Abstract
Here, we describe the isolation of a strain of the genus Pantoea encoding a VIM carbapenemase, the first to our knowledge. The strain, isolated from a rectal swab of a 10-day-old newborn admitted to a neonatal intensive care unit (NICU), was identified through whole-genome sequencing analyses as Pantoea brenneri. The strain harbored the carbapenemases gene blaVIM-1. The prompt application of contact measures and the isolation of the newborn prevented the dissemination of VIM-producing P. brenneri and of the plasmid carrying the VIM-1 gene to other newborns.
1. Introduction
Pantoea spp. is a well-known plant pathogen [1] belonging to the Enterobacteriaceae family. A taxonomical revision in the Pantoea agglomerans complex amended the description of the genus Pantoea and described Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov., Pantoea conspicua sp. nov., and Pantoea cypripedii comb. nov. [2]. Pantoea spp. rarely causes opportunistic infections [3], occurring mainly due to the contamination of wounds, or through hospital-acquired infection in individuals with a weak immune system such as newborns [4,5]. In preterm newborns, contamination of the parenteral nutrition with Pantoea spp. has been reported as a cause of bacteremia [6].
Carbapenem-resistant Enterobacteriaceae are increasingly common worldwide with growing numbers of people being infected by these organisms associated with significant morbidity and mortality. Knowledge of carbapenemase type is crucial for antibiotic treatment because not all β-lactamase inhibitor combinations are active against all carbapenemase types. Indeed, metallo-β-lactamases such as Verona integron metallo-β-lactamase (VIM) are not inhibited by beta-lactamase inhibitors such as clavulanic acid, tazobactam, avibactam, and vaborbactam, leaving few options for therapy. VIM-producing Enterobacterales are reported globally, and in Europe especially in the Mediterranean area [7,8,9]. Nevertheless, no Pantoea spp. bearing carbapenemase genes have been reported until now, while extended-spectrum beta-lactamase (ESBL)-producing strains bearing blaCTX-M and blaTEM-1 genes have been reported [10,11,12].
In this case report, we report the first isolation of Pantoea brenneri carbapenem-resistant bearing VIM carbapenemase. The strain was isolated from a newborn who was colonized.
2. Case Description
A pre-term male baby (34 weeks) born on 1 March 2022 by cesarean section from a diabetic mother was admitted to the Neonatal Intensive Care Unit (NICU) of tertiary hospital Fondazione IRCCS Policlinico San Matteo in Pavia (Italy) due to respiratory distress. The newborn underwent oxygen therapy with nasal continuous positive airway pressure (nCPAP) because a congenital pneumonia was suspected. Ampicillin and gentamycin were started as empirical treatment at admission as per internal procedures. Ceftazidime was also started and blood cultures collected due to the critical clinical condition of the patient and the suspicion of early-onset sepsis [13]. Total parenteral nutrition was administered by the central venous route until day 3, when enteral feeding through an orogastric tube was started. Ceftazidime was stopped after 72 h according to the American Academy of Pediatrics [14] while the administration of ampicillin and gentamycin was continued. Surveillance rectal swabs for the screening of ESBL-producing Enterobacterales collected on days 3 and 7 were negative.
On day 10, Pantoea spp. (6775PV) was cultured from the surveillance rectal swab. The patient was spatially isolated and contact isolation precautions were taken including dedicated materials such as gauzes, band-aids, syringes, and disinfectants, dedicated nursing personnel 24/7, and a more accurate environmental cleaning performed more frequently with sodium hypochlorite 1000 ppm with particular attention for surfaces that could be touched by multiple people such as door handles and electromedical equipment. Disposable materials were used when possible, and disinfecting bottles or wipes were placed near ready-to-use equipment such as the imaging ultrasound system. Hand hygiene practices were implemented for all of the personnel of the NICU, and ESBL surveillance for all of the other newborns of the NICU continued with rectal swabs performed at admission and weekly thereafter, as defined in the hospital operating procedures for multidrug resistance (MDR) bacteria surveillance. On day 12, ampicillin and gentamycin were stopped as well. Pantoea spp. was also grown from the rectal swab collected on day 14. On day 19, the newborn male was discharged in generally good health with normal cardio-respiratory parameters.
3. Microbiological Investigations
Surveillance rectal swabs were cultured on chromID ESBL Agar (BioMerieux, Marcy-l’Etoile, France). Flat blue-green colonies grew after incubation at 37 °C for 18 h (Figure 1). Species identification was performed through MALDI-TOF mass spectrometry (Bruker Daltonics GmbH, Bremen, Germany) equipped with Bruker Biotyper 3.1 database. The isolate 6775PV was identified as Pantoea agglomerans (score 1.828), but scores between 1.7 and 1.99 indicate “probable genus identification”, thus the strain was reported as Pantoea spp.
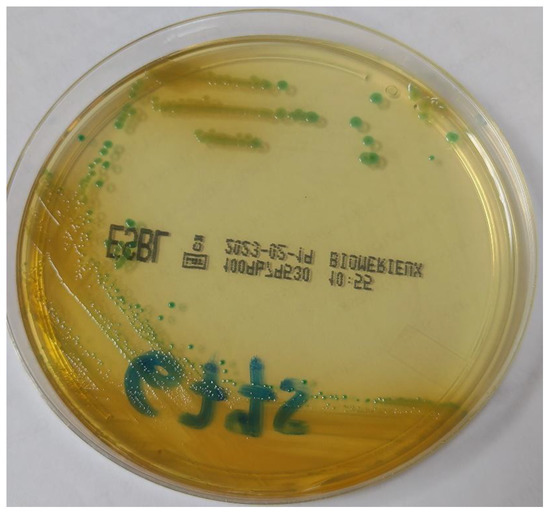
Figure 1.
Morphology of the colonies of Pantoea brenneri on chromID ESBL agar (BioMerieux, Marcy-l’Etoile, France).
The susceptibility profile determined using the NMIC-402 panel of the Phoenix M50 BD automated system (Becton Dickinson, Franklin Lakes, NJ, USA) and Sensititre DKMGN (ThermoFisher Scientific, Rodano, Italy) showed resistance to cephalosporins and carbapenems (Table 1). Fosfomycin was tested via fosfomycin agar dilution (Liofilchem, Roseto degli Abruzzi, Italy). The presence of VIM carbapenemase was assessed using NG test CARBA 5 (NG Biotech, Guipry, France) and confirmed with the Cepheid Xpert® Carba-R assay (Cepheid, Sunnyvale, CA, USA).

Table 1.
Antibiotic susceptibility profiles of the 6775PV isolate. Interpretation was performed according to EUCAST breakpoints (version 12, 2022) for Enterobacterales: S was used for susceptible, I for susceptible increased exposure and R for resistant. Breakpoints are displayed under each antibiotic name.
Genomic DNA of the isolate was extracted with a blood and tissue kit (Qiagen, Düsseldorf, Germany) following the manufacturer’s instructions for short-read sequencing. Short reads were obtained by an Illumina MiSeq platform with a 2 × 150 paired-end run, after a Nextera XT library preparation step (Illumina Inc., San Diego, CA, USA).
Genomes were assembled using Shovill 1.1. The genome of strain 6775PV is 4,756,675 bp in length with Ncontig (>200 bp) = 79 and N50 = 148,391. The software PGAP identified the genome as Pantoea brenneri, a species that is not present in the Bruker Biotyper 3.1 database. Thus, a phylogeny was performed using the software Jolytree on the genome of our isolate and all high-quality genomes of both P. agglomerans and P. brenneri, retrieved from the PATRIC database (https://www.patricbrc.org/, accessed on 22 June 2022). The resulting tree (Figure 2) indicates strain 6775PV clusters with the genomes of Pantoea brenneri. The closest genome to 6775PV in the phylogeny was isolated from a surface in the International Space Station in 2015 [15].
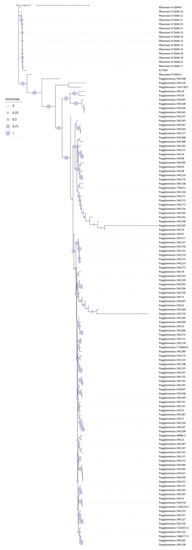
Figure 2.
Phylogeny of the genome of isolate 6775PV and all high-quality genomes of the P. agglomerans and P. brenneri species. The Phylogeny was obtained using the software JolyTree. Bootstrap values are represented with colored dots of variable size on the tree branches.
Resistance genes were searched using ResFinder 4.1 [16] with a coverage threshold of 90% and identity threshold of 100%, and plasmids were searched with PlasmidFinder 2.1 [17] with a coverage threshold of 100% and an identity threshold of 95%. The genome 6775PV harbored the resistance genes blaVIM-1, aa’(6’)-Il, and qnrS1 encoding, respectively, for resistance to carbapenems, aminoglycosides and fluoroquinolones, and IncN2 plasmid replicon type.
4. Discussion
Here, we describe the first isolation of a strain of the genus Pantoea bearing VIM-1 carbapenemase isolated from a rectal swab of a 10-day-old newborn The isolation of a strain of Pantoea spp. resistant to carbapenems, one of the last options to treat multidrug-resistant Gram-negative organisms, is particularly worrisome. Indeed, plasmids carrying blaVIM usually also carry other genes conferring resistance to antibiotics, including aminoglycosides, macrolides, and sulfamethoxazole, making these strains MDR, further limiting treatment options.
The colonization by Pantoea spp. as well as by other Enterobacterales carrying VIM carbapenemases, even if asymptomatic, can be considered a risk factor to develop infection. Hence, the role of surveillance in infection control programs is crucial since contact isolation measures reduce the chances of patient-to-patient transmissions, and patient outcomes improve due to the earlier availability of the susceptibility profile of the pathogens, resulting in prompt administration of appropriate antimicrobial therapy.
Bacteria belonging to the genus Pantoea spp. are considered the cause of opportunistic human infections, mostly wound infections but also bacteremia. Indeed, the only report of infections caused by wild-type P. brenneri was a nationwide sepsis outbreak in the USA in 1971 [2]. Bacteremia caused by Pantoea spp. has been described in adults in Japan [18] and the USA [19]. In newborns, bacteremia caused by wild-type Pantoea spp. has been previously described in Brazil and India [20,21]. Sepsis and wound infections caused by carbapenem-resistant P. agglomerans in newborns were reported in Turkey and in Yemen [10,22], but no indication of the class of resistance mechanism is given in either study.
The only case of Pantoea infection reported in Europe previously regarding ESBL-producing Pantoea agglomerans was described from clinical specimens of pediatric patients in a Bulgarian hospital [8], and recently, in the same NICU as this study, where ESBL-producing Pantoea calida colistin-susceptible but bearing the mcr-9 gene for colistin resistance was isolated from a newborn [23]. In Italy, wild-type strains of Pantoea spp. were reported from blood cultures of patients admitted to oncology and ICU [24,25] and from a contaminated port-a-cath [26]; but in both studies, the isolates were wild type.
Although Pantoea spp. is not considered a pathogen, horizontal transfer of resistance genes mediated by plasmids and other mobile elements occurs especially in healthcare settings where antimicrobial use increases selection pressure on bacterial populations. In this study, strain 6775PV harbored a plasmid that belongs to the IncN incompatibility group, one of the most common mobile genetic platforms for the spreading of resistance genes among Enterobacteriaceae. A multispecies cluster of Enterobacterales carrying VIM-1 carbapenemase was isolated in 2019 from rectal swabs and blood cultures of patients of a teaching hospital in Rome (Italy) [27]. However, this cluster is characterized by IncA plasmid.
The emergence and dissemination of carbapenem-resistance mechanisms represent a global public health concern, resulting in infections associated with poor outcomes, high mortality rates, and limited treatment options. The emergence of metallo-β-lactamase-producing isolates requires prompt surveillance especially of colonization by carbapenem-resistant isolates belonging also to species not usually considered pathogenic, which can act as reservoirs and can promote inter- and intra-care setting dissemination.
In this case, the strict surveillance and the prompt application of infection control measures have prevented the spreading of VIM-1-producing P. brenneri and the dissemination among other bacterial species of the blaVIM-1 carbapenemase gene. Infections caused by VIM-producing Enterobacterales are worrying especially for critical patients like newborns in NICU. In the future, epidemiologic studies should be performed to investigate the diffusion of carbapenem resistance among species not usually considered in surveillance, especially in critical patients, and bundled infection control measures, education, and training should be implemented to limit and control the spreading of carbapenem resistance.
Author Contributions
Conceptualization: C.M. and M.C.; formal analysis: S.G. (Stefano Gaiarsa); investigation: I.M., C.A. and S.G. (Stefano Ghirardello); supervision: F.B. and P.C.; writing—original draft: I.M., C.M., S.G. (Stefano Ghirardello) and M.C.; writing—review & editing: F.B. and P.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was designed and conducted in accordance with the Helsinki Declaration and approved by the Ethics Committee of Policlinico San Matteo in Pavia (internal project code: 08022213).
Informed Consent Statement
Informed consent was obtained for all subjects involved in the study.
Data Availability Statement
Genome assembly data are available at NCBI under BioProject ID PRJNA842925 (BioSample accession: SAMN28693404).
Acknowledgments
The authors would like to thank Edward Christopher Davis for the grammatical review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cruz, A.T.; Cazacu, A.C.; Allen, C.H. Pantoea agglomerans, a Plant Pathogen Causing Human Disease. J. Clin. Microbiol. 2007, 45, 1989–1992. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Cleenwerck, I.; Venter, S.; Engelbeen, K.; De Vos, P.; Coutinho, T.A. Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60 (Pt 10), 2430–2440. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Mackiewicz, B.; Kinga Lemieszek, M.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. Deleterious effects: Infections of humans, animals and plants. Ann. Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Aly, N.Y.A.; Salmeen, H.N.; Lila, R.A.A.; Nagaraja, P.A. Pantoea agglomerans Bloodstream Infection in Preterm Neonates. Med. Princ. Pract. 2008, 17, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Nair, J. Pantoea Infections in the Neonatal Intensive Care Unit. Cureus 2021, 13, e13103. [Google Scholar] [CrossRef] [PubMed]
- Habsah, H.; Zeehaida, M.; van Rostenberghe, H.; Noraida, R.; Pauzi, W.I.W.; Fatimah, I.; Rosliza, A.R.; Sharimah, N.Y.N.; Maimunah, H. An outbreak of Pantoea spp. in a neonatal intensive care unit secondary to contaminated parenteral nutrition. J. Hosp. Infect. 2005, 61, 213–218. [Google Scholar] [CrossRef]
- Curiao, T.; Morosini, M.I.; Ruiz-Garbajosa, P.; Robustillo, A.; Baquero, F.; Coque, T.M.; Cantón, R. Emergence of blaKPC-3-Tn4401a associated with a pKPN3/4-like plasmid within ST384 and ST388 Klebsiella pneumoniae clones in Spain. J. Antimicrob. Chemother. 2010, 65, 1608–1614. [Google Scholar] [CrossRef]
- Cantón, R.; Akóva, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: An Evolving Crisis of Global Dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef]
- Büyükcam, A.; Tuncer, Ö.; Gür, D.; Sancak, B.; Ceyhan, M.; Cengiz, A.B.; Kara, A. Clinical and microbiological characteristics of Pantoea agglomerans infection in children. J. Infect. Public Health 2018, 11, 304–309. [Google Scholar] [CrossRef]
- Markovska, R.D.; Stoeva, T.J.; Bojkova, K.D.; Mitov, I.G. Epidemiology and Molecular Characterization of Extended-Spectrum Beta-Lactamase-Producing Enterobacter spp., Pantoea agglomerans, and Serratia marcescens Isolates from a Bulgarian Hospital. Microb. Drug Resist. 2014, 20, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Selmi, R.; Tayh, G.; Srairi, S.; Mamlouk, A.; Ben Chehida, F.; Lahmar, S.; Bouslama, M.; Daaloul-Jedidi, M.; Messadi, L. Prevalence, risk factors and emergence of extended-spectrum β-lactamase producing-, carbapenem- and colistin-resistant Enterobacterales isolated from wild boar (Sus scrofa) in Tunisia. Microb. Pathog. 2022, 163, 105385. [Google Scholar] [CrossRef] [PubMed]
- Puopolo, K.M.; Benitz, W.E.; Zaoutis, T.E.; Committee on Fetus and Newborn; Committee on Infectious Diseases. Management of Neonates Born at ≤34 6/7 Weeks’ Gestation With Suspected or Proven Early-Onset Bacterial Sepsis. Pediatrics 2018, 142, e20182896. [Google Scholar] [CrossRef] [PubMed]
- Puopolo, K.M. American Academy of Pediatrics (AAP) Clinical Report. New Sepsis Guidance Addresses Epidemiology, Microbiology, Recommended Empiric Treatment. Available online: https://publications.aap.org/aapnews/news/12310/New-sepsis-guidance-addresses-epidemiology (accessed on 19 November 2018).
- Singh, N.K.; Wood, J.M.; Karouia, F.; Venkateswaran, K. Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome 2018, 6, 204. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Asai, N.; Koizumi, Y.; Yamada, A.; Sakanashi, D.; Watanabe, H.; Kato, H.; Shiota, A.; Hagihara, M.; Suematsu, H.; Yamagishi, Y.; et al. Pantoea dispersa bacteremia in an immunocompetent patient: A case report and review of the literature. J. Med Case Rep. 2019, 13, 33. [Google Scholar] [CrossRef]
- Yablon, B.R.; Dantes, R.; Tsai, V.; Lim, R.; Moulton-Meissner, H.; Arduino, M.; Jensen, B.; Patel, M.T.; Vernon, M.O.; Grant-Greene, Y.; et al. Outbreak of Pantoea agglomerans Bloodstream Infections at an Oncology Clinic—Illinois, 2012–2013. Infect. Control Hosp. Epidemiol. 2017, 38, 314–319. [Google Scholar] [CrossRef]
- Bicudo, E.L.; Macedo, V.O.; Carrara, M.A.; Castro, F.F.; Rage, R.I. Nosocomial outbreak of Pantoea agglomerans in a pediatric urgent care center. Braz. J. Infect. Dis. 2007, 11, 281–284. [Google Scholar] [CrossRef]
- Tiwari, S.; Beriha, S.S. Pantoea species causing early onset neonatal sepsis: A case report. J. Med. Case Rep. 2015, 9, 188. [Google Scholar] [CrossRef]
- Salah, A.; Al-Subol, I.; Hudna, A.; Alhaj, A.; Alqubaty, A.R.; Farie, W.; Sulieman, D.; Alnadhari, O.; Alwajeeh, T.; Alobathani, F.; et al. Neonatal sepsis in Sana’a city, Yemen: A predominance of Burkholderia cepacia. BMC Infect. Dis. 2021, 21, 1108. [Google Scholar] [CrossRef] [PubMed]
- Gaiarsa, S.; Merla, C.; Corbella, M.; Mariani, B.; Zatelli, M.; Sciabica, I.; Castelli, M.; Piazza, A.; Zecca, M.; Sassera, D.; et al. Isolation of a Colistin-Susceptible MDR Pantoea calida Harboring the mcr-9 Gene Suggests the Silent Spread of the Resistance Factor. Microb. Drug Resist. 2022, 28, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Liberto, M.C.; Matera, G.; Puccio, R.; Russo, T.L.; Colosimo, E.; Focà, E. Six cases of sepsis caused by Pantoea agglomerans in a teaching hospital. New Microbiol. 2009, 32, 119–123. [Google Scholar]
- Mirtella, D.; Fedeli, P.; Scendoni, R.; Cannovo, N.; Cingolani, M. A Case of Nosocomial Outbreak of Pantoea agglomerans Related to Parenteral Nutrition Procedures. Healthcare 2021, 9, 684. [Google Scholar] [CrossRef] [PubMed]
- Izzo, I.; Lania, D.; Castro, A.; Lanzini, F.; Bella, D.; Pagani, A.; Colombini, P. Sette casi di contaminazione di CVC a permanenza (tipo “Port-a-cath”) da Pantoea agglomerans in pazienti afferenti al Servizio di Oncologia del P.O. di Iseo, Brescia [Seven cases of port-a-cath contamination caused by Pantoea agglomerans in the Oncological Service of Iseo Hospital, Brescia (Italy)]. Infez. Med. 2014, 22, 152–155. (In Italian) [Google Scholar] [PubMed]
- Arcari, G.; Di Lella, F.M.; Bibbolino, G.; Mengoni, F.; Beccaccioli, M.; Antonelli, G.; Faino, L.; Carattoli, A. A Multispecies Cluster of VIM-1 Carbapenemase-Producing Enterobacterales Linked by a Novel, Highly Conjugative, and Broad-Host-Range IncA Plasmid Forebodes the Reemergence of VIM-1. Antimicrob. Agents Chemother. 2020, 64, e02435-19. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).