Abstract
Colistin resistance in bacteria has become a significant threat to food safety and public health, and its development was mainly attributed to the plasmid-mediated mcr genes. This study aimed to determine the global prevalence and molecular characteristics of mcr-producing Salmonella enterica isolates. A total of 2279 mcr-producing Salmonella genomes were obtained from the public database, which were disseminated in 37 countries from five continents worldwide, including Asia, Europe, America, Australia, and Africa. Human samples (39.5%; 900/2279) were the predominant sources of mcr-producing Salmonella isolates, followed by foods (32.6%), animals (13.7%), and environment (4.4%). Furthermore, 80 Salmonella serotypes were identified, and Typhimurium and 1,4,[5],12:i:- were the predominant serotypes, accounting for 18.3% and 18.7%, respectively. Twenty mcr variants were identified, and the most common ones were mcr-9.1 (65.2%) and mcr-1.1 (24.4%). Carbapenems-resistance gene blaNDM-1 and tigecycline-resistance gene tet(X4) were identified in one isolate, respectively. Phylogenetic results indicated that mcr-producing Salmonella fell into nine lineages (Lineages I-IX), and Salmonella Typhimurium, 1,4,[5],12:i:- and 4,[5],12:i:- isolates from different countries were mixed in Lineages I, II and III, suggesting that international spread occurred. These findings underline further challenges for the spread of Salmonella-bearing mcr genes.
1. Introduction
Colistin was considered the last resort drug in the treatment for multidrug-resistant (MDR) and even carbapenem-resistant Gram-negative bacteria. Colistin-resistant bacteria have spread across Asia, Africa, Europe, North America, South America, and Oceania [1,2,3]. The colistin resistance in bacteria was previously reported to result from mutations in chromosomes [4]. Until 2015, the novel plasmid-borne colistin resistance gene mcr-1 was firstly reported in an Escherichia coli isolate [5]. MCR-1 is a phosphoethanolamine (pEtN) transferase that is able to modify the phosphate groups in lipid A [5], the catalysis of which could be offered mainly by mcr-1 from plasmids but also encoded by chromosome in the Enterobacteriaceae family [6].
Among countries with mcr-producing Salmonella enterica isolates, China was the most frequent one, and countries in Europe harbored a wide diversity of mcr variants [1]. The mcr gene was frequently identified in animal- and human-borne Salmonella enterica isolates [1]. Among more than 2600 Salmonella serotypes, Salmonella Typhimurium was the top serovar to carry the mcr genes [1]. Besides Salmonella, Escherichia coli, Klebsiella Pneumoniae, Klebsiella oxytoca, Cronobacter sakazakii, Kluyvera ascorbata, Shigella sonnei, Citrobacter freundii, Citrobacter braakii, Raoultella ornithinolytica, Proteus mirabilis, Aeromonas, Moraxella, and Enterobacter species were also found to be mcr-producing Gram-negative bacteria [3]. Furthermore, the mcr genes were widely distributed in various sample sources, including humans, animals, animal-origin foods, and environment [3].
Currently, a variety of mcr genes, including mcr-1 to -10, have been reported in bacteria [6]. Furthermore, mcr genes were also found to co-exist with carbapenem resistance genes such as blaVIM-1, blaNDM-5, and blaNDM-9 [7,8,9]. Hence, the quick spread of plasmid-mediated mcr genes poses a significant threat to public health, requiring global surveillance.
Here, we characterized the global distribution of mcr-positive Salmonella using a data set of 2279 Salmonella genomes from humans, foods, animals, and environment. Salmonella serotypes, mcr variants, and phylogenomic characteristics were further analyzed.
2. Results
2.1. Widely Spread of mcr-Producing Salmonella Enterica in the Global and Sources Analysis
Currently, a total of 2279 mcr-producing Salmonella genomes are available in the NCBI database. It was found that these mcr-producing Salmonella isolates had spread in 37 countries on five continents, including Asia, Europe, America, Australia, and Africa (Figure 1A). The top six countries are the United States (n = 1187), China (n = 256), United Kingdom (n = 211), Germany (n = 184), Australia (n = 79), and Canada (n = 28) (Figure 1A).
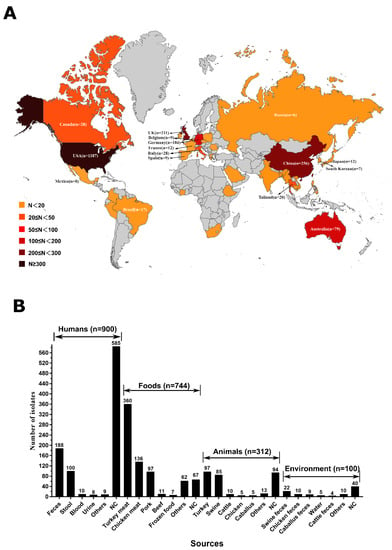
Figure 1.
Global spread of MCR-producing Salmonella enterica isolates (A) and sources analysis (B).
In this study, human samples (39.5%; 900/2279) were the predominant sources of mcr-producing Salmonella isolates (Figure 1B). It was further demonstrated that human samples were mainly composed of feces (n = 188), stool (n = 100), blood (n = 10), and urine (n = 8). A total of 744 (32.6%) isolates were recovered from foods. Turkey meats (n = 360) accounted for the largest portion, and then chicken meat (n = 136), pork (n = 97), beef (n = 11), and frozen foods (n = 7) (Figure 1B), which suggested that turkey meats were the main dissemination vehicle of mcr-producing Salmonella isolates. A total of 312 (13.7%) isolates were recovered from animals. Turkey (n = 97) accounted for the largest portion, and then swine (n = 85), cattle (n = 10), chicken (n = 5), and caballus (n = 5). In addition, 100 (4.4%) isolates were recovered from the environment, such as swine feces (n = 22), chicken feces (n = 10), caballus feces (n = 9), water (n = 5), and cattle feces (n = 4).
2.2. Typhimurium and 1,4,[5],12:i:- Were the Main Serotypes Carrying mcr Genes in Salmonella
We collected and analyzed Salmonella serotypes in these 2279 isolates, and the missing serotypes information in some isolates was compensated by the predicted results from SeqSero 1.2 software (UGA College of Agricultural and Environmental Sciences food science, Athens, GA, USA). A total of 80 Salmonella serotypes were identified among these Salmonella isolates. Typhimurium and 1,4,[5],12:i:- were the predominant serotypes, accounting for 18.3% and 18.7%, respectively (Figure 2A). Other serotypes were Saintpaul (12.4%), Heidelberg (9.1%), 4,[5],12:i:- (5.5%), Agona (4.1%), Paratyphi B var. d-tartrate+ (3.9%), Schwarzengrund (2.0%), Senftenberg (1.7%), Enteritidis (1.5%), Montevideo (1.5%), Mbandaka (1.4%), Cubana (1.2%), Ouakam (1.0%), Infantis (1.0%), Kentucky (0.9%), Thompson (0.9%), Anatum (0.9%), and Java (0.9%) (Figure 2A).
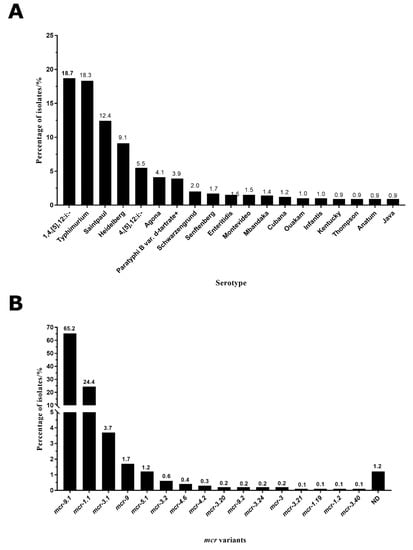
Figure 2.
The distribution of the serotypes (A) and mcr variants (B) in MCR-producing Salmonella.
It was found that the prevalent mcr-producing Salmonella serotypes varied in different countries (Figure 3A). In the United States, the most common Salmonella serotype was Saintpaul (21.4%), followed by Heidelberg (14.2%), 1,4,[5],12:i:- (12.7%), 4,[5],12:i:- (8.3%), Agona (5.9%), Schwarzengrund (3.0%), and Typhimurium (2.7%) (Figure 3A). In China, the most frequently identified Salmonella serotype was 1,4,[5],12:i:- (45.7%), followed by Typhimurium (38.7%), Anatum (3.5%), Thompson (2.0%), Goldcoast (2.0%), Indiana (1.2%), and Albany (1.2%). In the United Kingdom, the most common Salmonella serotype was Typhimurium (40.8%), followed by 1,4,[5],12:i:- (20.9%), Java (8.1%), Mbandaka (3.8%), Enteritidis (2.8%), Bovismorbificans (1.9%), and Choleraesuis (1.9%). In the Germany, the most frequently identified Salmonella serotype was Paratyphi B var. d-tartrate+ (45.7%), followed by Typhimurium (11.4%), 1,4,[5],12:i:- (10.9%), Paratyphi B (7.1%), Infantis (4.9%), Saintpaul (3.3%), and Ohio (3.3%). In Australia, the most common Salmonella serotype was Typhimurium (59.5%), followed by 1,4,[5],12:i:- (29.1%), Enteritidis (5.1%), Senftenberg (3.8%), and Havana (1.3%). In Canada, the most frequently identified Salmonella serotype was Heidelberg (50.0%), followed by 4,[5],12:i:- (14.3%), 1,4,[5],12:i:- (10.7%), Albany (7.1%), Typhimurium (7.1%), and Agona (3.6%).
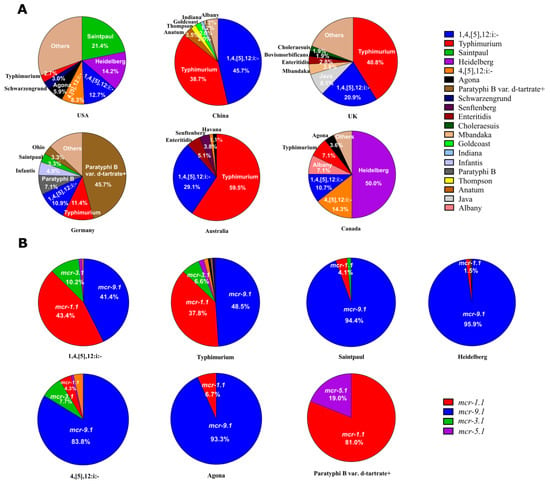
Figure 3.
(A) The distribution of mcr-positive Salmonella serotypes in the USA, China, UK, Germany, Australia, and Canada. (B) The distribution of mcr variants in serotypes 1,4,[5],12:i:-, Typhimurium, Saintpaul, Heidelberg, 4,[5],12:i:-, Agona, and Paratyphi B var. d-tartrate+.
2.3. mcr-9.1 and mcr-1.1 Were the Dominant Variants in Salmonella
We identified the mcr variants from genomes through ResFinder 4.1 (Lyngby, Denmark). A total of 20 mcr variants were identified, and the most common variant was mcr-9.1, accounting for 65.2% (Figure 2B). Other mcr variants were mcr-1.1 (24.4%), mcr-3.1 (3.7%), mcr-9 (1.7%), mcr-5.1 (1.2%), mcr-3.2 (0.6%), mcr-4.6 (0.4%), mcr-4.2 (0.3%), mcr-3.20 (0.2%), mcr-9.2 (0.2%), mcr-3.24 (0.2%), mcr-3 (0.2%), mcr-3.21 (0.1%), mcr-1.19 (0.1%), mcr-1.2 (0.1%), and mcr-3.40 (0.1%). The difference of mcr variant sequences could be found in Supplementary Materials. The partial or incomplete mcr genes could not be confirmed to a specific subtype, which were defined as not determined (ND) in Figure 2B, accounting for 1.2%.
Then, we analyzed the prevalence of mcr variants in the top seven Salmonella serotypes (1,4,[5],12:i:-, Typhimurium, Saintpaul, Heidelberg, 4,[5],12:i:-, Agona, and Paratyphi B var. d-tartrate+). It was found that mcr-9.1 was the most common variant in these serotypes except for 1,4,[5],12:i:- (Figure 3B). Gene mcr-1.1 (43.4%) was the most frequently identified variant in Salmonella 1,4,[5],12:i:-, followed by mcr-9.1 (41.4%), and mcr-3.1 (10.2%). Gene mcr-9.1 (48.5%) was the most common variant in Salmonella Typhimurium, followed by mcr-1.1 (37.8%), and mcr-3.1 (6.6%). Gene mcr-9.1 (94.4%) was the most frequently identified variant in Salmonella Saintpaul, and then mcr-1.1 (4.1%). Gene mcr-9.1 (95.9%) was the most common variant in Salmonella Heidelberg, and then mcr-1.1 (1.5%). Gene mcr-9.1 (83.8%) was the most frequently identified variant in Salmonella 4,[5],12:i:-, followed by mcr-3.1 (7.7%), and mcr-1.1 (4.3%). Gene mcr-9.1 (93.3%) was the most common variant in Salmonella Agona, and then mcr-1.1 (6.7%). Gene mcr-1.1 (81.0%) was the most frequently identified variant in Salmonella Paratyphi B var. d-tartrate+, and then mcr-5.1 (19.0%).
2.4. Antimicrobial Resistance (AMR) Genotypes in mcr-Producing Salmonella Isolates
A total of 68 acquired AMR genes were found in these mcr-producing Salmonella isolates (Table 1). Carbapenems-resistance gene blaNDM-1 was identified in one isolate. It was noted that a tigecycline-resistance gene tet(X4) was identified in one isolate. Furthermore, a total of 8 blaCTX-M variants were identified, blaCTX-M-14 (6.0%) of which were the most common one, followed by blaCTX-M-9 (4.8%), blaCTX-M-55 (3.9%), blaCTX-M-15 (0.8%), blaCTX-M-65 (0.6%), blaCTX-M-3 (0.6%), blaCTX-M-2 (0.2%), and blaCTX-M-130 (0.1%). Gene mph(A) (5.0%) was the most common macrolides-resistance gene, then erm(B) (0.4%), mef(B) (0.3%), and erm(42) (0.2%). Fosfomycin-resistance gene fosA7 and fosA3 accounted for 14.3% and 6.0%, respectively. Gene qnrB2 (9.4%) was the most common plasmid-mediated quinolones-resistance (PMQR) gene, followed by oqxAB (7.9%), qnrS1 (7.0%), qnrB19 (2.6%), qnrB4 (1.4%), and qnrS2 (0.4%). Tetracycline-resistance genes tet(B) (48.2%), tet(A) (25.4%), tet(D) (16.1%), tet(M) (3.1%), and tet(C) (1.5%) were identified. Mutations in genes associated with AMR from genomes were also identified. It was found that gyrA_D87G (5.6%) was the most common mutation, followed by gyrA_D87N (4.3%), gyrA_S83Y (2.5%), gyrA_S83F (2.5%), parC_S80I (0.5%), and parC_S80R (0.2%). In addition, gyrA_G81C and parE_S458P were identified in one isolate, respectively.

Table 1.
AMR genotypes in mcr-producing Salmonella isolates.
2.5. Phylogenomic Analysis of mcr-Producing Salmonella
To provide the evolutional characteristics of mcr-producing Salmonella, we compared 2145 genomes of different Salmonella serotypes from different countries. A total of 25,735 core SNPs were identified to construct a maximum likelihood tree (Figure 4). The evolutional strategy was based on serotypes. Currently, among 2145 isolates, the gene mcr (mcr-5.1) could be traced back to a Salmonella Typhimurium isolate in 1985, which was recovered from Bos taurus in Japan. It was found that Typhimurium and 1,4,[5],12:i:- as well as the close serotypes such as Saintpaul, Heidelberg and 4,[5],12:i:- were the main serotypes carrying mcr genes in Salmonella (Figure 4). The Salmonella Typhimurium, 1,4,[5],12:i:- and 4,[5],12:i:- isolates were distributed in Lineages I, II, and III. The phylogenetic tree showed that Lineages I and II were mixed clusters, suggesting their close genetic relationship. Lineage I was mainly composed of Salmonella 4,[5],12:i:- and 1,4,[5],12:i:- isolates. Lineage II was mainly composed of Salmonella 1,4,[5],12:i:- as well as some Salmonella 4,[5],12:i:- and Salmonella Typhimurium isolates. Lineage III was almost composed of Salmonella Typhimurium isolates. Furthermore, Salmonella Typhimurium isolates from the United Kingdom and Australia in Lineage III were divided into two sub-clades, respectively. It was found that most isolates of Lineage II were from China, but the base isolates were recovered from Germany, suggesting that mcr-producing Salmonella 1,4,[5],12:i:- isolates might be introduced into China from Germany. Similar results were also found in Lineage IV. Lineage IV was composed of Salmonella Saintpaul isolates from the USA, and their evolutionary path implied that a major introduction might occur in the USA from Germany and then nationwide spread. Indeed, the predominant serotype carrying mcr genes in Germany was Salmonella Paratyphi B var. d-tartrate+, which could be found in Lineage VI. We also found that isolates from Lineages I, II, V, VII, VIII, and IX were mostly isolated after 2013, but those from Lineages IV and VI were mainly isolated from 2002 to 2013. In addition, isolates from different sources shared a close genetic relationship, which suggested that clonal spread occurred. For example, most isolates of Lineage II were recovered from humans but also clustered with some isolates from animals. Similar results were also found in isolates of Lineages I and III.
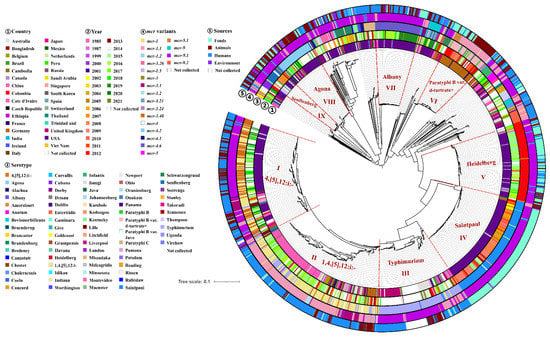
Figure 4.
Phylogenetic tree of 2145 Salmonella genomes from different countries. Circles ①–⑤ denote countries, years, serotypes, mcr variants, and sources, respectively. The detailed information in circles ①–⑤ using various colors shown in the key.
3. Discussion
Colistin had been considered a therapeutic drug and feed additive for animals since the early 1980s and was approved for clinical treatment of human beings in 2017 in China [10,11]. In a previous study, mcr-1-producing E. coli isolates were able to transfer from animals to humans through the chicken production chain [12]. E. coli, E. cloacae, Salmonella spp., and K. pneumonia were reported to be the dominant mcr-1-producing bacteria members [1,5,11,13]. China’s government has banned colistin as a feed additive to prevent colistin resistance in clinics, and then the drug production decreased markedly from 27,170 tons in 2015 to 2,497 tons in 2018, and the corresponding sale also decreased from USD 71.5 million in 2015 to USD 8.0 million in 2018 in China [14]. Furthermore, the median relative abundance of mcr-1 per 16S RNA in E. coli from animals and humans decreased significantly from 0.0009 in 2017 to 0.0002 in 2018 [14]. Therefore, the ban on colistin in animal farming causes a significant decrease in mcr-1 of E. coli isolates from animals and humans.
The emergence of plasmid-borne mcr genes has attracted significant attention worldwide [3,5]. In a previous study about the prevalence of mcr-producing bacterial isolates, it was found that a significant increase occurred in 2009 [3]. The chicken-borne mcr-1-producing E. coli isolates increased from 5.2% in 2009 to 30.0% in 2014 [15]. In a retrospective survey from China, the earliest chicken-borne mcr-1-producing E. coli isolate was found in the 1980s, which corresponded to the start use of colistin in animal husbandry as a feed additive [15]. In this study, the earliest emergence of mcr was identified from a Salmonella Typhimurium isolate from Bos taurus in 1985 (Japan, mcr-5.1), and the earliest mcr gene from humans was identified in a Salmonella 1,4,[5],12:i:- isolate in 2001 (the USA, mcr-9.1). Up to 1 January 2022, a total of 2279 mcr-positive Salmonella genomes were available from the NCBI database and were widely spread in 37 countries. Therefore, it was necessary to monitor the prevalence of mcr-1 in Salmonella to evaluate its burden on human health.
In this study, 80 Salmonella serotypes were identified to be carrying mcr genes, among which Typhimurium (18.3%) and 1,4,[5],12:i:- (18.7%) were the predominant ones. S. Typhimurium and 1,4,[5],12:i:- generally exhibited high-level prevalence and strong MDR features and spread worldwide [16]. Furthermore, the top three mcr variants in both Typhimurium and 1,4,[5],12:i:- were mcr-1.1, mcr-9.1, and mcr-3.1. Typhimurium and 1,4,[5],12:i:- were the predominant serotypes carrying mcr genes in China, the UK, and Australia. Furthermore, the phylogenetic relationship of S. Typhimurium and 1,4,[5],12:i:- isolates from these three countries was close, suggesting the possibility of the international spread among them. Unlike China, 1,4,[5],12:i:- (12.7%) was the third serotype carrying mcr genes in the USA, lower than Saintpaul (21.4%) and Heidelberg (14.2%). In Germany, Paratyphi B var. d-tartrate+ was the predominant serotype carrying mcr genes, and the top 2 mcr variants were mcr-1.1 and mcr-5.1. Therefore, the prevalence of Salmonella serotypes and their mcr variants showed a wide variety in different geographical regions.
Here are some limitations of this study. First, the numerous genomes used in this study were associated with the development of sequencing technology. The genome information used was hard to represent the real prevalence of mcr-producing isolates. Second, the majority of the mcr-producing Salmonella genomes originated from the United States, China, the United Kingdom, and Germany, but data were limited from other countries. Therefore, the above limitations would bias the deduced results.
In conclusion, our study highlighted that mcr-producing Salmonella isolates have spread in five continents (Asia, Europe, America, Australia, and Africa) globally. Typhimurium and 1,4,[5],12:i:- were the predominant serotypes carrying mcr genes in Salmonella, and mcr-9.1 and mcr-1.1 were the predominant variants. The spread of such various mcr variants is of great concern for food safety and public health, and it is urgent to enhance surveillance and control the spread of mcr genes among Salmonella.
4. Materials and Methods
4.1. Salmonella Genomes Collected
We searched two important words, “mcr” and “Salmonella”, in the public NCBI database on 1 January 2022. A total of 2279 Salmonella isolates were positive for mcr genes, and the detailed information of isolates, including years, countries, host, and serotypes, were then downloaded. However, 2145 out of 2279 genomes were available because the rest had not been released. These genomes were used for phylogenetic analysis to explore the implied spread of MCR-producing Salmonella worldwide.
4.2. The Identification of mcr Variants and Serotypes
The genomes used were further to identify the variant types of mcr genes through ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/, accessed on 1 June 2022). Salmonella serotypes were available from the uploaded information, and the serotypes that were not submitted by the uploader were predicted by SeqSero 1.2 (https://cge.cbs.dtu.dk/services/SeqSero/, accessed on 1 June 2022).
4.3. Phylogenetic Analysis
A total of 2145 Salmonella genomes from different countries were used for phylogenetic analysis. Single-nucleotide polymorphisms (SNPs) were extracted using Snippy (https://github.com/tseemann/snippy, accessed on 1 June 2022) to generate core genomic alignment. Gubbins [17] was then used to identify and remove recombination regions using an algorithm that iteratively identifies loci containing elevated densities of base substitutions, and then resulting pairwise SNP differences were calculated. The core SNP alignment was used to generate a maximum-likelihood phylogeny using RAxML v8.1.23 (Karlsruhe, Germany) [18] with the GTR nucleotide substitution model. The display, annotation, and management of phylogenetic trees were performed by the ITOL tool [19]. The antibiotic resistance genes in all genomes were identified by ResFinder 4.1 (Lyngby, Denmark) [20].
4.4. Data Analysis
The bar and circle charts used to analyze the prevalence of mcr serotypes were generated with GraphPad Prism 7 software (San Diego, California, CA, USA). The world map marked with the distribution of mcr-positive Salmonella was drawn by Inkscape 0.92 (New York, NY, USA).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11080998/s1, Comparison of mcr variant sequences from Salmonella (.fas profile).
Author Contributions
Conceptualization, Z.Z.; methodology, Z.Z. and X.T.; validation, Z.Z.; investigation, Z.Z.; data curation, Z.Z.; writing—original draft preparation, Z.Z.; writing—review and editing, Z.Z. and C.S.; supervision, C.S.; project administration, C.S.; funding acquisition, C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Shanghai Agriculture Applied Technology Development Program, China (grant number 2020-02-08-00-08-F01487) and the National Natural Science Foundation of China (grant number 31972169).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Portes, A.B.; Rodrigues, G.; Leitão, M.P.; Ferrari, R.; Conte Junior, C.A.; Panzenhagen, P. Global Distribution of Plasmid-Mediated Colistin Resistance Mcr Gene in Salmonella: A Systematic Review. J. Appl. Microbiol. 2022, 132, 872–889. [Google Scholar] [CrossRef]
- Wang, R.; van Dorp, L.; Shaw, L.P.; Bradley, P.; Wang, Q.; Wang, X.; Jin, L.; Zhang, Q.; Liu, Y.; Rieux, A.; et al. The Global Distribution and Spread of the Mobilized Colistin Resistance Gene Mcr-1. Nat. Commun. 2018, 9, 1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nang, S.C.; Li, J.; Velkov, T. The Rise and Spread of Mcr Plasmid-Mediated Polymyxin Resistance. Crit. Rev. Microbiol. 2019, 45, 131–161. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Morand, S.; Rolain, J.-M. Mechanisms of Polymyxin Resistance: Acquired and Intrinsic Resistance in Bacteria. Front. Microbiol. 2014, 5, 643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Hussein, N.H.; AL-Kadmy, I.M.S.; Taha, B.M.; Hussein, J.D. Mobilized Colistin Resistance (Mcr) Genes from 1 to 10: A Comprehensive Review. Mol. Biol. Rep. 2021, 48, 2897–2907. [Google Scholar] [CrossRef]
- Poirel, L.; Kieffer, N.; Liassine, N.; Thanh, D.; Nordmann, P. Plasmid-Mediated Carbapenem and Colistin Resistance in a Clinical Isolate of Escherichia coli. Lancet Infect. Dis. 2016, 16, 281. [Google Scholar] [CrossRef] [Green Version]
- Du, H.; Chen, L.; Tang, Y.-W.; Kreiswirth, B.N. Emergence of the Mcr-1 Colistin Resistance Gene in Carbapenem-Resistant Enterobacteriaceae. Lancet Infect. Dis. 2016, 16, 287–288. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Doi, Y.; Zeng, L.; Lv, L.; Liu, J.-H. Carbapenem-Resistant and Colistin-Resistant Escherichia coli Co-Producing NDM-9 and MCR-1. Lancet Infect. Dis. 2016, 16, 288–289. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Zhou, H.; Xu, J.; Wang, Y.; Zhang, Q.; Walsh, T.R.; Shao, B.; Wu, C.; Hu, Y.; Yang, L.; et al. Anthropogenic and Environmental Factors Associated with High Incidence of Mcr-1 Carriage in Humans across China. Nat. Microbiol. 2018, 3, 1054–1062. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, G.-B.; Zhang, R.; Shen, Y.; Tyrrell, J.M.; Huang, X.; Zhou, H.; Lei, L.; Li, H.-Y.; Doi, Y.; et al. Prevalence, Risk Factors, Outcomes, and Molecular Epidemiology of Mcr-1-Positive Enterobacteriaceae in Patients and Healthy Adults from China: An Epidemiological and Clinical Study. Lancet Infect. Dis. 2017, 17, 390–399. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive Resistome Analysis Reveals the Prevalence of NDM and MCR-1 in Chinese Poultry Production. Nat. Microbiol. 2017, 2, 16260. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Li, X.; Chen, Y.; Jiang, Y.; Zhou, Z.; Zhang, H.; Sun, L.; Ruan, Z.; Feng, Y.; Akova, M.; et al. Prevalence of Mcr-1 in Escherichia coli and Klebsiella Pneumoniae Recovered from Bloodstream Infections in China: A Multicentre Longitudinal Study. Lancet Infect. Dis. 2017, 17, 400–410. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in Colistin Resistance and Mcr-1 Abundance in Escherichia coli of Animal and Human Origins Following the Ban of Colistin-Positive Additives in China: An Epidemiological Comparative Study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Shen, Y.; Shen, J.; Wu, C. Early Emergence of Mcr-1 in Escherichia coli from Food-Producing Animals. Lancet Infect. Dis. 2016, 16, 293. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Wan, Y.; Du, P.; Bai, L. The Epidemiology of Monophasic Salmonella Typhimurium. Foodborne Pathog. Dis. 2020, 17, 87–97. [Google Scholar] [CrossRef]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).