The Generation of an Artificial ATP Deficit Triggers Antibiotic Production in Streptomyces lividans
Abstract
:1. Introduction
2. Results
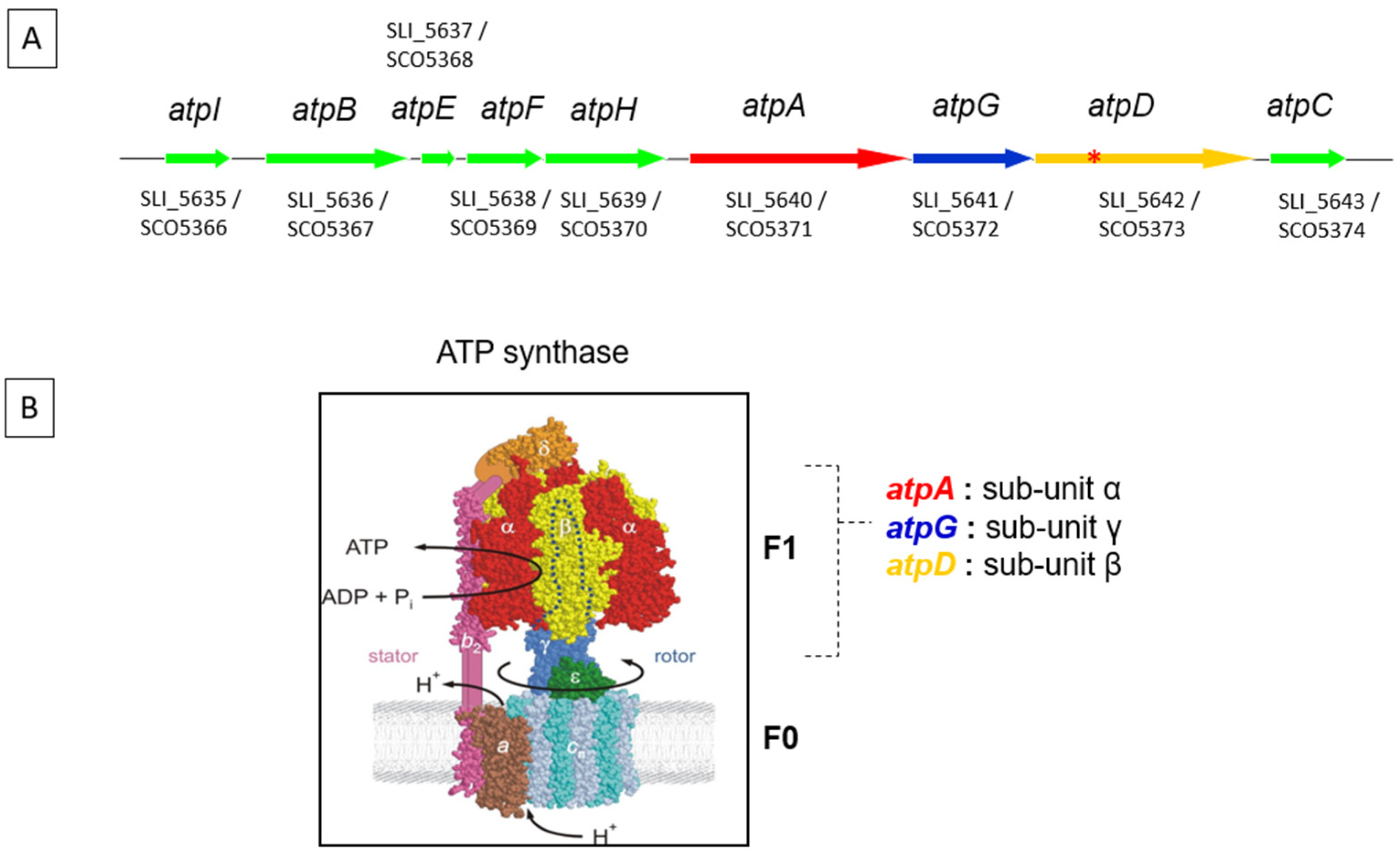
2.1. Quantification of Growth, ATPase Expression and Intracellular ATP Content in S. lividans/pOSV206_atpAGD and S. lividans/pOSV206_atpAGDΔ
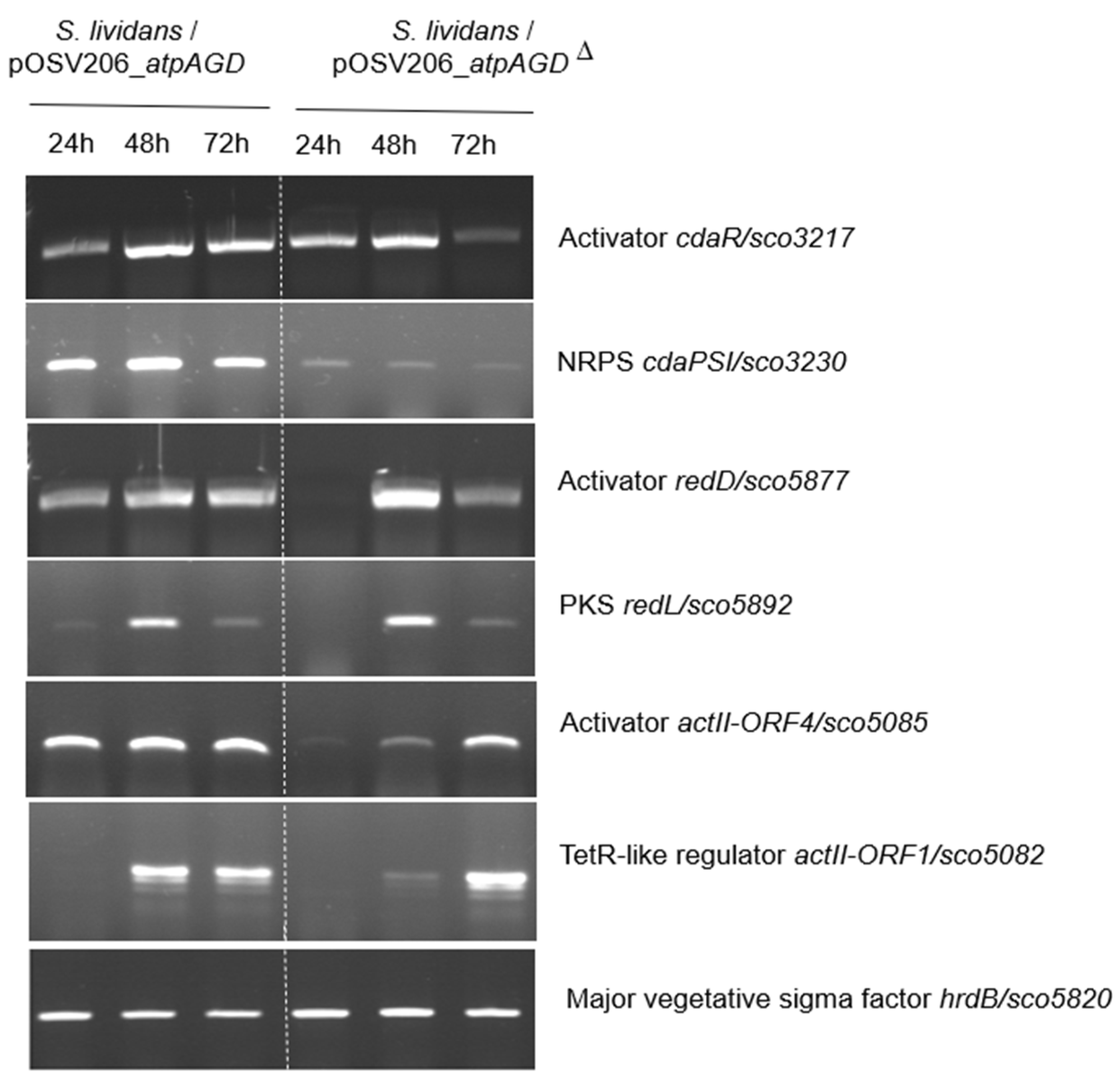
2.2. Analysis in RT-PCT of the Transcriptional Level of Expression of Genes of the CDA, RED and ACT Clusters in S. lividans/pOSV206_atpAGD and S. lividans/pOSV206_atpAGDΔ
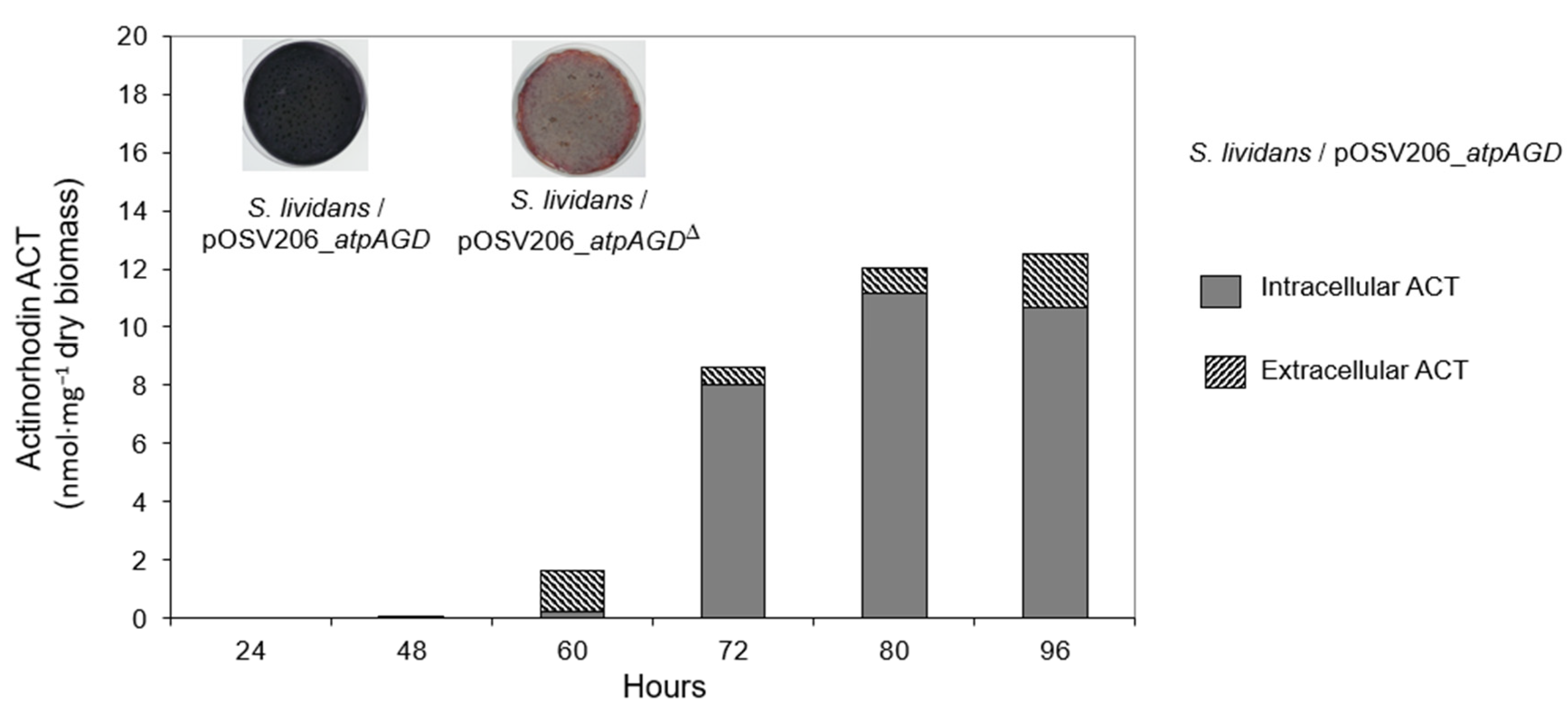
2.3. Assay of CDA, RED and ACT Production in S. lividans/pOSV206_atpAGD and S. lividans pOSV206_atpAGDΔ Grown in HT or R2YE
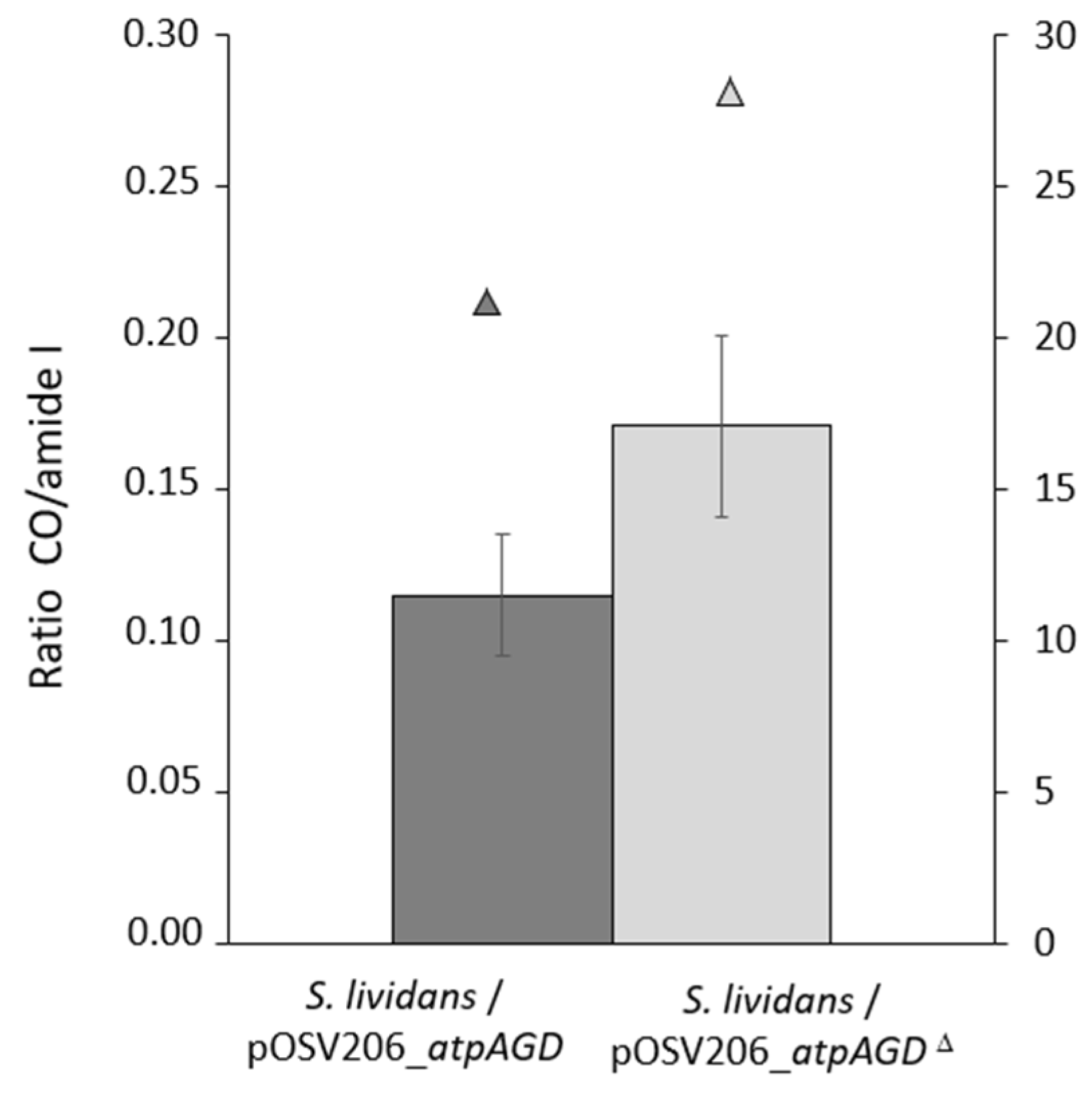
2.4. Analysis of the Total Lipid Content of the Strains S. lividans/pOSV206_atpAGD and S. lividans/pOSV206_atpAGDΔ
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction
4.2. Strains and Culture Conditions
4.3. Extraction and Assay of Intracellular ATP Concentrations
4.4. RNA Preparation
4.5. Determination of the Level of Expression of Genes of Various Biosynthetic Pathways by RT-PCR
4.6. Assay of CDA, RED and ACT Production
4.7. Determination of Total Lipid Content Using Attenuated Total Reflectance-Fourier Transform Infra Red Spectroscopy (ATR-FTIRS) Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pham, J.V.; Yilma, M.A.; Feliz, A.; Majid, M.T.; Maffetone, N.; Walker, J.R.; Kim, E.; Cho, H.J.; Reynolds, J.M.; Song, M.C.; et al. A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 2019, 10, 1404. [Google Scholar] [CrossRef] [PubMed]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; van Wezel, G.P. Correction for Barka et al., Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, iii. [Google Scholar] [CrossRef]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular Regulation of Antibiotic Biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.F. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: An unfinished story. J. Bacteriol. 2004, 186, 5197–5201. [Google Scholar] [CrossRef] [PubMed]
- Esnault, C.; Dulermo, T.; Smirnov, A.; Askora, A.; David, M.; Deniset-Besseau, B.; Holland, I.B.; Virolle, M.J. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci. Rep. 2017, 7, 200. [Google Scholar] [CrossRef]
- Lejeune, C.; Sago, L.; Cornu, D.; Redeker, V.; Virolle, M.J. A Proteomic Analysis Indicates That Oxidative Stress Is the Common Feature Triggering Antibiotic Production in Streptomyces coelicolor and in the pptA Mutant of Streptomyces lividans. Front. Microbiol. 2021, 12, 813993. [Google Scholar] [CrossRef] [PubMed]
- Millan-Oropeza, A.; Henry, C.; Lejeune, C.; David, M.; Virolle, M.-J. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci. Rep. 2020, 10, 8492. [Google Scholar] [CrossRef]
- Lakey, J.H.; Lea, E.J.; Rudd, B.A.; Wright, H.M.; Hopwood, D.A. A new channel-forming antibiotic from Streptomyces coelicolor A3(2) which requires calcium for its activity. J. Gen. Microbiol. 1983, 129, 3565–3573. [Google Scholar] [CrossRef]
- Adamidis, T.; Riggle, P.; Champness, W. Mutations in a new Streptomyces coelicolor locus which globally block antibiotic biosynthesis but not sporulation. J. Bacteriol. 1990, 172, 2962–2969. [Google Scholar] [CrossRef] [PubMed]
- Shikura, N.; Darbon, E.; Esnault, C.; Deniset-Besseau, A.; Xu, D.; Lejeune, C.; Jacquet, E.; Nhiri, N.; Sago, L.; Cornu, D.; et al. The Phosin PptA Plays a Negative Role in the Regulation of Antibiotic Production in Streptomyces lividans. Antibiotics 2021, 10, 325. [Google Scholar] [CrossRef]
- David, M.; Lejeune, C.; Abreu, S.; Thibessard, A.; Leblond, P.; Chaminade, P.; Virolle, M.J. Negative Correlation between Lipid Content and Antibiotic Activity in Streptomyces: General Rule and Exceptions. Antibiotics 2020, 9, 280. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lih, C.-J.; Pan, K.-H.; Cohen, S.N. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 2001, 15, 3183–3192. [Google Scholar] [CrossRef] [PubMed]
- Redenbach, M.; Kieser, H.M.; Denapaite, D.; Eichner, A.; Cullum, J.; Kinashi, H.; Hopwood, D.A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996, 21, 77–96. [Google Scholar] [CrossRef] [PubMed]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Tenconi, E.; Traxler, M.; Hoebreck, C.; Van Wezel, G.P.; Rigali, S. Production of Prodiginines Is Part of a Programmed Cell Death Process in Streptomyces coelicolor. Front. Microbiol. 2018, 9, 1742. [Google Scholar] [CrossRef]
- Mazodier, P.; Petter, R.; Thompson, C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 1989, 171, 3583–3585. [Google Scholar] [CrossRef]
- Wehmeier, U.F. New multifunctional Escherichia coli-Streptomyces shuttle vectors allowing blue-white screening on XGal plates. Gene 1995, 165, 149–150. [Google Scholar] [CrossRef]
- Thompson, C.J.; Kieser, T.; Ward, J.M.; Hopwood, D.A. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene 1982, 20, 51–62. [Google Scholar] [CrossRef]
- Skeggs, P.A.; Holmes, D.J.; Cundliffe, E. Cloning of aminoglycoside-resistance determinants from Streptomyces tenebrarius and comparison with related genes from other actinomycetes. J. Gen. Microbiol. 1987, 133, 915–923. [Google Scholar] [CrossRef]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. (Eds.) Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Hopwood, D.A.; Chater, K.F.; Kieser, T.; Bruton, C.J.; Kieser, H.M.; Lydiate, D.J.; Smith, C.P.; Ward, J.M.; Schrempf, H. Genetic Manipulation of Streptomyces: A laboratory Manual; John Innes Foundation: Norwich, UK, 1985. [Google Scholar]
- Simon, R.; Priefer, U.; Pülher, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotechnol. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Millan-Oropeza, A.; Rebois, R.; David, M.; Moussa, F.; Dazzi, A.; Bleton, J.; Virolle, M.J.; Deniset-Besseau, A. Attenuated Total Reflection Fourier Transform Infrared (ATR FT-IR) for Rapid Determination of Microbial Cell Lipid Content: Correlation with Gas Chromatography-Mass Spectrometry (GC-MS). Appl. Spectrosc. 2017, 71, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Deniset-Besseau, A.; Prater, C.B.; Virolle, M.-J.; Dazzi, A. Monitoring TriAcylGlycerols Accumulation by Atomic Force Microscopy Based Infrared Spectroscopy in Streptomyces Species for Biodiesel Applications. J. Phys. Chem. Lett. 2014, 5, 654–658. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seghezzi, N.; Darbon, E.; Martel, C.; David, M.; Lejeune, C.; Esnault, C.; Virolle, M.-J. The Generation of an Artificial ATP Deficit Triggers Antibiotic Production in Streptomyces lividans. Antibiotics 2022, 11, 1157. https://doi.org/10.3390/antibiotics11091157
Seghezzi N, Darbon E, Martel C, David M, Lejeune C, Esnault C, Virolle M-J. The Generation of an Artificial ATP Deficit Triggers Antibiotic Production in Streptomyces lividans. Antibiotics. 2022; 11(9):1157. https://doi.org/10.3390/antibiotics11091157
Chicago/Turabian StyleSeghezzi, Nicolas, Emmanuelle Darbon, Cécile Martel, Michelle David, Clara Lejeune, Catherine Esnault, and Marie-Joelle Virolle. 2022. "The Generation of an Artificial ATP Deficit Triggers Antibiotic Production in Streptomyces lividans" Antibiotics 11, no. 9: 1157. https://doi.org/10.3390/antibiotics11091157
APA StyleSeghezzi, N., Darbon, E., Martel, C., David, M., Lejeune, C., Esnault, C., & Virolle, M.-J. (2022). The Generation of an Artificial ATP Deficit Triggers Antibiotic Production in Streptomyces lividans. Antibiotics, 11(9), 1157. https://doi.org/10.3390/antibiotics11091157







