Systematic Review and Meta-Analysis on the Frequency of Antibiotic-Resistant Clostridium Species in Saudi Arabia
Abstract
1. Introduction
2. Results
2.1. Literature Search
2.2. Characteristics of the Study
2.3. Antibiotic Resistance Frequencies in C. difficile
2.4. Antibiotic Resistance Frequencies in C. perfringens
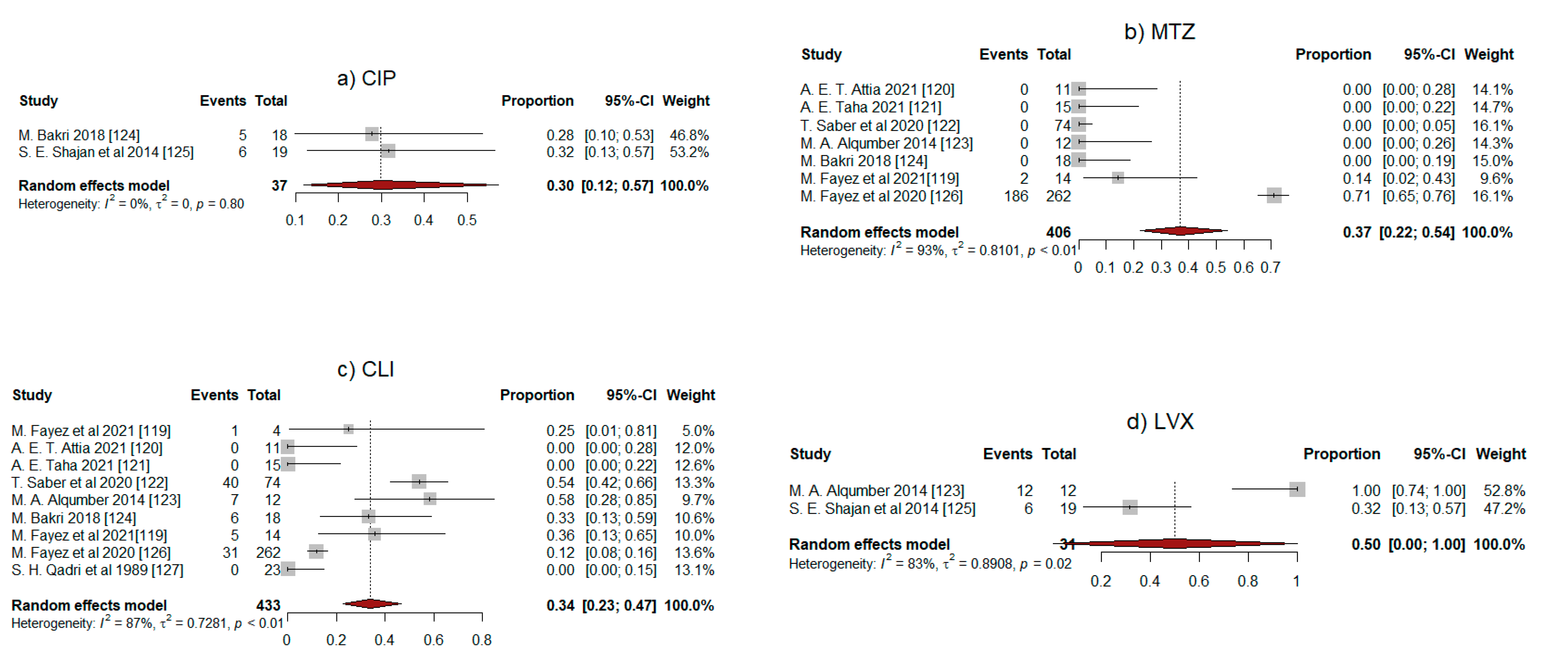
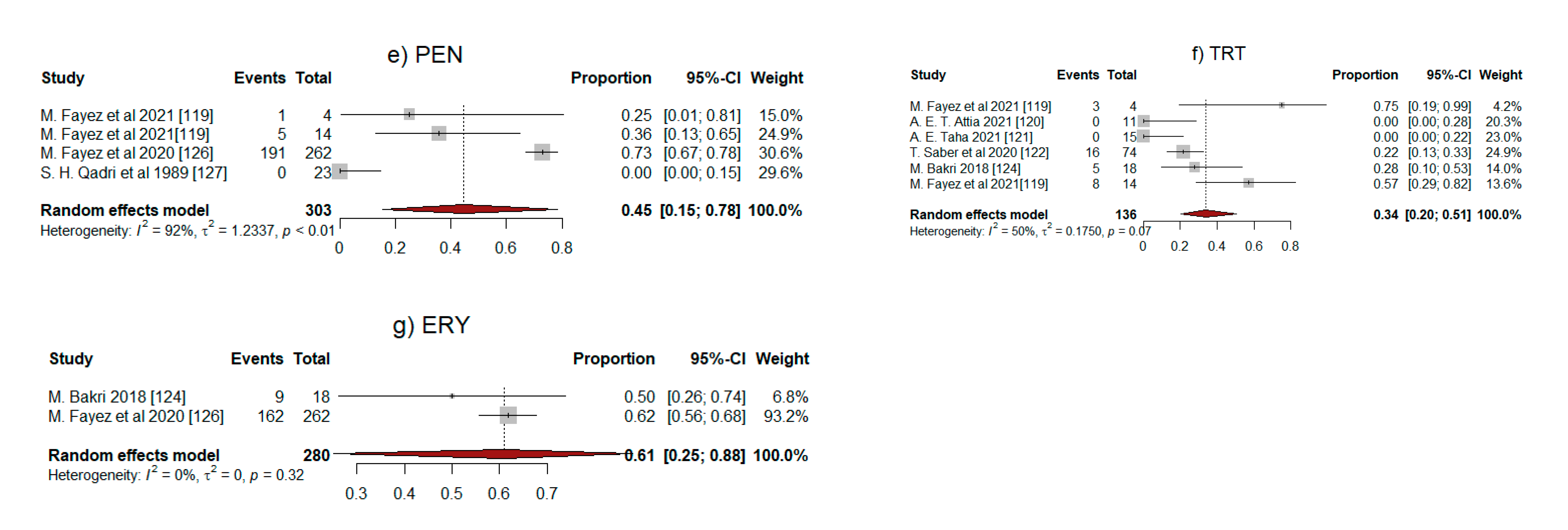
2.5. Meta-Analysis Results
3. Discussion
4. Materials and Methods
4.1. Literature Search
4.2. Selection Criteria
4.3. Data Extraction
4.4. Statistical Analysis
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAD | antibiotic-associated diarrhea |
| CDI | C. difficile infection |
| CPE | C. perfringens enterotoxin |
| FP | food poisoning |
| GI | gastrointestinal |
| NFB | non-foodborne |
References
- McClane, B.A.; Robertson, S.L.; Li, J. Clostridium perfringens. In Food Microbiology: Fundamentals and Frontiers, 4th ed.; Doyle, M.P., Buchanan, R., Eds.; ASM Press: Washington, DC, USA, 2013; pp. 465–486. [Google Scholar]
- Murray, R.; Rosenthal, S.; Pfaller, A. Medical Microbiology, 9th ed.; Elsevier Health Sciences: Philadelphia, PA, USA, 2020. [Google Scholar]
- WHO. Antibiotic Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (accessed on 31 July 2020).
- Aly, M.; Balkhy, H.H. The prevalence of antimicrobial resistance in clinical isolates from Gulf Corporation Council countries. Antimicrob. Resist. Infect. Control. 2012, 1, 1–5. [Google Scholar]
- Paredes-Sabja, D.; Shen, A.; Sorg, J.A. Clostridium difficile spore biology: Sporulation, germination, and spore structural proteins. Trends Microbiol. 2014, 22, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Obaid, N.A.; Alhifany, A.A. Clostridioides difficile infections in Saudi Arabia: Where are we standing? Saudi Pharm. J. 2020, 28, 1118. [Google Scholar] [CrossRef]
- Banawas, S.S. Clostridium difficile infections: A global overview of drug sensitivity and resistance mechanisms. Biomed. Res. Int. 2018, 2018, 8414257. [Google Scholar] [CrossRef] [PubMed]
- Pike, C.M.; Theriot, C.M. Mechanisms of colonization resistance against Clostridioides difficile. J. Infect. Dis. 2020, 223, S194–S200. [Google Scholar] [CrossRef]
- Goudarzi, M.; Seyedjavadi, S.S.; Goudarzi, H.; Mehdizadeh Aghdam, E.; Nazeri, S. Clostridium difficile infection: Epidemiology, pathogenesis, risk factors, and therapeutic options. Scientifica 2014, 2014, 916826. [Google Scholar] [CrossRef]
- Evans, C.T.; Safdar, N. Current trends in the epidemiology and outcomes of Clostridium difficile infection. Clin. Infect. Dis. 2015, 60, S66–S71. [Google Scholar] [CrossRef]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2019.
- Cohen, S.H.; Gerding, D.N.; Johnson, S.; Kelly, C.P.; Loo, V.G.; McDonald, L.C.; Pepin, J.; Wilcox, M.H.; Society for Healthcare Epidemiology of America; Infectious Diseases Society of America. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect. Control. Hosp. Epidemiol. 2010, 31, 431–455. [Google Scholar] [CrossRef]
- Rupnik, M.; Wilcox, M.H.; Gerding, D.N. Clostridium difficile infection: New developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 526–536. [Google Scholar] [CrossRef]
- Aljafel, N.A.; Al-Shaikhy, H.H.; Alnahdi, M.A.; Thabit, A.K. Incidence of Clostridioides difficile infection at a Saudi tertiary academic medical center and compliance with IDSA/SHEA, ACG, and ESCMID guidelines for treatment over a 10-year period. J. Infect. Public Health 2020, 13, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Abed, M.S. Clostridium difficile-associated disease among patients in Dhahran, Saudi Arabia. Travel Med. Infect. Dis. 2010, 8, 373–376. [Google Scholar] [CrossRef] [PubMed]
- Qutub, M.; Govindan, P.; Vattappillil, A. Effectiveness of a two-step testing algorithm for reliable and cost-effective detection of Clostridium difficile infection in a tertiary care hospital in Saudi Arabia. Med. Sci. (Basel) 2019, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.A.; Hawkey, P.M.; Riley, T.V. Epidemiology of Clostridium difficile infection in Asia. Antimicrob. Resist. Infect. Control. 2013, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Smith, A. Outbreak of Clostridium difficile infection in an English hospital linked to hypertoxin-producing strains in Canada and the US. Wkly. Releases (1997–2007) 2005, 10, 2735. [Google Scholar] [CrossRef]
- Alzahrani, N.; Johani, S.A. Emergence of a highly resistant Clostridium difficile strain (NAP/BI/027) in a tertiary care center in Saudi Arabia. Ann. Saudi Med. 2013, 33, 198–199. [Google Scholar] [CrossRef]
- Mastrantonio, P.; Rupnik, M. Erratum to: Updates on Clostridium difficile in Europe. In Updates on Clostridium Difficile in Europe; Springer: Cham, Switzerland, 2018; Volume 8, p. E1. [Google Scholar]
- Tickler, I.A.; Goering, R.V.; Whitmore, J.D.; Lynn, A.N.; Persing, D.H.; Tenover, F.C.; Healthcare Associated Infection Consortium. Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob. Agents Chemother. 2014, 58, 4214–4218. [Google Scholar] [CrossRef]
- Reil, M.; Hensgens, M.P.; Kuijper, E.J.; Jakobiak, T.; Gruber, H.; Kist, M.; Borgmann, S. Seasonality of Clostridium difficile infections in Southern Germany. Epidemiol. Infect. 2012, 140, 1787–1793. [Google Scholar] [CrossRef]
- Tenover, F.C.; Tickler, I.A.; Persing, D.H. Antimicrobial-resistant strains of Clostridium difficile from North America. Antimicrob. Agents Chemother. 2012, 56, 2929–2932. [Google Scholar] [CrossRef]
- Obuch-Woszczatynski, P.; Lachowicz, D.; Schneider, A.; Mol, A.; Pawlowska, J.; Ozdzenska-Milke, E.; Pruszczyk, P.; Wultanska, D.; Mlynarczyk, G.; Harmanus, C.; et al. Occurrence of Clostridium difficile PCR-ribotype 027 and it’s closely related PCR-ribotype 176 in hospitals in Poland in 2008-2010. Anaerobe 2014, 28, 13–17. [Google Scholar] [CrossRef]
- Zhou, Y.; Burnham, C.A.; Hink, T.; Chen, L.; Shaikh, N.; Wollam, A.; Sodergren, E.; Weinstock, G.M.; Tarr, P.I.; Dubberke, E.R. Phenotypic and genotypic analysis of Clostridium difficile isolates: A single-center study. J. Clin. Microbiol. 2014, 52, 4260–4266. [Google Scholar] [CrossRef] [PubMed]
- Lachowicz, D.; Pituch, H.; Obuch-Woszczatynski, P. Antimicrobial susceptibility patterns of Clostridium difficile strains belonging to different polymerase chain reaction ribotypes isolated in Poland in 2012. Anaerobe 2015, 31, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Pelaez, T.; Alcala, L.; Alonso, R.; Martin-Lopez, A.; Garcia-Arias, V.; Marin, M.; Bouza, E. In vitro activity of ramoplanin against Clostridium difficile, including strains with reduced susceptibility to vancomycin or with resistance to metronidazole. Antimicrob. Agents Chemother. 2005, 49, 1157–1159. [Google Scholar] [CrossRef]
- Surawicz, C.M.; Brandt, L.J.; Binion, D.G.; Ananthakrishnan, A.N.; Curry, S.R.; Gilligan, P.H.; McFarland, L.V.; Mellow, M.; Zuckerbraun, B.S. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am. J. Gastroenterol. 2013, 108, 478–498, quiz 499. [Google Scholar] [CrossRef]
- McDonald, L.C.; Killgore, G.E.; Thompson, A.; Owens, R.C., Jr.; Kazakova, S.V.; Sambol, S.P.; Johnson, S.; Gerding, D.N. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 2005, 353, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Muto, C.A.; Blank, M.K.; Marsh, J.W.; Vergis, E.N.; O’leary, M.M.; Shutt, K.A.; Pasculle, A.W.; Pokrywka, M.; Garcia, J.G.; Posey, K. Control of an outbreak of infection with the hypervirulent Clostridium difficile BI strain in a university hospital using a comprehensive “bundle” approach. Clin. Infect. Dis. 2007, 45, 1266–1273. [Google Scholar] [CrossRef]
- Clements, A.C.; Magalhães, R.J.S.; Tatem, A.J.; Paterson, D.L.; Riley, T.V. Clostridium difficile PCR ribotype 027: Assessing the risks of further worldwide spread. Lancet Infect. Dis. 2010, 10, 395–404. [Google Scholar] [CrossRef]
- Krutova, M.; Matejkova, J.; Tkadlec, J.; Nyc, O. Antibiotic profiling of Clostridium difficile ribotype 176—A multidrug resistant relative to C. difficile ribotype 027. Anaerobe 2015, 36, 88–90. [Google Scholar] [CrossRef]
- Goorhuis, A.; Bakker, D.; Corver, J.; Debast, S.B.; Harmanus, C.; Notermans, D.W.; Bergwerff, A.A.; Dekker, F.W.; Kuijper, E.J. Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 2008, 47, 1162–1170. [Google Scholar] [CrossRef]
- Goorhuis, A.; Van der Kooi, T.; Vaessen, N.; Dekker, F.W.; Van den Berg, R.; Harmanus, C.; van den Hof, S.; Notermans, D.W.; Kuijper, E.J. Spread and epidemiology of Clostridium difficile polymerase chain reaction ribotype 027/toxinotype III in The Netherlands. Clin. Infect. Dis. 2007, 45, 695–703. [Google Scholar] [CrossRef]
- Bauer, M.P.; Notermans, D.W.; van Benthem, B.H.; Brazier, J.S.; Wilcox, M.H.; Rupnik, M.; Monnet, D.L.; van Dissel, J.T.; Kuijper, E.J.; Group, E.S. Clostridium difficile infection in Europe: A hospital-based survey. Lancet 2011, 377, 63–73. [Google Scholar] [CrossRef]
- Limbago, B.M.; Long, C.M.; Thompson, A.D.; Killgore, G.E.; Hannett, G.E.; Havill, N.L.; Mickelson, S.; Lathrop, S.; Jones, T.F.; Park, M.M.; et al. Clostridium difficile strains from community-associated infections. J. Clin. Microbiol. 2009, 47, 3004–3007. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Keel, K.; Brazier, J.S.; Post, K.W.; Weese, S.; Songer, J.G. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J. Clin. Microbiol. 2007, 45, 1963–1964. [Google Scholar] [CrossRef]
- Kelly, C.P.; LaMont, J.T. Clostridium difficile—more difficult than ever. N. Engl. J. Med. 2008, 359, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Wu, S.; Wang, M.; Zhang, Y.; Fang, H.; Palmgren, A.C.; Weintraub, A.; Nord, C.E. Molecular and clinical characteristics of Clostridium difficile infection in a University Hospital in Shanghai, China. Clin. Infect. Dis. 2008, 47, 1606–1608. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, Y.; Lee, K.; Riley, T.V.; Kim, H. The changes of PCR ribotype and antimicrobial resistance of Clostridium difficile in a tertiary care hospital over 10 years. J. Med. Microbiol. 2014, 63, 819–823. [Google Scholar] [CrossRef]
- Kim, J.; Kang, J.O.; Pai, H.; Choi, T.Y. Association between PCR ribotypes and antimicrobial susceptibility among Clostridium difficile isolates from healthcare-associated infections in South Korea. Int. J. Antimicrob. Agents 2012, 40, 24–29. [Google Scholar] [CrossRef]
- Gerding, D.N.; Muto, C.A.; Owens, R.C., Jr. Measures to control and prevent Clostridium difficile infection. Clin. Infect. Dis. 2008, 46, S43–S49. [Google Scholar] [CrossRef]
- Barbut, F.; Petit, J.C. Epidemiology of Clostridium difficile-associated infections. Clin. Microbiol. Infect. 2001, 7, 405–410. [Google Scholar] [CrossRef]
- Mylonakis, E.; Ryan, E.T.; Calderwood, S.B. Clostridium difficile—Associated diarrhea: A review. Arch. Intern. Med. 2001, 161, 525–533. [Google Scholar] [CrossRef]
- Wistrom, J.; Norrby, S.R.; Myhre, E.B.; Eriksson, S.; Granstrom, G.; Lagergren, L.; Englund, G.; Nord, C.E.; Svenungsson, B. Frequency of antibiotic-associated diarrhoea in 2462 antibiotic-treated hospitalized patients: A prospective study. J. Antimicrob. Chemother. 2001, 47, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Noren, T.; Tang-Feldman, Y.J.; Cohen, S.H.; Silva, J., Jr.; Olcen, P. Clindamycin resistant strains of Clostridium difficile isolated from cases of C. difficile associated diarrhea (CDAD) in a hospital in Sweden. Diagn. Microbiol. Infect. Dis. 2002, 42, 149–151. [Google Scholar] [CrossRef]
- Johnson, S.; Samore, M.H.; Farrow, K.A.; Killgore, G.E.; Tenover, F.C.; Lyras, D.; Rood, J.I.; DeGirolami, P.; Baltch, A.L.; Rafferty, M.E.; et al. Epidemics of diarrhea caused by a clindamycin-resistant strain of Clostridium difficile in four hospitals. N. Engl. J. Med. 1999, 341, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.; Heffernan, H.; Al Anbuky, N.; Pope, C.; Paviour, S.; Camp, T.; Swager, T. Molecular epidemiology and susceptibility profiles of Clostridium difficile in New Zealand, 2009. N. Z. Med. J. 2011, 124, 45–51. [Google Scholar]
- Huang, H.; Weintraub, A.; Fang, H.; Nord, C.E. Antimicrobial resistance in Clostridium difficile. Int. J. Antimicrob. Agents 2009, 34, 516–522. [Google Scholar] [CrossRef]
- Imwattana, K.; Knight, D.R.; Kullin, B.; Collins, D.A.; Putsathit, P.; Kiratisin, P.; Riley, T.V. Antimicrobial resistance in Clostridium difficile ribotype 017. Expert Rev. Anti. Infect. Ther. 2020, 18, 17–25. [Google Scholar] [CrossRef]
- Evans, M.E.; Simbartl, L.A.; Kralovic, S.M.; Jain, R.; Roselle, G.A. Clostridium difficile infections in Veterans Health Administration acute care facilities. Infect. Control. Hosp. Epidemiol. 2014, 35, 1037–1042. [Google Scholar] [CrossRef]
- Reeves, J.S.; Evans, M.E.; Simbartl, L.A.; Kralovic, S.M.; Kelly, A.A.; Jain, R.; Roselle, G.A. Clostridium difficile infections in Veterans Health Administration long-term care facilities. Infect. Control. Hosp. Epidemiol. 2016, 37, 295–300. [Google Scholar] [CrossRef]
- Bakken, T.L.; Sageng, H. Mental health nursing of adults with intellectual disabilities and mental illness: A review of empirical studies 1994–2013. Arch. Psychiatr. Nurs. 2016, 30, 286–291. [Google Scholar] [CrossRef]
- Bouza, E. Consequences of Clostridium difficile infection: Understanding the healthcare burden. Clin. Microbiol. Infect. 2012, 18, 5–12. [Google Scholar] [CrossRef]
- Pelaez, T.; Alcala, L.; Alonso, R.; Rodriguez-Creixems, M.; Garcia-Lechuz, J.M.; Bouza, E. Reassessment of Clostridium difficile susceptibility to metronidazole and vancomycin. Antimicrob. Agents Chemother. 2002, 46, 1647–1650. [Google Scholar] [CrossRef] [PubMed]
- Teasley, D.; Olson, M.; Gebhard, R.; Gerding, D.N.; Peterson, L.R.; Schwartz, M.; Lee, J., Jr. Prospective randomised trial of metronidazole versus vancomycin for Clostridium-difficile-associated diarrhoea and colitis. Lancet 1983, 322, 1043–1046. [Google Scholar] [CrossRef]
- Dworczyński, A.; Sokół, B.; Meisel-Mikołajczyk, F. Antibiotic resistance of Clostridium difficile isolates. Cytobios 1990, 65, 149–153. [Google Scholar]
- Tkhawkho, L.; Nitzan, O.; Pastukh, N.; Brodsky, D.; Jackson, K.; Peretz, A. Antimicrobial susceptibility of Clostridium difficile isolates in Israel. J. Glob. Antimicrob. Resist. 2017, 10, 161–164. [Google Scholar] [CrossRef]
- Fraga, E.G.; Nicodemo, A.C.; Sampaio, J.L. Antimicrobial susceptibility of Brazilian Clostridium difficile strains determined by agar dilution and disk diffusion. Braz. J. Infect. Dis. 2016, 20, 476–481. [Google Scholar] [CrossRef]
- Baghani, A.; Ghourchian, S.; Aliramezani, A.; Yaseri, M.; Mesdaghinia, A.; Douraghi, M. Highly antibiotic-resistant Clostridium difficile isolates from Iranian patients. J. Appl. Microbiol. 2018, 125, 1518–1525. [Google Scholar] [CrossRef]
- Kouzegaran, S.; Ganjifard, M.; Tanha, A. Detection, ribotyping and antimicrobial resistance properties of Clostridium difficile strains isolated from the cases of diarrhea. Mater. Sociomed. 2016, 28, 324. [Google Scholar] [CrossRef]
- Goudarzi, M.; Goudarzi, H.; Alebouyeh, M.; Rad, M.; Mehr, F.; Zali, M.R.; Aslani, M.M. Antimicrobial susceptibility of Clostridium difficile clinical isolates in Iran. Iran. Red. Crescent. Med. J. 2013, 15, 704. [Google Scholar] [CrossRef]
- Banawas, S.; Sarker, M.R. l-lysine (pH 6.0) induces germination of spores of Clostridium perfringens type F isolates carrying chromosomal or plasmid-borne enterotoxin gene. Microb. Pathog. 2018, 123, 227–232. [Google Scholar] [CrossRef]
- Olguin-Araneda, V.; Banawas, S.; Sarker, M.R.; Paredes-Sabja, D. Recent advances in germination of Clostridium spores. Res. Microbiol. 2015, 166, 236–243. [Google Scholar] [CrossRef]
- Uzal, F.A.; Freedman, J.C.; Shrestha, A.; Theoret, J.R.; Garcia, J.; Awad, M.M.; Adams, V.; Moore, R.J.; Rood, J.I.; McClane, B.A. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014, 9, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Rood, J.I. General physiological and virulence properties of the pathogenic Clostridia. In Clostridial Diseases of Animals, 1st ed.; Uzal, F.A., Songer, J.G., Prescott, J.F., Popoff, M.R., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2016; Volume 7, pp. 7–12. [Google Scholar]
- Li, J.; Paredes-Sabja, D.; Sarker, M.R.; McClane, B.A. Clostridium perfringens sporulation and sporulation-associated toxin production. Microbiol. Spectr. 2016, 4, 4-3. [Google Scholar] [CrossRef]
- Banawas, S.; Paredes-Sabja, D.; Setlow, P.; Sarker, M.R. Characterization of germinants and their receptors for spores of non-food-borne Clostridium perfringens strain F4969. Microbiology 2016, 162, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Banawas, S.; Paredes-Sabja, D.; Korza, G.; Li, Y.; Hao, B.; Setlow, P.; Sarker, M.R. The Clostridium perfringens germinant receptor protein GerKC is located in the spore inner membrane and is crucial for spore germination. J. Bacteriol. 2013, 195, 5084–5091. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; McClane, B.A. Comparative effects of osmotic, sodium nitrite-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl. Environ. Microbiol. 2006, 72, 7620–7625. [Google Scholar] [CrossRef]
- Songer, J.G. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 1996, 9, 216–234. [Google Scholar] [CrossRef]
- Rood, J.I.; Adams, V.; Lacey, J.; Lyras, D.; McClane, B.A.; Melville, S.B.; Moore, R.J.; Popoff, M.R.; Sarker, M.R.; Songer, J.G.; et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 2018, 53, 5–10. [Google Scholar] [CrossRef]
- McClane, B.; Uzal, F.A.; Miyakawa, M.F.; Lyerly, D.; Wilkins, T. The enterotoxic clostridia. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2004; pp. 698–752. [Google Scholar]
- Revitt-Mills, S.A.; Rood, J.I.; Adams, V. Clostridium perfringens extracellular toxins and enzymes: 20 and counting. Microbiol. Aust. 2015, 36, 114–117. [Google Scholar] [CrossRef]
- Asha, N.J.; Wilcox, M.H. Laboratory diagnosis of Clostridium perfringens antibiotic-associated diarrhoea. J. Med. Microbiol. 2002, 51, 891–894. [Google Scholar] [CrossRef]
- Sarker, M.R.; Carman, R.J.; McClane, B.A. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 1999, 33, 946–958. [Google Scholar] [CrossRef]
- Carman, R.J. Clostridium perfringens spontaneous and antibiotic associated diarrhoea of man and other animals. Rev. Med. Microbiol. 1997, 8, S43–S45. [Google Scholar] [CrossRef]
- Borriello, S.P.; Larson, H.E.; Welch, A.R.; Barclay, F.; Stringer, M.F.; Bartholomew, B.A. Enterotoxigenic Clostridium perfringens: A possible cause of antibiotic-associated diarrhoea. Lancet 1984, 1, 305–307. [Google Scholar] [CrossRef]
- Banaszkiewicz, A.; Kadzielska, J.; Gawronska, A.; Pituch, H.; Obuch-Woszczatynski, P.; Albrecht, P.; Mlynarczyk, G.; Radzikowski, A. Enterotoxigenic Clostridium perfringens infection and pediatric patients with inflammatory bowel disease. J. Crohns Colitis 2014, 8, 276–281. [Google Scholar] [CrossRef]
- Freedman, J.C.; Shrestha, A.; McClane, B.A. Clostridium perfringens Enterotoxin: Action, Genetics, and Translational Applications. Toxins (Basel) 2016, 8, 73. [Google Scholar] [CrossRef]
- Collie, R.E.; Kokai-Kun, J.F.; McClane, B.A. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe 1998, 4, 69–79. [Google Scholar] [CrossRef]
- Cornillot, E.; Saint-Joanis, B.; Daube, G.; Katayama, S.; Granum, P.E.; Canard, B.; Cole, S.T. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 1995, 15, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Sarker, M.R.; Shivers, R.P.; Sparks, S.G.; Juneja, V.K.; McClane, B.A. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 2000, 66, 3234–3240. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, S.; Batz, M.B.; Morris, J.G., Jr. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J. Food Prot. 2012, 75, 1292–1302. [Google Scholar] [CrossRef]
- Lynch, M.; Painter, J.; Woodruff, R.; Braden, C. Surveillance for Foodborne-disease Outbreaks: United States, 1998–2002. MMWR Surveill. Summ. 2006, 55, 1–42. [Google Scholar]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Brynestad, S.; Granum, P.E. Evidence that Tn 5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. FEMS Microbiol. Lett. 1999, 170, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.L.; Derby, P.; Abratt, V.R. In-vitro antibiotic susceptibility and molecular analysis of anaerobic bacteria isolated in Cape Town, South Africa. J. Antimicrob. Chemother. 1998, 42, 245–248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Camacho, N.; Espinoza, C.; Rodríguez, C.; Rodríguez, E. Isolates of Clostridium perfringens recovered from Costa Rican patients with antibiotic-associated diarrhoea are mostly enterotoxin-negative and susceptible to first-choice antimicrobials. J. Med. Microbiol. 2008, 57, 343347. [Google Scholar] [CrossRef] [PubMed]
- Akhi, M.T.; Bidar Asl, S.; Pirzadeh, T.; Naghili, B.; Yeganeh, F.; Memar, Y.; Mohammadzadeh, Y. Antibiotic sensitivity of Clostridium perfringens isolated from faeces in Tabriz, Iran. Jundishapur. J. Microbiol. 2015, 8, e20863. [Google Scholar] [CrossRef] [PubMed]
- Tansuphasiri, U.; Matra, W.; Sangsuk, L. Antimicrobial resistance among Clostridium perfringens isolated from various sources in Thailand. Southeast. Asian J. Trop. Med. Public Health 2005, 36, 954–961. [Google Scholar] [PubMed]
- Osman, K.M.; Elhariri, M. Antibiotic resistance of Clostridium perfringens isolates from broiler chickens in Egypt. Rev. Sci. Tech. 2013, 32, 841–850. [Google Scholar] [CrossRef]
- Martel, A.; Devriese, L.A.; Cauwerts, K.; De Gussem, K.; Decostere, A.; Haesebrouck, F. Susceptibility of Clostridium perfringens strains from broiler chickens to antibiotics and anticoccidials. Avian Pathol. 2004, 33, 3–7. [Google Scholar] [CrossRef]
- Milton, A.A.P.; Sanjukta, R.; Gogoi, A.P.; Momin, K.M.; Priya, G.B.; Das, S.; Ghatak, S.; Sen, A.; Kandpal, B.K. Prevalence, molecular typing and antibiotic resistance of Clostridium perfringens in free range ducks in Northeast India. Anaerobe 2020, 64, 102242. [Google Scholar] [CrossRef]
- Nigam, P.K.; Nigam, A. Botulinum toxin. Indian J. Dermatol. 2010, 55, 8–14. [Google Scholar] [CrossRef]
- Arnon, S.S.; Schechter, R.; Inglesby, T.V.; Henderson, D.A.; Bartlett, J.G.; Ascher, M.S.; Eitzen, E.; Fine, A.D.; Hauer, J.; Layton, M. Botulinum toxin as a biological weapon: Medical and public health management. JAMA 2001, 285, 1059–1070. [Google Scholar] [CrossRef]
- Peck, M.W. Clostridium botulinum and the safety of minimally heated, chilled foods: An emerging issue? J. Appl. Microbiol. 2006, 101, 556–570. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J. Botulism. Clin. Infect. Dis. 2005, 41, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- CDC. Botulism Annual Summary; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2017.
- Arias, C.A.; Murray, B.E. Antibiotic-resistant bugs in the 21st century—A clinical super-challenge. N. Engl. J. Med. 2009, 360, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Boyanova, L.; Kolarov, R.; Mitov, I. Antimicrobial resistance and the management of anaerobic infections. Rev. Anti. Infect. Ther. 2007, 5, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Dezfulian, M.; Dowell, V.R. Cultural and physiological characteristics and antimicrobial susceptibility of Clostridium botulinum isolates from foodborne and infant botulism cases. Clin. Microbiol. Infect. 1980, 11, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.E.; Dolin, R.; Blaser, M.J. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases: 2-Volume Set, 8th ed.; Elsevier Health Sciences: Amsterdam, The Netherlands, 2014; Volume 2. [Google Scholar]
- Sharma, D.S.; Shah, M.B. A Rare Case of Localized Tetanus. Indian J. Crit. Care Med. 2018, 22, 678–679. [Google Scholar] [CrossRef]
- Cohen, J.E.; Wang, R.; Shen, R.F.; Wu, W.W.; Keller, J.E. Comparative pathogenomics of Clostridium tetani. PLoS ONE 2017, 12, e0182909. [Google Scholar] [CrossRef]
- ECDC. Disease Factsheet about Tetanus. European Centre for Disease Prevention and Control, 2021. Available online: https://www.ecdc.europa.eu/en/tetanus/facts (accessed on 7 July 2022).
- WHO. Tetanus vaccines: WHO position paper, February 2017–recommendations. Vaccine 2018, 36, 3573–3575. [Google Scholar] [CrossRef]
- Hanif, H.; Anjum, A.; Ali, N.; Jamal, A.; Imran, M.; Ahmad, B.; Ali, M.I. Isolation and antibiogram of Clostridium tetani from clinically diagnosed tetanus patients. Am. J. Trop. Med. Hyg. 2015, 93, 752–756. [Google Scholar] [CrossRef]
- Fayez, M.; El-Ghareeb, W.R.; Elmoslemany, A.; Alsunaini, S.J.; Alkafafy, M.; Alzahrani, O.M.; Mahmoud, S.F.; Elsohaby, I. Genotyping and antimicrobial susceptibility of Clostridium perfringens and Clostridioides difficile in camel minced meat. Pathogens 2021, 10, 1640. [Google Scholar] [CrossRef]
- Attia, A.E.T. Retail chicken meats as potential sources of Clostridioides difficile in Al-Jouf, Saudi Arabia. J. Infect. Dev. Ctries. 2021, 15, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.E. Raw animal meats as potential sources of Clostridium difficile in Al-Jouf, Saudi Arabia. Food Sci. Anim. Resour. 2021, 41, 883. [Google Scholar] [CrossRef] [PubMed]
- Saber, T.; Hawash, Y.A.; Ismail, K.A.; Khalifa, A.S.; Alsharif, K.F.; Alghamdi, S.A.; Saber, T.; Eed, E.M. Prevalence, toxin gene profile, genotypes and antibiotic susceptibility of Clostridium difficile in a tertiary care hospital in Taif, Saudi Arabia. Indian J. Med. Microbiol. 2020, 38, 176–182. [Google Scholar] [CrossRef]
- Alqumber, M.A. Clostridium difficile in retail baskets, trolleys, conveyor belts, and plastic bags in Saudi Arabia. Saudi Med. J. 2014, 35, 1274–1277. [Google Scholar]
- Bakri, M. Prevalence of Clostridium difficile in raw cow, sheep, and goat meat in Jazan, Saudi Arabia. Saudi J. Biol. Sci. 2018, 25, 783–785. [Google Scholar] [CrossRef] [PubMed]
- Shajan, S.E.; Hashim, M.F.; Michael, A. Prevalence of Clostridium difficile toxin in diarrhoeal stool samples of patients from a general hospital in eastern province, Saudi Arabia. Int. J. Med. Health Res. 2014, 3, 302–308. [Google Scholar]
- Fayez, M.; Elsohaby, I.; Al-Marri, T.; Zidan, K.; Aldoweriej, A.; El-Sergany, E.; Elmoslemany, A. Genotyping and antimicrobial susceptibility of Clostridium perfringens isolated from dromedary camels, pastures and herders. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101460. [Google Scholar] [CrossRef]
- Qadri, S.H.; Ueno, Y.; Ostrawski, S. Anaerobic infections at a referral center: Incidence, etiology, and antimicrobial susceptibility pattern. Ann. Saudi Med. 1989, 9, 551–555. [Google Scholar] [CrossRef]
- Owens, R.C., Jr.; Donskey, C.J.; Gaynes, R.P.; Loo, V.G.; Muto, C.A. Antimicrobial-associated risk factors for Clostridium difficile infection. Clin. Infect. Dis. 2008, 46, S19–S31. [Google Scholar] [CrossRef]
- Peng, Z.; Jin, D.; Kim, H.B.; Stratton, C.W.; Wu, B.; Tang, Y.W.; Sun, X. Update on antimicrobial resistance in Clostridium difficile: Resistance mechanisms and antimicrobial susceptibility testing. J. Clin. Microbiol. 2017, 55, 1998–2008. [Google Scholar] [CrossRef]
- Ofosu, A. Clostridium difficile infection: A review of current and emerging therapies. Ann. Gastroenterol. 2016, 29, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Khademi, F.; Sahebkar, A. The prevalence of antibiotic-resistant Clostridium species in Iran: A meta-analysis. Pathog. Glob. Health 2019, 113, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, J.G.; Onderdonk, A.B.; Cisneros, R.L.; Kasper, D.L. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J. Infect. Dis. 1977, 136, 701–705. [Google Scholar] [CrossRef] [PubMed]
- Salvarani, F.M.; Silva, R.O.S.; Pires, P.S.; Cruz, E.C.D.; Albefaro, I.S.; Guedes, R.M.D.; Lobato, F.C.F. Antimicrobial susceptibility of Clostridium Perfringens isolated from piglets with or without diarrhea in Brazil. Braz. J. Microbiol. 2012, 43, 1030–1033. [Google Scholar] [CrossRef]
- Li, J.; Zhou, Y.; Yang, D.; Zhang, S.; Sun, Z.; Wang, Y.; Wang, S.; Wu, C. Prevalence and antimicrobial susceptibility of Clostridium perfringens in chickens and pigs from Beijing and Shanxi, China. Vet. Microbiol. 2020, 252, 108932. [Google Scholar] [CrossRef]
- Tassanaudom, U.; Toorisut, Y.; Tuitemwong, K.; Jittaprasartsin, C.; Wangroongsarb, P.; Mahakarnchanakul, W. Prevalence of toxigenic Clostridium perfringens strains isolated from dried spur pepper in Thailand. Int. Food Res. J. 2017, 24, 955–962. [Google Scholar]
- Silva, R.O.; Ribeiro, M.G.; Palhares, M.S.; Borges, A.S.; Maranhao, R.P.; Silva, M.X.; Lucas, T.M.; Olivo, G.; Lobato, F.C. Detection of A/B toxin and isolation of Clostridium difficile and Clostridium perfringens from foals. Equine Vet. J. 2013, 45, 671–675. [Google Scholar] [CrossRef]
- Roberts, S.A.; Shore, K.P.; Paviour, S.D.; Holland, D.; Morris, A.J. Antimicrobial susceptibility of anaerobic bacteria in New Zealand: 1999–2003. J. Antimicrob. Chemother. 2006, 57, 992–998. [Google Scholar] [CrossRef][Green Version]
- Leal, J.; Gregson, D.B.; Ross, T.; Church, D.L.; Laupland, K.B. Epidemiology of Clostridium species bacteremia in Calgary, Canada, 2000–2006. J. Infect. 2008, 57, 198–203. [Google Scholar] [CrossRef]
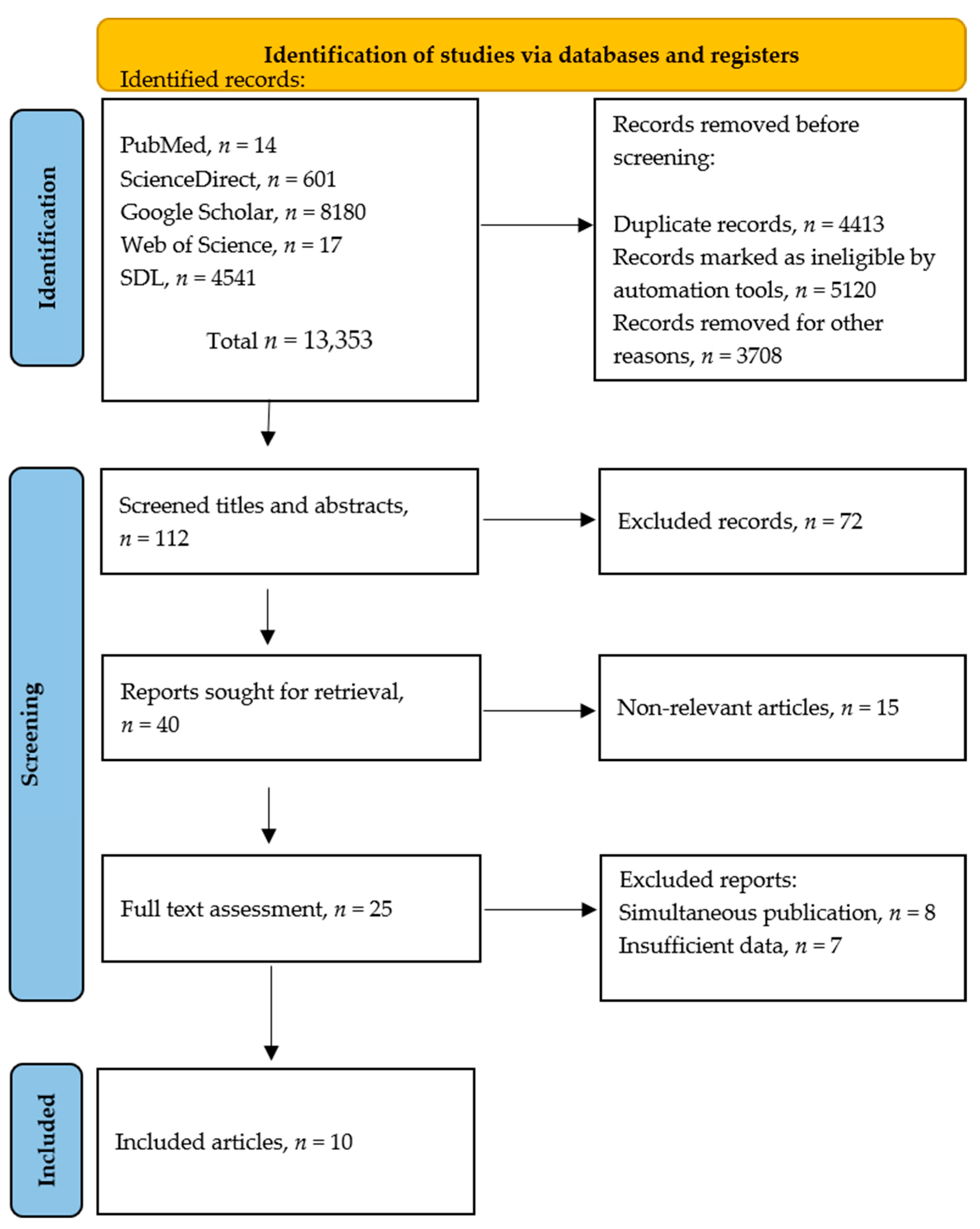
| Inclusion Criterium | Search Terms |
|---|---|
| Antibiotic resistance | Drug resistance, Antimicrobial resistance |
| Clostridium spp. | C. difficile, C. botulinum, C. tetani, C. perfringens |
| Saudi Arabia | Kingdom of Saudi Arabia, SA, KSA, Saudi Arabia |
| Province/ City | Year | Sample Type (Origin) | Strain (n) | Clostridium spp. | AST | Antibiotic Resistance (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | MTZ | CEF | GEN | CLI | LVX | PEN | VAN | TRT | ERY | OXY | AMP | MXF | LIN | Reference | ||||||
| Al-Ahsa | 2019–2020 | Camels | 4 | C. difficile | Broth dilution | ND | ND | ND | ND | 25 | ND | 25 | ND | 75 | ND | ND | ND | 25 | ND | [110] |
| Al-Jouf | 2019 | Chickens | 11 | C. difficile | E-tests | ND | 0 | ND | ND | 0 | ND | ND | 0 | 0 | ND | ND | ND | 18 | ND | [111] |
| Al-Jouf | 2019 | Camels, cows, sheep, and goats | 15 | C. difficile | E-tests | ND | 0 | ND | ND | 0 | ND | ND | 0 | 0 | ND | ND | ND | 20 | ND | [112] |
| Al-Taif | 2019 | Stools | 74 | C. difficile | E-tests | ND | 0 | ND | ND | 54 | ND | ND | 0 | 21.6 | ND | ND | ND | 48.6 | ND | [113] |
| Al-Bahah and Al-Taif | 2011 | Baskets, trolleys, conveyor belts, and plastic bags | 12 | C. difficile | E-tests | ND | 0 | ND | ND | 58 | 100 | ND | 0 | ND | ND | ND | ND | ND | ND | [114] |
| Jazan | 2015 | Cow, sheep, and goat meat | 18 | C. difficile | Disk diffusion | 27 | 0 | ND | 83 | 33 | ND | ND | 0 | 28 | 50 | ND | 72 | ND | ND | [115] |
| Eastern province | 2011–2012 | Stools | 19 | C. difficile | E-tests | 30 | ND | ND | ND | ND | 30 | ND | ND | ND | ND | ND | ND | 30 | ND | [116] |
| Al-Ahsa | 2019–2020 | Camels | 14 | C. perfringens | Broth dilution | ND | 14 | ND | ND | 35 | ND | 35 | ND | 56 | ND | ND | ND | ND | ND | [110] |
| Eastern province | 2018 | Dromedary camels, pastures, and herders | 262 | C. perfringens | Broth dilution | ND | 27 | 83 | ND | 12 | ND | 73 | ND | ND | 62 | 47 | ND | ND | 47 | [117] |
| Riyadh | 1988 | Wounds, blood, body fluids, and female genitalia | 23 | C. perfringens | Disk elution | ND | ND | ND | ND | 0 | ND | 0 | 0 | ND | ND | ND | ND | ND | ND | [118] |
| Antibiotic | No. of Studies | Pooled Proportion of Resistance (95% CI) | I2 (%) * | p |
|---|---|---|---|---|
| CIP | 2 | 0.30 (0.12–0.57) | 0 | 0.8 |
| MTZ | 7 | 0.37 (0.22–0.54) | 93 | <0.01 |
| CLI | 9 | 0.34 (0.23–0.47) | 87 | <0.01 |
| LVX | 2 | 0.50 (0.00–1.00) | 83 | 0.02 |
| PEN | 4 | 0.45 (0.15–0.78) | 92 | <0.01 |
| TRT | 6 | 0.34 (0.20–0.51) | 50 | 0.07 |
| ERY | 2 | 0.61 (0.25–0.88) | 0 | 0.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banawas, S.S. Systematic Review and Meta-Analysis on the Frequency of Antibiotic-Resistant Clostridium Species in Saudi Arabia. Antibiotics 2022, 11, 1165. https://doi.org/10.3390/antibiotics11091165
Banawas SS. Systematic Review and Meta-Analysis on the Frequency of Antibiotic-Resistant Clostridium Species in Saudi Arabia. Antibiotics. 2022; 11(9):1165. https://doi.org/10.3390/antibiotics11091165
Chicago/Turabian StyleBanawas, Saeed S. 2022. "Systematic Review and Meta-Analysis on the Frequency of Antibiotic-Resistant Clostridium Species in Saudi Arabia" Antibiotics 11, no. 9: 1165. https://doi.org/10.3390/antibiotics11091165
APA StyleBanawas, S. S. (2022). Systematic Review and Meta-Analysis on the Frequency of Antibiotic-Resistant Clostridium Species in Saudi Arabia. Antibiotics, 11(9), 1165. https://doi.org/10.3390/antibiotics11091165







