Differentiation of Escherichia fergusonii and Escherichia coli Isolated from Patients with Inflammatory Bowel Disease/Ischemic Colitis and Their Antimicrobial Susceptibility Patterns
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients with IBD/IC and Healthy Controls (HCs)
2.2. Isolation and Preservation of Strains from Disease-Associated Patients and HCs
2.3. Identification & Characterization of Strains
2.4. PCR Amplification of adk
2.5. Phylogenetic Analyses
2.6. Antimicrobial Susceptibility Testing
3. Results
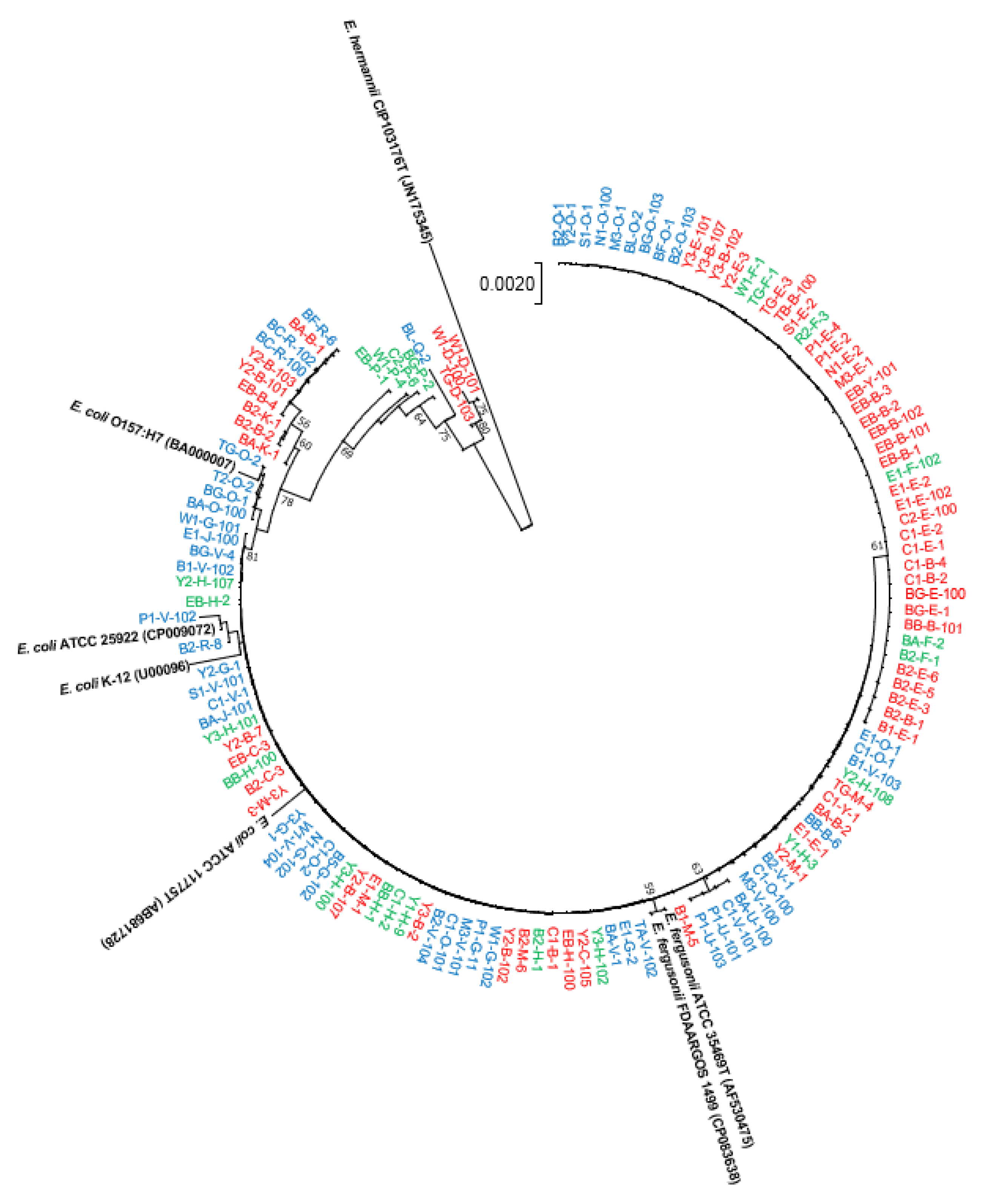
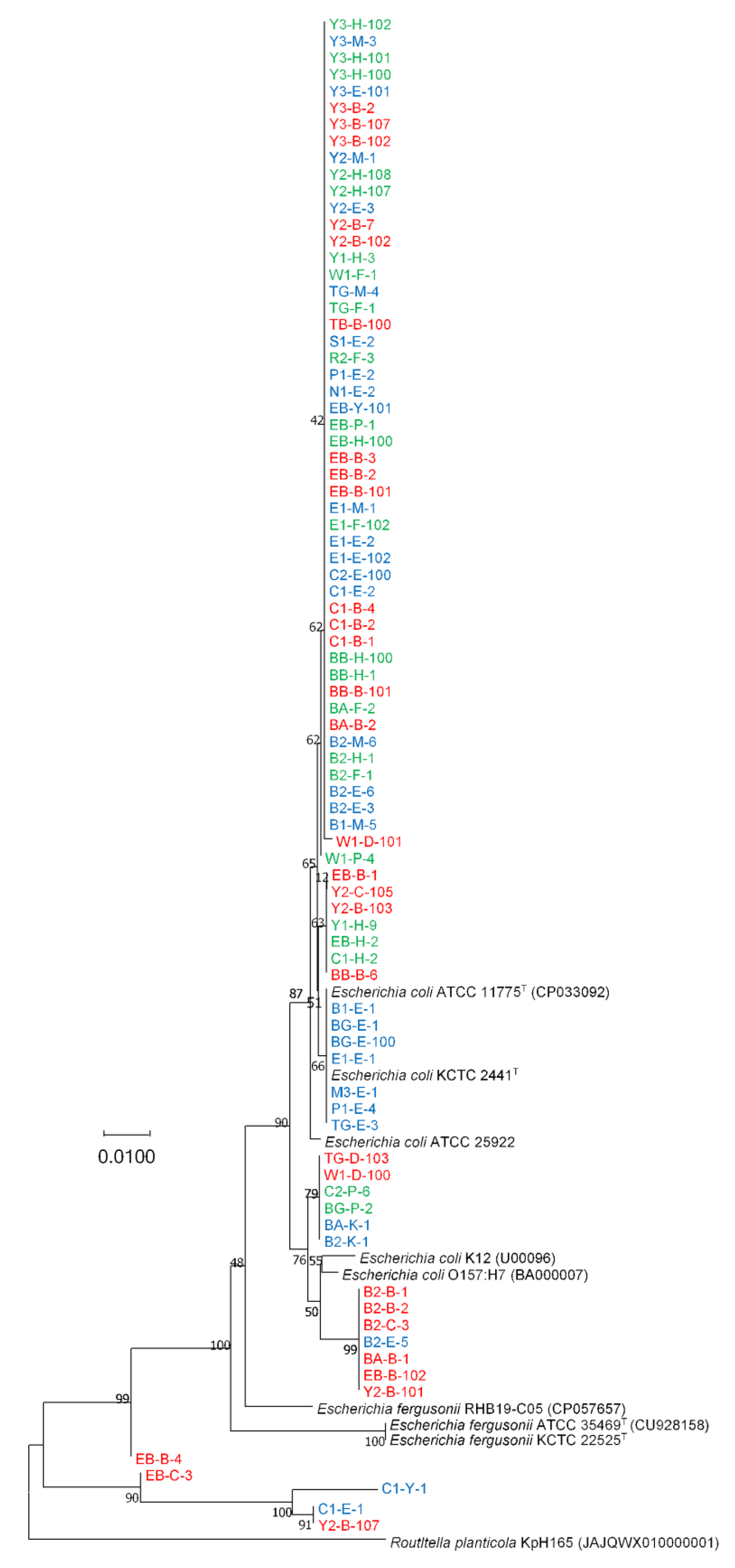
3.1. Phylogenetic Analyses
3.2. Differentiation of E. coli and E. fergusonii Based on adk Gene Sequences
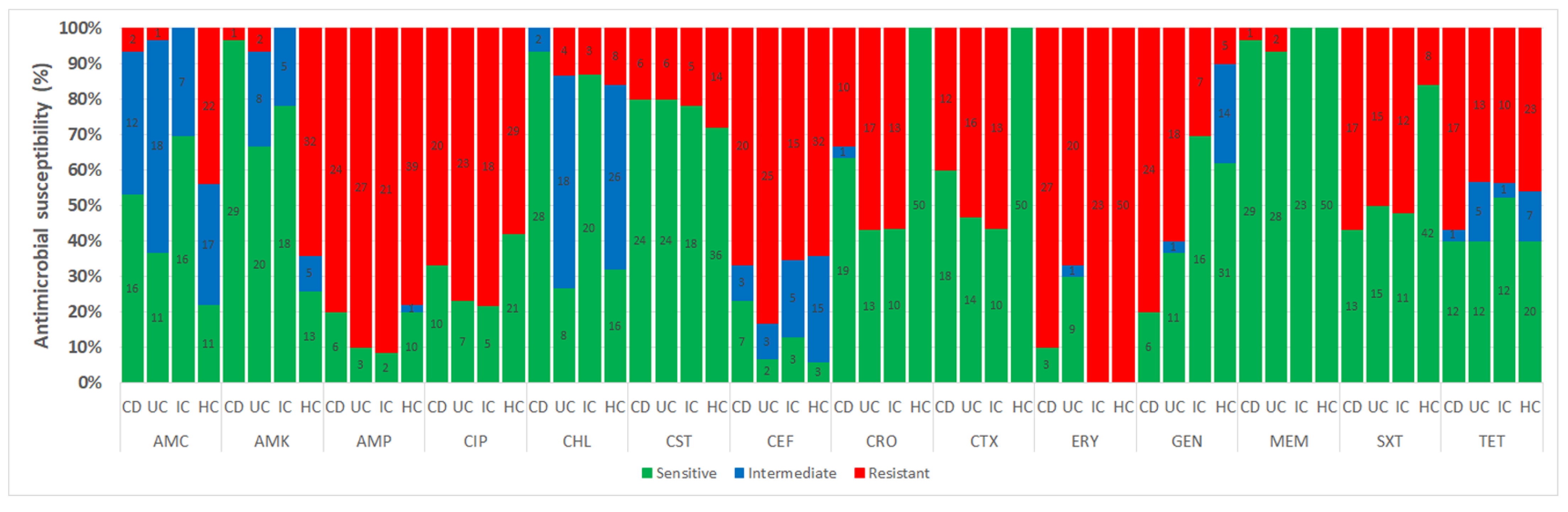
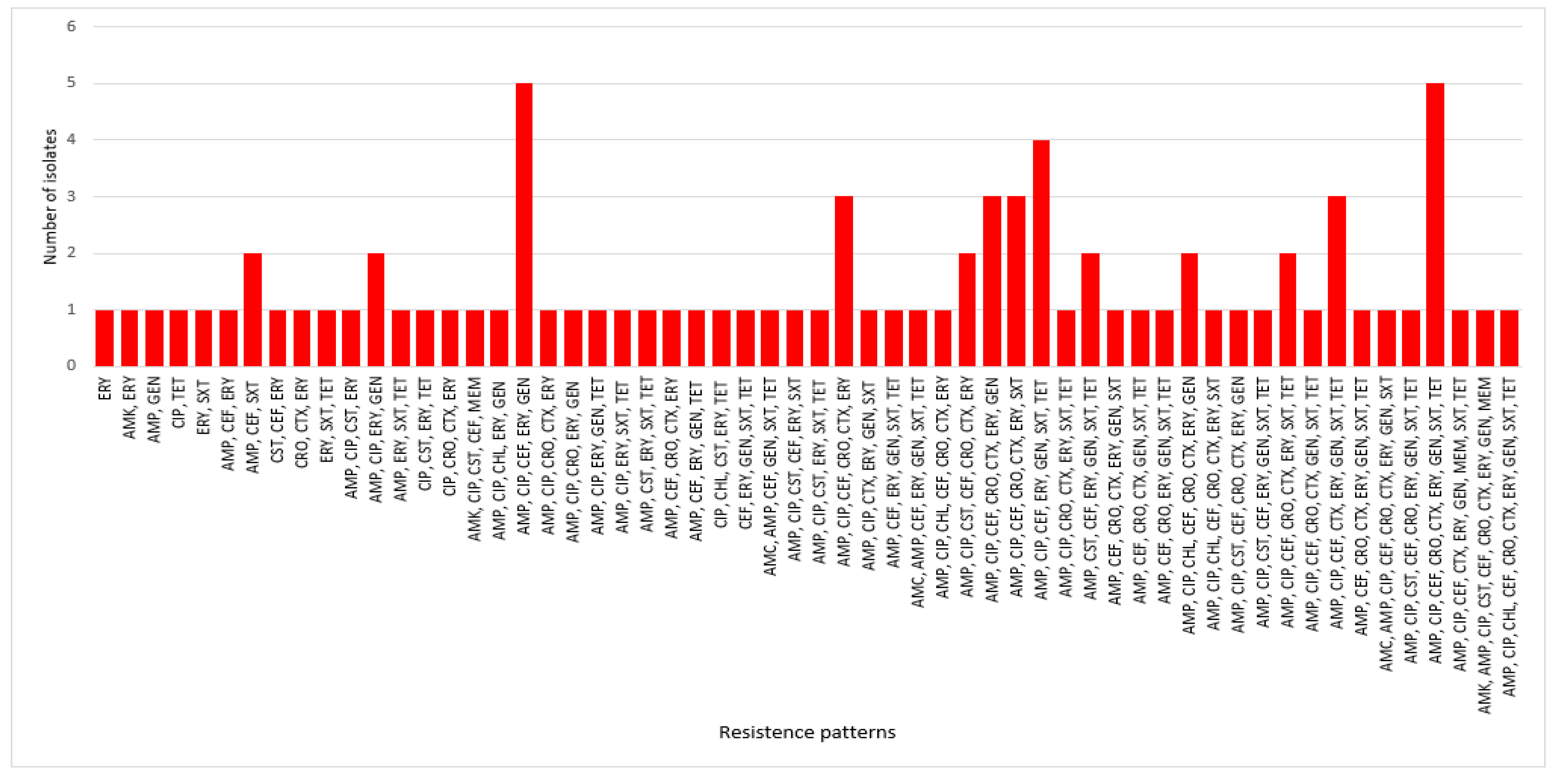
3.3. Antimicrobial Susceptibility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Maloy, K.J.; Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 2011, 474, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Xavier, R.J.; Podolsky, D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007, 448, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Li, Y.Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, D.C.; Carding, S.R. Inflammatory bowel disease: Cause and immunobiology. Lancet 2007, 369, 1627–1640. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.G.; Chang, E.B. The intestinal microbiota in the pathogenesis of inflammatory bowel diseases: New insights into complex disease. Clin. Sci. 2018, 132, 2013. [Google Scholar] [CrossRef]
- Mirsepasi-Lauridsen, H.C.; Vallance, B.A.; Krogfelt, K.A.; Petersen, A.M. Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 2019, 32, e00060-18. [Google Scholar] [CrossRef]
- Rashid, T.; Ebringer, A.; Wilson, C. The role of Klebsiella in Crohn’s disease with a potential for the use of antimicrobial measures. Int. J. Rheumatol. 2013, 2013, 610393. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef]
- Schultz, B.M.; Paduro, C.A.; Salazar, G.A.; Salazar-Echegarai, F.J.; Sebastián, V.P.; Riedel, C.A.; Kalergis, A.M.; Alvarez-Lobos, M.; Bueno, S.M. A Potential Role of Salmonella Infection in the Onset of Inflammatory Bowel Diseases. Front. Immunol. 2017, 8, 191. [Google Scholar] [CrossRef]
- Zhang, Q.; Su, X.; Zhang, C.; Chen, W.; Wang, Y.; Yang, X.; Liu, D.; Zhang, Y.; Yang, R. Klebsiella pneumoniae induces inflammatory bowel disease through caspase-11–mediated IL18 in the gut epithelial cells. Cell. Mol. Gastroenterol. Hepatol. 2022, in press. [Google Scholar] [CrossRef]
- Zhang, J.; Hoedt, E.C.; Liu, Q.; Berendsen, E.; Teh, J.J.; Hamilton, A.; O’ Brien, A.W.; Ching, J.Y.L.; Wei, H.; Yang, K.; et al. Elucidation of Proteus mirabilis as a Key Bacterium in Crohn’s Disease Inflammation. Gastroenterology 2021, 160, 317–330.e11. [Google Scholar] [CrossRef] [PubMed]
- Le Baut, G.; O’brien, C.; Pavli, P.; Roy, M.; Seksik, P.; Tréton, X.; Nancey, S.; Barnich, N.; Bezault, M.; Auzolle, C.; et al. Prevalence of Yersinia Species in the Ileum of Crohn’s Disease Patients and Controls. Front. Cell. Infect. Microbiol. 2018, 8, 336. [Google Scholar] [CrossRef] [PubMed]
- Kamali Dolatabadi, R.; Feizi, A.; Halaji, M.; Fazeli, H.; Adibi, P. The prevalence of adherent-invasive Escherichia coli and its association with inflammatory bowel diseases: A systematic review and meta-analysis. Front. Med. 2021, 8, 2221. [Google Scholar] [CrossRef]
- Dogan, B.; Scherl, E.; Bosworth, B.; Yantiss, R.; Altier, C.; McDonough, P.L.; Jiang, Z.D.; DuPont, H.L.; Garneau, P.; Harel, J.; et al. Multidrug resistance is common in Escherichia coli associated with ileal Crohn’s disease. Inflamm. Bowel Dis. 2013, 19, 141–150. [Google Scholar] [CrossRef]
- FitzGerald, J.F.; Hernandez, L.O. Ischemic Colitis. Clin. Colon Rectal Surg. 2015, 28, 93. [Google Scholar] [PubMed]
- Nimmons, D.; Limdi, J.K. Elderly patients and inflammatory bowel disease. World J. Gastrointest. Pharmacol. Ther. 2016, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Griffin, P.M.; Olmstead, L.C.; Petras, R.E. Escherichia coli O157:H7-associated colitis: A clinical and histological study of 11 cases. Gastroenterology 1990, 99, 142–149. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Migula, W. Bactreiaceae (Stäbchenbacterien). In Die Natürlichen Pflanzenfamilien; Engler, A., Prantl, K., Teil, I., Abteilung Ia, W., Eds.; Engelmann: Leipzig, Germany, 1895; pp. 20–30. [Google Scholar]
- Castellani, A.; Chalmers, A.J. Manual of Tropical Medicine, 3rd ed.; Williams Wood and Co.: New York, NY, USA, 1919. [Google Scholar]
- Jang, J.; Hur, H.G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications—A review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef]
- Yu, D.; Banting, G.; Neumann, N.F. A review of the taxonomy, genetics, and biology of the genus Escherichia and the type species Escherichia coli. Can. J. Microbiol. 2021, 67, 553–571. [Google Scholar] [CrossRef] [PubMed]
- Gaastra, W.; Kusters, J.G.; van Duijkeren, E.; Lipman, L.J.A. Escherichia fergusonii. Vet. Microbiol. 2014, 172, 7–12. [Google Scholar] [CrossRef]
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef]
- Farmer, J.J.; Fanning, G.R.; Davis, B.R.; O’Hara, C.M.; Riddle, C.; Hickman-Brenner, F.W.; Asbury, M.A.; Lowery, V.A.; Brenner, D.J. Escherichia fergusonii and Enterobacter taylorae, two new species of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 1985, 21, 77–81. [Google Scholar] [CrossRef]
- Maheux, A.F.; Boudreau, D.K.; Bergeron, M.G.; Rodriguez, M.J. Characterization of Escherichia fergusonii and Escherichia albertii isolated from water. J. Appl. Microbiol. 2014, 117, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Dahal, R.H.; Chaudhary, D.K. Microbial infections and antimicrobial resistance in Nepal: Current trends and recommendations. Open Microbiol. J. 2018, 12, 230–242. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Medina, M.; Garcia-Gil, L.J. Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity. World J. Gastrointest. Pathophysiol. 2014, 5, 213. [Google Scholar] [CrossRef]
- Tanquilut, C.D.; Jung, C.W.; Nelson, A.W.; Lau, S.K. Infection due to Shiga toxin-producing enterohemorrhagic Escherichia coli (EHEC) presenting as ischemic colitis. IDCases 2019, 18, e00629. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Örtqvist, A.K.; Cao, Y.; Simon, T.G.; Roelstraete, B.; Song, M.; Joshi, A.D.; Staller, K.; Chan, A.T.; Khalili, H.; et al. Antibiotic use and the development of inflammatory bowel disease: A national case-control study in Sweden. Lancet Gastroenterol. Hepatol. 2020, 5, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Tzelepi, E.; Giakkoupi, P.; Sofianou, D.; Loukova, V.; Kemeroglou, A.; Tsakris, A. Detection of extended-spectrum β-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J. Clin. Microbiol. 2000, 38, 542–546. [Google Scholar] [CrossRef]
- Pasteran, F.; Gonzalez, L.J.; Albornoz, E.; Bahr, G.; Vila, A.J.; Corso, A. Triton Hodge Test: Improved Protocol for Modified Hodge Test for Enhanced Detection of NDM and Other Carbapenemase Producers. J. Clin. Microbiol. 2016, 54, 640–649. [Google Scholar] [CrossRef]
- Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Kibret, M.; Abera, B. Antimicrobial susceptibility patterns of E. coli from clinical sources in northeast Ethiopia. Afr. Health Sci. 2011, 11 (Suppl. 1), S40–S45. [Google Scholar] [CrossRef]
- Scott, H.M.; Acuff, G.; Bergeron, G.; Bourassa, M.W.; Gill, J.; Graham, D.W.; Kahn, L.H.; Morley, P.S.; Salois, M.J.; Simjee, S.; et al. Critically important antibiotics: Criteria and approaches for measuring and reducing their use in food animal agriculture. Ann. N. Y. Acad. Sci. 2019, 1441, 8. [Google Scholar] [CrossRef]
- Skuja, V.; Pekarska, K.; Straume, Z.; Rudzīte, D.; Lavrinoviča, E.; Piekuse, L.; Dauvarte, H.; Vašuka, E.; Dobelniece, L.; Zalizko, P.; et al. P783 Ciprofloxacin resistance in ESBL producing Enterobacteriaceae colonizing the gut in IBD patients. J. Crohn’s Colitis 2017, 11, S481. [Google Scholar] [CrossRef]
- Park, S.B.; Park, Y.K.; Ha, M.W.; Thompson, K.D.; Jung, T.S. Antimicrobial resistance, pathogenic, and molecular characterization of Escherichia coli from diarrheal patients in South Korea. Pathogens 2022, 11, 385. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Park, J.H.; Han, D.S.; Lee, H.L.; Park, C.H.; Eun, C.S. The uncertain effect of antimicrobial therapy in the treatment of patients with ischemic colitis. J. Clin. Med. 2020, 9, 2182. [Google Scholar] [CrossRef] [PubMed]
- Skuja, V.; Derovs, A.; Pekarska, K.; Rudzite, D.; Lavrinovica, E.; Piekuse, L.; Kempa, I.; Straume, Z.; Eglite, J.; Lejnieks, A.; et al. Gut colonization with extended-spectrum β-lactamase-producing Enterobacteriaceae may increase disease activity in biologic-naive outpatients with ulcerative colitis: An interim analysis. Eur. J. Gastroenterol. Hepatol. 2018, 30, 92–100. [Google Scholar] [CrossRef]
- Meheissen, M.A.; Header, D.; Abdelaty, K. Phylogenetic and pathotype analysis of Escherichia coli stool isolates from Egyptian patients with inflammatory bowel disease. Germs 2019, 9, 172. [Google Scholar] [CrossRef]
- Rolhion, N.; Darfeuille-Michaud, A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm. Bowel Dis. 2007, 13, 1277–1283. [Google Scholar] [CrossRef]


| Loci | adk Gene Sequence Nucleotide | |
|---|---|---|
| E. coli | E. fergusonii | |
| 93 | T | G |
| 96 | C | T |
| 477 | T | C |
| 549 | C | A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dahal, R.H.; Choi, Y.-J.; Kim, S.; Kim, J. Differentiation of Escherichia fergusonii and Escherichia coli Isolated from Patients with Inflammatory Bowel Disease/Ischemic Colitis and Their Antimicrobial Susceptibility Patterns. Antibiotics 2023, 12, 154. https://doi.org/10.3390/antibiotics12010154
Dahal RH, Choi Y-J, Kim S, Kim J. Differentiation of Escherichia fergusonii and Escherichia coli Isolated from Patients with Inflammatory Bowel Disease/Ischemic Colitis and Their Antimicrobial Susceptibility Patterns. Antibiotics. 2023; 12(1):154. https://doi.org/10.3390/antibiotics12010154
Chicago/Turabian StyleDahal, Ram Hari, Yoon-Jung Choi, Shukho Kim, and Jungmin Kim. 2023. "Differentiation of Escherichia fergusonii and Escherichia coli Isolated from Patients with Inflammatory Bowel Disease/Ischemic Colitis and Their Antimicrobial Susceptibility Patterns" Antibiotics 12, no. 1: 154. https://doi.org/10.3390/antibiotics12010154
APA StyleDahal, R. H., Choi, Y.-J., Kim, S., & Kim, J. (2023). Differentiation of Escherichia fergusonii and Escherichia coli Isolated from Patients with Inflammatory Bowel Disease/Ischemic Colitis and Their Antimicrobial Susceptibility Patterns. Antibiotics, 12(1), 154. https://doi.org/10.3390/antibiotics12010154







