IncFIB(AP001918), IncFIC(FII), IncX1, IncY |
| ARS-CC11224 | dry cows | B1 | 1049 | S | |
| ARS-CC11225 | dry cows | B1 | 2521 | S | IncFIB(AP001918), IncX1 |
| ARS-CC11226 | preweaned calves | C | 23 | MDR | ColRNAI, IncFIB(AP001918) |
| ARS-CC11227 | postweaned calves | B1 | 1172 | S | Col156, ColRNAI, IncFIB(AP001918), IncFIC(FII), IncX1, IncX3 |
| ARS-CC11228 | lactating cattle | B1 | 101 | MDR | IncFIA(HI1)_HI1, IncFIB(pB171)_pB171 |
| ARS-CC11229 | preweaned calves | B2 | 4260 | S | IncFIB(AP001918) |
| ARS-CC11230 | dry cows | A | 8935 | S | ColRNAI, IncFIC(FII), IncI1_Alpha |
| ARS-CC11231 | lactating cattle | A | 1101 | MDR | ColRNAI, IncFIB(AP001918), IncFIC(FII), IncFII |
| ARS-CC11232 | lactating cattle | B1 | 56 | MDR | |
| ARS-CC11233 | preweaned calves | B1 | 58 | MDR | ColRNAI, IncI2_Delta |
| ARS-CC11234 | postweaned calves | E | 1140 | MDR | ColRNAI, IncA/C2 |
| ARS-CC11235 | dry cows | B2 | 95 | R | ColRNAI, IncFIB(AP001918), IncX1 |
| ARS-CC11236 | lactating cattle | B1 | 937 | MDR | IncFIB(AP001918), IncFIC(FII), IncI1_Alpha |
| ARS-CC11237 | preweaned calves | D | 973 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC11238 | lactating cattle | B1 | 442 | MDR | IncFIA, IncFIB(AP001918), IncX1 |
| ARS-CC11239 | postweaned calves | A | 329 | MDR | IncFIB(AP001918) |
| ARS-CC11240 | postweaned calves | B1 | 101 | MDR | Col440I, ColRNAI, IncFIB(AP001918), IncFII(pHN7A8)_pHN7A8 |
| ARS-CC11241 | preweaned calves | A | 10 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC11242 | postweaned calves | B1 | 2522 | MDR | Col156 |
| ARS-CC11243 | preweaned calves | E | 57 | MDR | IncA/C2 |
| ARS-CC11244 | postweaned calves | B1 | 446 | MDR | Col440I, IncFIB(AP001918) |
| ARS-CC11245 | dry cows | B1 | 56 | MDR | |
| ARS-CC11246 | postweaned calves | B1 | 1844 | S | |
| ARS-CC11247 | post-weaned calves | B1 | 278 | MDR | Col440I, ColRNAI |
| ARS-CC11248 | lactating cattle | B1 | 58 | MDR | Col8282, ColRNAI, ColpVC |
| ARS-CC11249 | preweaned calves | A | 10 | MDR | IncFIA, IncFIB(AP001918), IncFII, IncI1_Alpha |
| ARS-CC11250 | preweaned calves | A | 9191 | MDR | Col440I, IncFIA, IncFIB(pB171)_pB171, IncI_Gamma, IncX1 |
| ARS-CC11251 | preweaned calves | A | 93 | MDR | Col156, ColRNAI, IncB/O/K/Z, IncFIA, IncFIB(AP001918) |
| ARS-CC11252 | postweaned calves | A | 744 | MDR | IncFIA, IncFIB(AP001918), IncFII(pAMA1167-NDM-5)_pAMA1167-NDM-5 |
| ARS-CC11253 | preweaned calves | C | 23 | MDR | ColRNAI, IncFIB(AP001918) |
| ARS-CC11254 | postweaned calves | B1 | 58 | MDR | IncFIB(AP001918) |
| ARS-CC11255 | postweaned calves | A | 206 | MDR | Col156, IncA/C2, IncFII(pSE11)_pSE11, IncHI2A, IncHI2, RepA_pKPC-CAV1321, p0111 |
| ARS-CC11256 | postweaned calves | B1 | 155 | MDR | IncFIB(AP001918) |
| ARS-CC11257 | preweaned calves | G | 657 | S | ColRNAI, IncY |
| ARS-CC11258 | lactating cattle | B1 | 164 | MDR | Col440I, ColRNAI, IncFIA, IncFIB(AP001918), IncI2_Delta |
| ARS-CC11260 | postweaned calves | B1 | 155 | S | IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11261 | dry cows | E | 4175 | S | ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC11262 | dry cows | B1 | 2163 | S | |
| ARS-CC11263 | dry cows | B1 | 278 | MDR | ColRNAI, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC11264 | preweaned calves | B1 | 8185 | S | IncFIA, IncFIB(AP001918), IncX1 |
| ARS-CC11265 | dry cows | B1 | 13 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC11266 | dry cows | B1 | 4481 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC11267 | preweaned calves | B1 | 21 | MDR | IncA/C2, IncB/O/K/Z, IncFIB(AP001918), IncY |
| ARS-CC11268 | postweaned calves | A | 6927 | MDR | Col440I, IncA/C2, IncFIA, IncFIB(pB171)_pB171 |
| ARS-CC11269 | postweaned calves | B2 | 95 | R | IncFIB(AP001918), IncX1, IncY |
| ARS-CC11270 | dry cows | B1 | 56 | S | Col(MG828), IncFIA, IncFIB(AP001918), IncFIC(FII), IncI1_Alpha |
| ARS-CC11271 | preweaned calves | B1 | 6559 | S | |
| ARS-CC11272 | dry cows | G | 9192 | MDR | ColRNAI, IncFIA, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11273 | lactating cattle | G | 9192 | MDR | ColRNAI, IncFIA, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC11274 | lactating cattle | B1 | 316 | MDR | ColRNAI, IncFIB(AP001918) |
| ARS-CC7050 | lactating cattle | A | 617 | MDR | ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC9092 | preweaned calves | A | 10 | MDR | IncFII, IncR |
| ARS-CC9095 | preweaned calves | C | 88 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC9098 | postweaned calves | C | 88 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC9100 | postweaned calves | B1 | 641 | MDR | IncA/C2, IncX1, IncX3, IncX4, IncY |
| ARS-CC9105 | dry cows | A | 48 | MDR | IncA/C2 |
| ARS-CC9108 | preweaned calves | B1 | 9190 | MDR | Col440I, ColRNAI, IncA/C2, IncFIB(AP001918) |
| ARS-CC9117 | dry cows | B1 | 58 | MDR | IncFIB(AP001918), IncY |
| ARS-CC9119 | preweaned calves | B1 | 2522 | MDR | IncA/C2 |
| ARS-CC9127 | postweaned calves | F | 457 | MDR | ColRNAI, IncFII, IncI2_Delta |
| ARS-CC9128 | postweaned calves | B1 | 297 | R | ColRNAI, IncA/C2, IncI1_Alpha |
| ARS-CC9129 | dry cows | B1 | 297 | R | ColRNAI, IncI1_Alpha |
| ARS-CC9131 | preweaned calves | D | 69 | MDR | ColRNAI, IncFIA, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC9545 | preweaned calves | A | 10 | R | Col156, IncFIA, IncFIB(AP001918), IncI2_Delta, IncX3 |
| ARS-CC9546 | preweaned calves | B1 | 58 | R | IncFIA(HI1)_HI1, IncFIB(pB171)_pB171 |
| ARS-CC9550 | preweaned calves | E | 9188 | S | IncI1_Alpha |
| ARS-CC9554 | preweaned calves | B1 | 21 | MDR | Col(MG828), ColRNAI, IncB/O/K/Z, IncFIB(AP001918), p0111 |
| ARS-CC9555 | preweaned calves | B1 | 58 | R | IncFIA(HI1)_HI1, IncFIB(pB171)_pB171, IncI1_Alpha |
| ARS-CC9557 | preweaned calves | B1 | 86 | MDR | IncA/C2, IncY |
| ARS-CC9561 | preweaned calves | B1 | 4086 | MDR | ColRNAI, IncI2_Delta |
| ARS-CC9564 | preweaned calves | D | 106 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC9565 | preweaned calves | A | 10 | S | ColRNAI, IncFIB(pB171)_pB171 |
| ARS-CC9567 | preweaned calves | E | 9195 | R | IncFIB(AP001918) |
| ARS-CC9568 | preweaned calves | E | 5597 | S | IncFIB(AP001918) |
| ARS-CC9570 | preweaned calves | A | 10 | MDR | Col(MG828), Col440I, ColRNAI, IncA/C2, IncFIA, IncFIB(AP001918) |
| ARS-CC9573 | preweaned calves | B1 | 17 | R | ColRNAI, IncFIB(AP001918), IncFII |
| ARS-CC9575 | preweaned calves | A | 744 | MDR | ColRNAI, IncX1 |
| ARS-CC9705 | lactating cattle | B1 | 58 | S | IncFIB(AP001918), IncFII(pHN7A8)_pHN7A8 |
| ARS-CC9706 | postweaned calves | A | 10 | MDR | ColRNAI, IncFIA, IncFIB(AP001918), IncFII(pHN7A8)_pHN7A8, IncFII |
| ARS-CC9707 | dry cows | A | 10 | MDR | ColRNAI, IncFIA, IncFIB(AP001918), IncFII(pHN7A8)_pHN7A8, IncFII |
| ARS-CC9708 | preweaned calves | E | 1131 | S | Col440I |
| ARS-CC9709 | lactating cattle | B1 | 58 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9710 | postweaned calves | A | 540 | S | |
| ARS-CC9711 | lactating cattle | A | 548 | S | ColRNAI, IncFIB(AP001918) |
| ARS-CC9712 | dry cows | B1 | 2280 | S | |
| ARS-CC9713 | preweaned calves | B1 | 765 | S | IncFIC(FII) |
| ARS-CC9714 | lactating cattle | B1 | 8393 | S | |
| ARS-CC9715 | lactating cattle | B1 | 4038 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9716 | lactating cattle | B1 | 109 | R | |
| ARS-CC9717 | preweaned calves | A | 4087 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9718 | dry cows | E | 9198 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9719 | dry cows | B1 | 164 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9720 | preweaned calves | D | 2485 | S | Col440I, ColRNAI, IncFIA, IncFIB(pB171)_pB171 |
| ARS-CC9721 | lactating cattle | B1 | 603 | S | ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC9722 | lactating cattle | B1 | 1079 | S | ColRNAI |
| ARS-CC9723 | preweaned calves | A | 342 | R | Col(MG828), Col156, ColRNAI, IncFIB(AP001918) |
| ARS-CC9724 | dry cows | B1 | 7289 | S | ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC9725 | postweaned calves | B1 | 6189 | S | IncFIA, IncFIB(AP001918), IncFIC(FII), IncY |
| ARS-CC9726 | lactating cattle | B1 | 164 | S | IncFIA |
| ARS-CC9727 | postweaned calves | B1 | 162 | S | ColRNAI, IncFIB(AP001918) |
| ARS-CC9728 | dry cows | B1 | 5221 | R | IncFIA, IncFIB(AP001918), IncFIC(FII), IncY |
| ARS-CC9730 | dry cows | B1 | 442 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9731 | preweaned calves | F | 1280 | R | ColRNAI |
| ARS-CC9732 | lactating cattle | B1 | 1727 | S | IncFIB(AP001918) |
| ARS-CC9733 | postweaned calves | A | 685 | S | IncI1_Alpha |
| ARS-CC9734 | lactating cattle | B1 | 58 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9735 | postweaned calves | B1 | 58 | S | IncFIB(AP001918) |
| ARS-CC9737 | postweaned calves | G | 657 | S | IncFIB(AP001918) |
| ARS-CC9738 | lactating cattle | B1 | 1123 | S | IncFIA(HI1)_HI1, IncFIB(K)_Kpn3 |
| ARS-CC9739 | lactating cattle | G | 657 | S | Col(MG828), IncB/O/K/Z, IncFIB(AP001918) |
| ARS-CC9741 | lactating cattle | B1 | 327 | S | IncFIB(AP001918) |
| ARS-CC9742 | preweaned calves | B1 | 21 | S | Col440I, IncB/O/K/Z, IncFIB(AP001918) |
| ARS-CC9743 | lactating cattle | B1 | 154 | S | IncFIA, IncFIC(FII) |
| ARS-CC9744 | postweaned calves | A | 329 | S | ColRNAI, IncFIB(AP001918), IncX1, IncY |
| ARS-CC9745 | lactating cattle | B1 | 847 | S | Col(MG828), ColRNAI, ColpVC, IncI1_Alpha |
| ARS-CC9746 | preweaned calves | D | 137 | S | IncFIB(AP001918), IncY |
| ARS-CC9747 | lactating cattle | B1 | 1611 | S | IncFIB(AP001918), IncFII(pHN7A8)_pHN7A8 |
| ARS-CC9748 | postweaned calves | B1 | 9193 | R | ColRNAI, IncFIB(AP001918) |
| ARS-CC9749 | dry cows | D | 6599 | S | IncI1_Alpha |
| ARS-CC9750 | preweaned calves | B1 | 711 | S | IncFIB(AP001918) |
| ARS-CC9751 | dry cows | C | 423 | S | ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC9752 | postweaned calves | B1 | 154 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9753 | lactating cattle | B1 | 155 | S | IncFIB(AP001918), IncI1_Alpha, IncX4 |
| ARS-CC9755 | postweaned calves | B1 | 336 | S | IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9756 | lactating cattle | B1 | 278 | S | IncFIB(AP001918), IncFIC(FII), IncFII, IncI1_Alpha |
| ARS-CC9757 | preweaned calves | D | 32 | S | IncFIB(AP001918) |
| ARS-CC9758 | dry cows | B1 | 2602 | S | ColRNAI, IncFIB(AP001918), IncI2_Delta, IncY |
| ARS-CC9759 | postweaned calves | A | 409 | S | IncFIB(K)_Kpn3, IncY |
| ARS-CC9760 | postweaned calves | C | 23 | S | Col440I, IncFIA, IncFIB(pB171)_pB171 |
| ARS-CC9761 | lactating cattle | B1 | 58 | S | IncFIB(AP001918), IncFIC(FII), IncX1 |
| ARS-CC9762 | dry cows | B1 | 75 | R | ColRNAI, IncFIA, IncFIB(AP001918), IncFIC(FII), IncFII(pCoo)_pCoo, |
| ARS-CC9763 | dry cows | B1 | 164 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9764 | postweaned calves | B1 | 1308 | S | IncFIB(AP001918) |
| ARS-CC9765 | lactating cattle | A | 10 | S | |
| ARS-CC9766 | lactating cattle | B1 | 58 | S | Col440I, IncI1_Alpha, IncI2_Delta |
| ARS-CC9767 | lactating cattle | B2 | 95 | R | IncFIB(AP001918), IncX1 |
| ARS-CC9768 | preweaned calves | D | 32 | S | IncB/O/K/Z, IncFIB(AP001918) |
| ARS-CC9769 | lactating cattle | B1 | 937 | S | |
| ARS-CC9770 | preweaned calves | B1 | 17 | S | IncFIB(AP001918) |
| ARS-CC9771 | dry cows | B1 | 1704 | S | ColRNAI, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC9772 | preweaned calves | E | 1087 | R | IncFIB(AP001918), IncY |
| ARS-CC9773 | lactating cattle | D | 1204 | S | ColRNAI, IncFIB(AP001918) |
| ARS-CC9774 | postweaned calves | B1 | 2521 | S | Col440I, IncFIA, IncFIB(AP001918), IncX1, IncX4 |
| ARS-CC9775 | dry cows | B1 | 711 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9776 | lactating cattle | B1 | 847 | S | Col440II, ColRNAI, ColpVC, IncI1_Alpha, p0111 |
| ARS-CC9777 | postweaned calves | B1 | 2521 | S | ColRNAI, IncFIB(AP001918) |
| ARS-CC9778 | lactating cattle | D | 1204 | R | IncFIB(AP001918), IncI2_Delta, IncY |
| ARS-CC9779 | postweaned calves | B1 | 392 | S | IncFIA(HI1)_HI1, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9780 | lactating cattle | A | 10 | S | ColRNAI, IncFIC(FII) |
| ARS-CC9781 | preweaned calves | B1 | 58 | S | IncFIA(HI1)_HI1, IncFIB(pB171)_pB171, IncY |
| ARS-CC9782 | lactating cattle | B1 | 9197 | S | Col440I, ColRNAI, IncFIA, IncFIB(AP001918), IncFIC(FII), IncI2_Delta |
| ARS-CC9783 | postweaned calves | B1 | 5730 | R | Col(MG828), Col440I, Col8282, ColRNAI, IncFII, IncN |
| ARS-CC9784 | dry cows | D | 4624 | S | Col156, ColRNAI, IncFIA, IncFIB(AP001918), IncFIC(FII), IncI2_Delta |
| ARS-CC9785 | preweaned calves | A | 361 | S | |
| ARS-CC9786 | lactating cattle | A | 10 | S | ColRNAI, IncFIB(pB171)_pB171 |
| ARS-CC9788 | postweaned calves | B1 | 711 | S | ColRNAI, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC9789 | dry cows | D | 38 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9790 | preweaned calves | B1 | 29 | S | ColRNAI, IncFIB(AP001918) |
| ARS-CC9791 | lactating cattle | B1 | 1246 | S | ColRNAI, IncFIA, IncI1_Alpha |
| ARS-CC9792 | preweaned calves | B1 | 1308 | R | IncFIB(pB171)_pB171 |
| ARS-CC9793 | dry cows | B1 | 300 | S | Col156, IncFIB(AP001918) |
| ARS-CC9794 | postweaned calves | B1 | 392 | S | IncFIA(HI1)_HI1, IncFIB(AP001918), IncFIC(FII), IncI1_Alpha |
| ARS-CC9795 | lactating cattle | B1 | 1246 | S | ColRNAI, IncFIA, IncI1_Alpha |
| ARS-CC9796 | lactating cattle | B1 | 9196 | R | IncFIB(AP001918), pENTAS02 |
| ARS-CC9798 | lactating cattle | A | 398 | R | IncFIA, IncFII |
| ARS-CC9799 | postweaned calves | D | 3509 | S | ColpVC, IncA/C2, IncFII(pCoo)_pCoo, IncHI2A, IncHI2, RepA_pKPC-CAV1321 |
| ARS-CC9800 | dry cows | B1 | 155 | S | ColRNAI |
| ARS-CC9801 | preweaned calves | B1 | 22 | S | |
| ARS-CC9802 | lactating cattle | B1 | 101 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9803 | preweaned calves | E | 7244 | S | IncFIB(AP001918) |
| ARS-CC9804 | postweaned calves | E | 118 | S | ColRNAI, IncFIB(AP001918) |
| ARS-CC9805 | lactating cattle | B1 | 5973 | S | ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC9806 | postweaned calves | B1 | 2521 | S | IncFIA(HI1)_HI1, IncFIB(pB171)_pB171 |
| ARS-CC9807 | lactating cattle | B1 | 187 | S | IncFIA, IncFIB(AP001918), IncY |
| ARS-CC9808 | dry cows | A | 10 | S | Col440I |
| ARS-CC9809 | dry cows | B1 | 58 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9810 | postweaned calves | A | 329 | S | ColRNAI, IncFIB(AP001918), IncI2_Delta |
| ARS-CC9811 | lactating cattle | A | 206 | R | |
| ARS-CC9812 | postweaned calves | B1 | 7812 | S | ColRNAI, IncFIB(AP001918), IncY |
| ARS-CC9813 | dry cows | B1 | 8860 | S | IncFIA, IncFIB(AP001918), IncFIC(FII) |
| ARS-CC9830 | dry cows | E | 4151 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9832 | lactating cattle | B1 | 603 | S | Col156, Col440I, ColRNAI, IncFIB(AP001918), IncFIC(FII), IncI2_Delta |
| ARS-CC9834 | lactating cattle | B1 | 297 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9835 | lactating cattle | B1 | 58 | S | ColRNAI, IncFIA, IncFIB(AP001918), IncFIC(FII), IncI_Gamma |
| ARS-CC9837 | lactating cattle | B1 | 58 | S | Col440I, IncY |
| ARS-CC9840 | lactating cattle | B1 | 4481 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9842 | dry cows | D | 8651 | S | IncY |
| ARS-CC9843 | lactating cattle | B1 | 336 | S | IncFIA, IncFIB(AP001918) |
| ARS-CC9861 | preweaned calves | B1 | 2539 | S | IncFIA, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC9862 | preweaned calves | A | 216 | S | Col440II, Col440I, IncFIA(HI1)_HI1, IncFIB(K)_Kpn3, IncHI1A, IncHI1B(R27)_R27 |
| ARS-CC9022 | preweaned calves | D | 362 | MDR | Col156, ColRNAI, IncA/C2, IncFIA, IncFIB(AP001918) |
| ARS-CC9023 | preweaned calves | A | 10 | MDR | IncFIB(AP001918), IncI1_Alpha |
| ARS-CC9024 | preweaned calves | B1 | 101 | MDR | IncA/C2, IncFIA(HI1)_HI1, IncFIB(pB171)_pB171 |
| ARS-CC9025 | preweaned calves | C | 88 | MDR | ColRNAI, IncFIA, IncFIB(AP001918) |
| ARS-CC9026 | preweaned calves | E | 219 | R | IncY |
| ARS-CC9027 | preweaned calves | A | 1703 | R | IncFII, IncQ1 |
| ARS-CC9028 | lactating cattle | B1 | 515 | MDR | IncFIB(AP001918), IncI1_Alpha |
| ARS-CC9029 | preweaned calves | C | 1083 | MDR | IncFIB(AP001918), IncI2_Delta |
| ARS-CC9032 | preweaned calves | D | 973 | MDR | IncB/O/K/Z, IncFIA, IncFIB(AP001918) |
| ARS-CC9033 | lactating cattle | C | 88 | MDR | ColRNAI, IncFII, IncI1_Alpha |
| ARS-CC9034 | lactating cattle | B1 | 101 | R | IncFIB(AP001918), IncFIC(FII), IncX1, IncX3 |
| ARS-CC9036 | postweaned calves | D | 973 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC9038 | postweaned calves | B1 | 56 | MDR | IncFIB(AP001918), IncHI2A, IncHI2, RepA_pKPC-CAV1321 |
| ARS-CC9039 | preweaned calves | A | 10 | MDR | IncA/C2, IncFIB(AP001918), IncI2_Delta, IncX1 |
| ARS-CC9040 | postweaned calves | B1 | 6345 | MDR | IncY |
| ARS-CC9041 | preweaned calves | D | 106 | MDR | IncFIA, IncFIB(AP001918), IncI1_Alpha, IncN |
| ARS-CC9042 | lactating cattle | B1 | 1252 | MDR | IncFIB(AP001918), IncFIC(FII), IncI1_Alpha |
| ARS-CC9043 | preweaned calves | B1 | 602 | MDR | IncFIB(AP001918), IncY |
| ARS-CC9044 | postweaned calves | G | 117 | MDR | Col(MG828), Col440I, ColRNAI, IncFIA, IncFIB(AP001918), IncFII |
| ARS-CC9068 | preweaned calves | G | 117 | MDR | Col8282, ColRNAI, IncFIA, IncFIB(AP001918), IncFII |
| ARS-CC9046 | preweaned calves | A | 10 | MDR | IncFIA, IncFIB(AP001918), IncI1_Alpha |
| ARS-CC9049 | postweaned calves | D | 714 | R | IncFIB(AP001918), IncFII, IncI2_Delta |
| ARS-CC9050 | preweaned calves | F | 967 | MDR | IncFIA, IncFIB(AP001918), IncI1_Alpha, IncI2_Delta |
| ARS-CC9051 | dry cows | D | 106 | R | ColRNAI, IncI1_Alpha |
| ARS-CC9052 | lactating cattle | D | 973 | MDR | IncFIA, IncFIB(AP001918), IncHI2A, IncHI2, IncI1_Alpha, RepA_pKPC-CAV1321 |
| ARS-CC9053 | lactating cattle | C | 88 | MDR | IncFIA, IncFIB(AP001918) |
| ARS-CC9055 | postweaned calves | A | 48 | MDR | IncFIB(AP001918), IncFIC(FII), IncI1_Alpha, IncR, IncY |
| ARS-CC9056 | dry cows | A | 167 | MDR | Col(MG828), Col156, ColRNAI, IncA/C2, IncFIA, IncFIB(AP001918), IncY |
| ARS-CC9057 | postweaned calves | A | 167 | MDR | Col(MG828), Col156, ColRNAI, IncA/C2, IncFIA, IncFIB(AP001918), IncY |
| ARS-CC9058 | lactating cattle | D | 9199 | MDR | IncA/C2 |
| ARS-CC9059 | postweaned calves | B1 | 17 | MDR | Col(MG828), IncA/C2, IncFIB(AP001918) |
| ARS-CC9060 | preweaned calves | D | 32 | MDR | IncA/C2, IncB/O/K/Z, IncFIB(AP001918) |
| ARS-CC9061 | dry cows | F | 1280 | MDR | ColRNAI, IncA/C2 |
| ARS-CC9062 | preweaned calves | A | 34 | MDR | Col(MG828), IncFIA, IncFIB(AP001918) |
| ARS-CC9063 | lactating cattle | C | 88 | MDR | IncFIA, IncFIB(AP001918), IncX4 |
| ARS-CC9064 | preweaned calves | B1 | 940 | MDR | ColE10, ColRNAI, IncFII |
| ARS-CC9065 | lactating cattle | B1 | 940 | MDR | ColE10, ColRNAI, IncFII |
| Operon | Function | Carried by MDR Strains (%) | Carried by Susceptible Strains (%) | q-Values |
|---|---|---|---|---|
| iucABCD-iutA | Aerobactin synthesis/receptor | 48% | 6% | 1.61 × 10−9 |
| sitACD | Iron ABC transporter | 28 to 42% | 8% | 1.71 × 10−6 |
| papABCDEGHIJK | P fimbriae | 30% to 53% | 11% | 7.99 × 10−5 |
| fecABCDE | Ferric citrate transport system | 60 to 62% | 24% | 0.00016 |
| iolABCDEG-iatA | myo-inositol transport and utilization | 22% | 3% | 0.0023 |
| alsABC | D-allose transport system | 49% | 21% | 0.0087 |
| ulaABC | Ascorbate transport | 50 to 56% | 26% | 0.013 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haley, B.J.; Kim, S.W.; Salaheen, S.; Hovingh, E.; Van Kessel, J.A.S. Genome-Wide Analysis of Escherichia coli Isolated from Dairy Animals Identifies Virulence Factors and Genes Enriched in Multidrug-Resistant Strains. Antibiotics 2023, 12, 1559. https://doi.org/10.3390/antibiotics12101559
Haley BJ, Kim SW, Salaheen S, Hovingh E, Van Kessel JAS. Genome-Wide Analysis of Escherichia coli Isolated from Dairy Animals Identifies Virulence Factors and Genes Enriched in Multidrug-Resistant Strains. Antibiotics. 2023; 12(10):1559. https://doi.org/10.3390/antibiotics12101559
Chicago/Turabian StyleHaley, Bradd J., Seon Woo Kim, Serajus Salaheen, Ernest Hovingh, and Jo Ann S. Van Kessel. 2023. "Genome-Wide Analysis of Escherichia coli Isolated from Dairy Animals Identifies Virulence Factors and Genes Enriched in Multidrug-Resistant Strains" Antibiotics 12, no. 10: 1559. https://doi.org/10.3390/antibiotics12101559
APA StyleHaley, B. J., Kim, S. W., Salaheen, S., Hovingh, E., & Van Kessel, J. A. S. (2023). Genome-Wide Analysis of Escherichia coli Isolated from Dairy Animals Identifies Virulence Factors and Genes Enriched in Multidrug-Resistant Strains. Antibiotics, 12(10), 1559. https://doi.org/10.3390/antibiotics12101559






