Analysis of a Library of Escherichia coli Transporter Knockout Strains to Identify Transport Pathways of Antibiotics
Abstract
:1. Introduction
2. Results
2.1. Antimicrobial Selection
2.2. Determination of Antibiotic Concentrations for High-Throughput Screening
2.3. Baseline Characteristics of Transporter Library
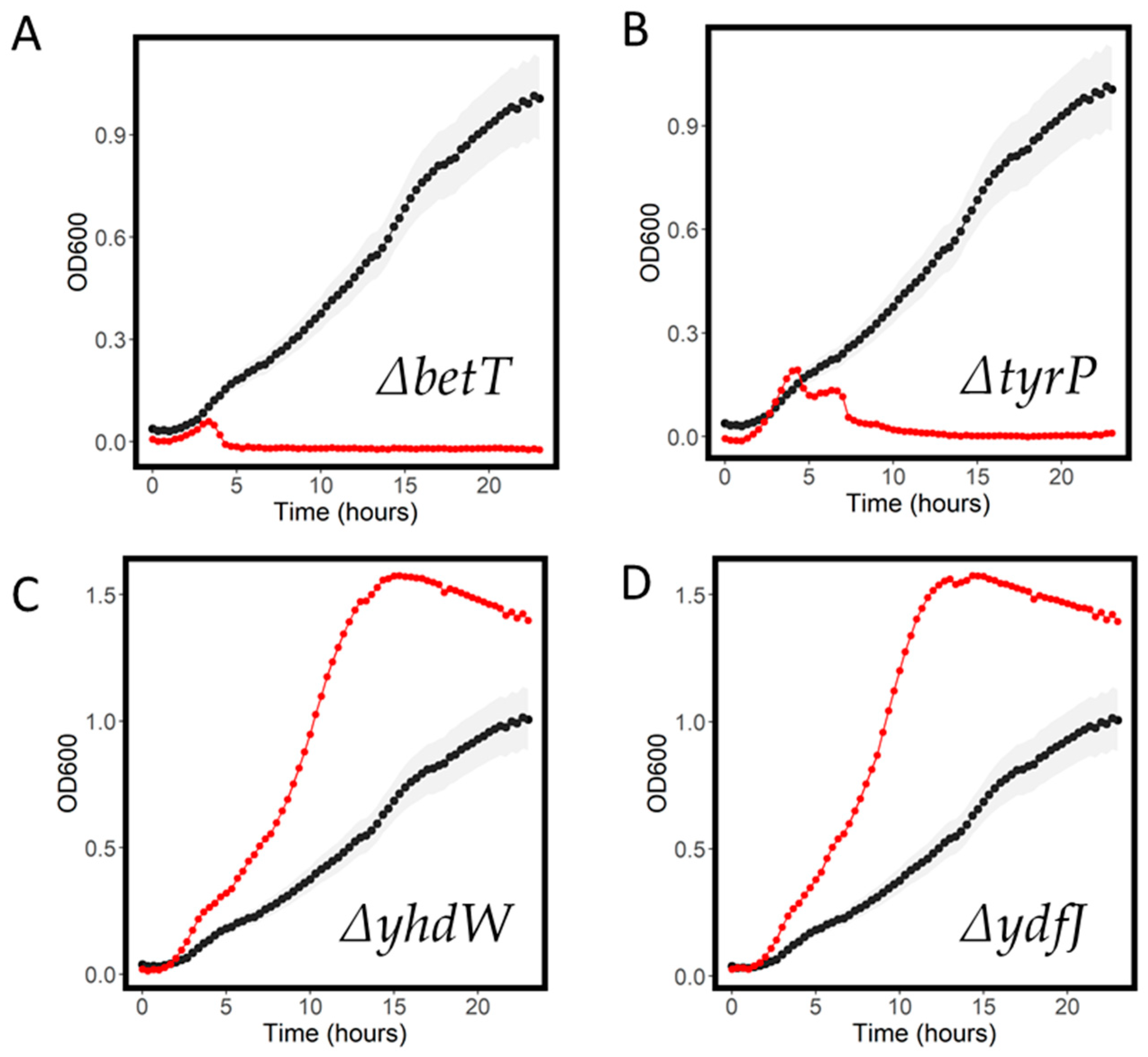
2.4. Resistant and Sensitive Transporter Results
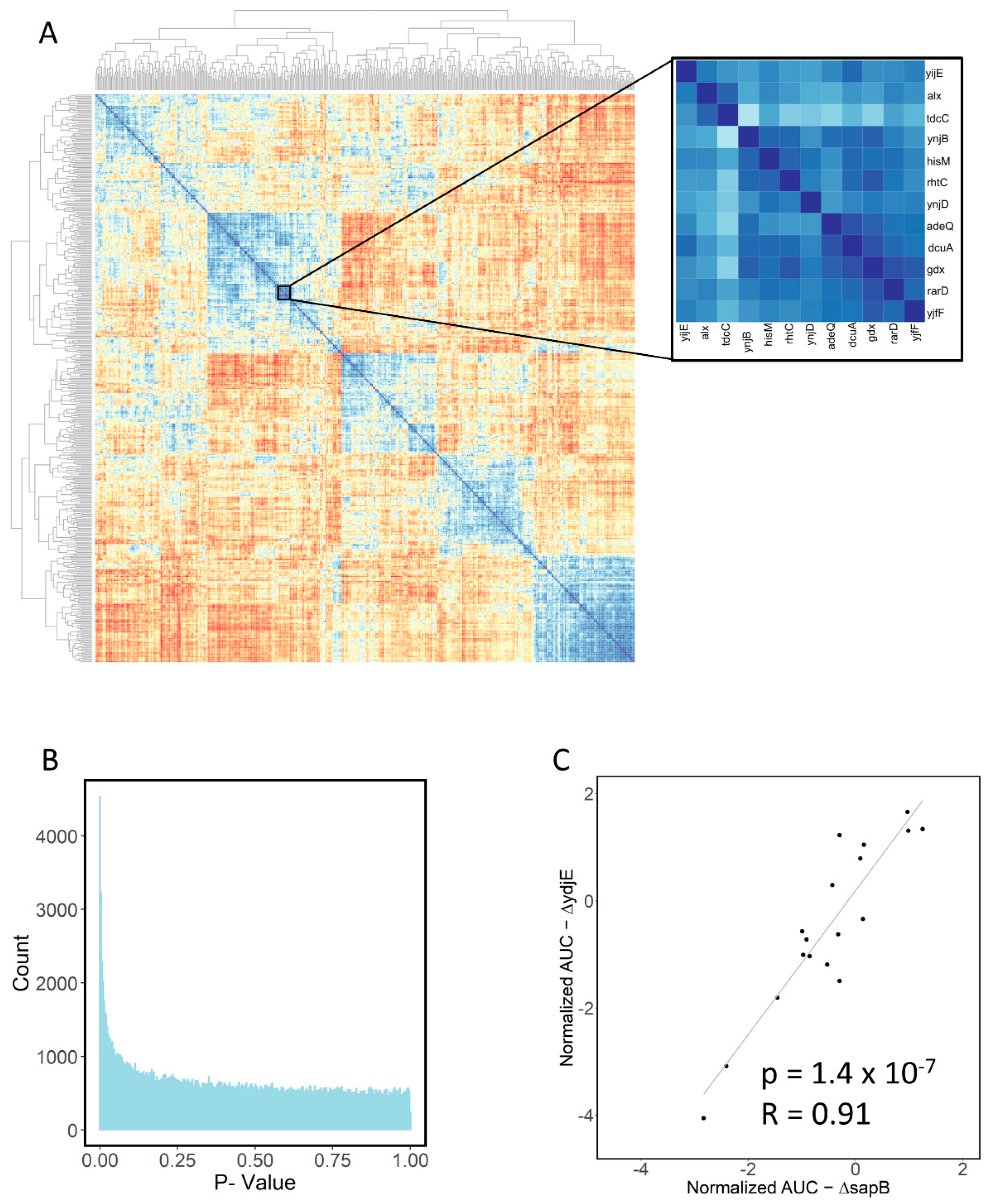
2.5. Large-Scale Data Analysis
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- WHO. Ten Threats to Global Health in 2019. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 8 July 2022).
- Spellberg, B.; Powers, J.H.; Brass, E.P.; Miller, L.G.; Edwards, J.J.E. Trends in Antimicrobial Drug Development: Implications for the Future. Clin. Infect. Dis. 2004, 38, 1279–1286. [Google Scholar] [CrossRef] [PubMed]
- Kalra, B.S.; Batta, A.; Khirasaria, R. Trends in FDA drug approvals over last 2 decades: An observational study. J. Fam. Med. Prim. Care 2020, 9, 105–114. [Google Scholar] [CrossRef]
- Glover, R.E.; Manton, J.; Willcocks, S.; Stabler, R.A. Subscription model for antibiotic development. BMJ 2019, 366, l5364. [Google Scholar] [CrossRef]
- Dobson, P.D.; Kell, D. Carrier-mediated cellular uptake of pharmaceutical drugs: An exception or the rule? Nat. Rev. Drug Discov. 2008, 7, 205–220. [Google Scholar] [CrossRef]
- Kell, D.B.; Oliver, S.G. How drugs get into cells: Tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front. Pharmacol. 2014, 5, 231. [Google Scholar] [CrossRef]
- Kell, D.B.; Dobson, P.D.; Bilsland, E.; Oliver, S.G. The promiscuous binding of pharmaceutical drugs and their transporter-mediated uptake into cells: What we (need to) know and how we can do so. Drug Discov. Today 2013, 18, 218–239. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Poole, K. Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 2007, 39, 162–176. [Google Scholar] [CrossRef]
- Willers, C.; Wentzel, J.; du Plessis, L.; Gouws, C.; Hamman, J.H. Efflux as a mechanism of antimicrobial drug resistance in clinical relevant microorganisms: The role of efflux inhibitors. Expert Opin. Ther. Targets 2017, 21, 23–36. [Google Scholar] [CrossRef]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Hayashi, K.; Nagata, C.; Hoshino, K.; Onodera, Y.; Nishino, K.; Yamaguchi, A. Structural basis for the inhibition of bacterial multidrug exporters. Nature 2013, 500, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Nishino, K.; Yamasaki, S.; Nakashima, R.; Zwama, M.; Hayashi-Nishino, M. Function and Inhibitory Mechanisms of Multidrug Efflux Pumps. Front. Microbiol. 2021, 12, 737288. [Google Scholar] [CrossRef] [PubMed]
- Mendes, P.; Girardi, E.; Superti-Furga, G.; Kell, D.B. Why Most Transporter Mutations That Cause Antibiotic Resistance Are to Efflux Pumps Rather than to Import Transporters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Castañeda-García, A.; Blázquez, J.; Rodríguez-Rojas, A. Molecular Mechanisms and Clinical Impact of Acquired and Intrinsic Fosfomycin Resistance. Antibiotics 2013, 2, 217–236. [Google Scholar] [CrossRef] [PubMed]
- Taber, H.W.; Mueller, J.P.; Miller, P.F.; Arrow, A.S. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 1987, 51, 439–457. [Google Scholar] [CrossRef] [PubMed]
- Prabhala, B.K.; Aduri, N.G.; Sharma, N.; Shaheen, A.; Sharma, A.; Iqbal, M.; Hansen, P.R.; Brasen, C.; Gajhede, M.; Rahman, M.; et al. The prototypical proton-coupled oligopeptide transporter YdgR from Escherichia coli facilitates chloramphenicol uptake into bacterial cells. J. Biol. Chem. 2018, 293, 1007–1017. [Google Scholar] [CrossRef]
- Elbourne, L.D.H.; Tetu, S.G.; Hassan, K.A.; Paulsen, I.T. TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 2017, 45, D320–D324. [Google Scholar] [CrossRef]
- Jindal, S.; Yang, L.; Day, P.J.; Kell, D.B. Involvement of multiple influx and efflux transporters in the accumulation of cationic fluorescent dyes by Escherichia coli. BMC Microbiol. 2019, 19, 195. [Google Scholar] [CrossRef]
- Yang, L.; Malla, S.; Özdemir, E.; Kim, S.H.; Lennen, R.; Christensen, H.B.; Christensen, U.; Munro, L.J.; Herrgård, M.J.; Kell, D.B.; et al. Identification and Engineering of Transporters for Efficient Melatonin Production in Escherichia coli. Front. Microbiol. 2022, 13, 880847. [Google Scholar] [CrossRef]
- Yamamoto, N.; Nakahigashi, K.; Nakamichi, T.; Yoshino, M.; Takai, Y.; Touda, Y.; Furubayashi, A.; Kinjyo, S.; Dose, H.; Hasegawa, M.; et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009, 5, 335. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed]
- Kobylka, J.; Kuth, M.S.; Müller, R.T.; Geertsma, E.R.; Pos, K.M. AcrB: A mean, keen, drug efflux machine. Ann. N. Y. Acad. Sci. 2020, 1459, 38–68. [Google Scholar] [CrossRef] [PubMed]
- Kahan, F.M.; Kahan, J.S.; Cassidy, P.J.; Kropp, H. The mechanism of action of fosfomycin (phosphonomycin). Ann. N. Y. Acad. Sci. 1974, 235, 364–386. [Google Scholar] [CrossRef] [PubMed]
- Silver, L.L. Fosfomycin: Mechanism and Resistance. Cold Spring Harb. Perspect. Med. 2017, 7, a025262. [Google Scholar] [CrossRef]
- Kobayashi, N.; Nishino, K.; Yamaguchi, A. Novel Macrolide-Specific ABC-Type Efflux Transporter in Escherichia coli. J. Bacteriol. 2001, 183, 5639–5644. [Google Scholar] [CrossRef]
- Nichols, R.J.; Sen, S.; Choo, Y.J.; Beltrao, P.; Zietek, M.; Chaba, R.; Lee, S.; Kazmierczak, K.M.; Lee, K.J.; Wong, A.; et al. Phenotypic Landscape of a Bacterial Cell. Cell 2011, 144, 143–156. [Google Scholar] [CrossRef]
- Ogasawara, H.; Ohe, S.; Ishihama, A. Role of transcription factor NimR (YeaM) in sensitivity control of Escherichia coli to 2-nitroimidazole. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef]
- Nandineni, M.R.; Gowrishankar, J. Evidence for an Arginine Exporter Encoded by yggA (argO) That Is Regulated by the LysR-Type Transcriptional Regulator ArgP in Escherichia coli. J. Bacteriol. 2004, 186, 3539–3546. [Google Scholar] [CrossRef]
- Carr, R.J.; Bilton, R.F.; Atkinson, T. Toxicity of paraquat to microorganisms. Appl. Environ. Microbiol. 1986, 52, 1112–1116. [Google Scholar] [CrossRef]
- Broadhurst, D.I.; Kell, D.B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2006, 2, 171–196. [Google Scholar] [CrossRef]
- O’Hagan, S.; Kell, D.B. Structural Similarities between Some Common Fluorophores Used in Biology, Marketed Drugs, Endogenous Metabolites, and Natural Products. Mar. Drugs 2020, 18, 582. [Google Scholar] [CrossRef] [PubMed]
- Featherstone, D.E.; Broadie, K. Wrestling with pleiotropy: Genomic and topological analysis of the yeast gene expression network. BioEssays 2002, 24, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Ara, T.; Arifuzzaman, M.; Ioka-Nakamichi, T.; Inamoto, E.; Toyonaga, H.; Mori, H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Res. 2005, 12, 291–299. [Google Scholar] [CrossRef]
- Salcedo-Sora, J.E.; Robison, A.T.R.; Zaengle-Barone, J.; Franz, K.J.; Kell, D.B. Membrane Transporters Involved in the Antimicrobial Activities of Pyrithione in Escherichia coli. Molecules 2021, 26, 5826. [Google Scholar] [CrossRef] [PubMed]
- Salcedo-Sora, J.E.; Jindal, S.; O’Hagan, S.; Kell, D.B. A palette of fluorophores that are differentially accumulated by wild-type and mutant strains of Escherichia coli: Surrogate ligands for profiling bacterial membrane transporters. Microbiology 2021, 167, 001016. [Google Scholar] [CrossRef] [PubMed]
- Almeida, L.D.; Silva, A.S.F.; Mota, D.C.; Vasconcelos, A.A.; Camargo, A.P.; Pires, G.S.; Furlan, M.; da Cunha Freire, H.M.R.; Klippel, A.H.; Silva, S.F.; et al. Yeast Double Transporter Gene Deletion Library for Identification of Xenobiotic Carriers in Low or High Throughput. mBio 2021, 12, e0322121. [Google Scholar] [CrossRef]
- French, S.; Mangat, C.; Bharat, A.; Côté, J.-P.; Mori, H.; Brown, E.D. A robust platform for chemical genomics in bacterial systems. Mol. Biol. Cell 2016, 27, 1015–1025. [Google Scholar] [CrossRef]
- Takeuchi, R.; Tamura, T.; Nakayashiki, T.; Tanaka, Y.; Muto, A.; Wanner, B.L.; Mori, H. Colony-live—A high-throughput method for measuring microbial colony growth kinetics—Reveals diverse growth effects of gene knockouts in Escherichia coli. BMC Microbiol. 2014, 14, 171. [Google Scholar] [CrossRef]
- Vines, T.H.; Albert, A.; Andrew, R.; Débarre, F.; Bock, D.G.; Franklin, M.T.; Gilbert, K.; Moore, J.-S.; Renaut, S.; Rennison, D. The Availability of Research Data Declines Rapidly with Article Age. Curr. Biol. 2014, 24, 94–97. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef] [PubMed]
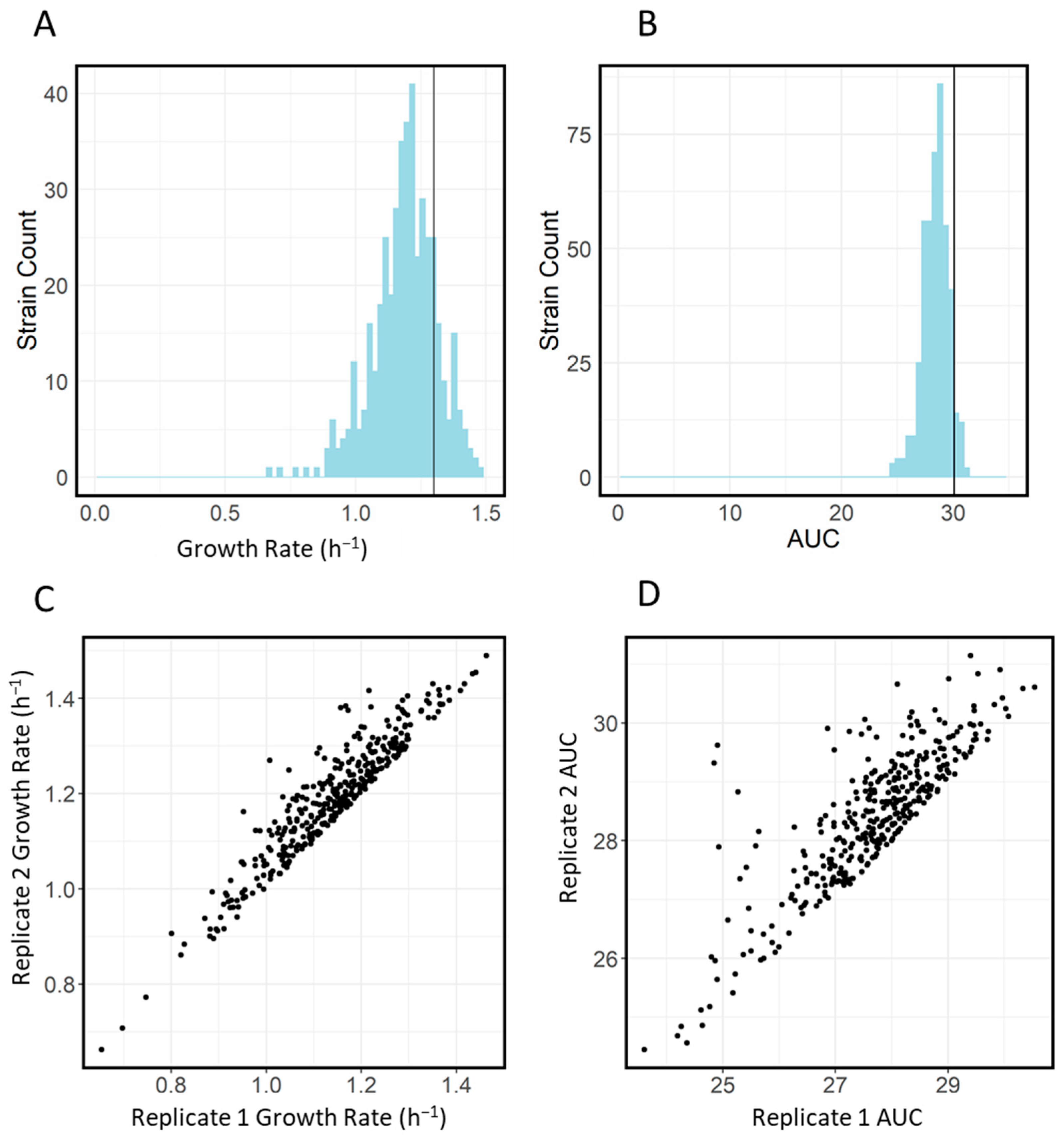

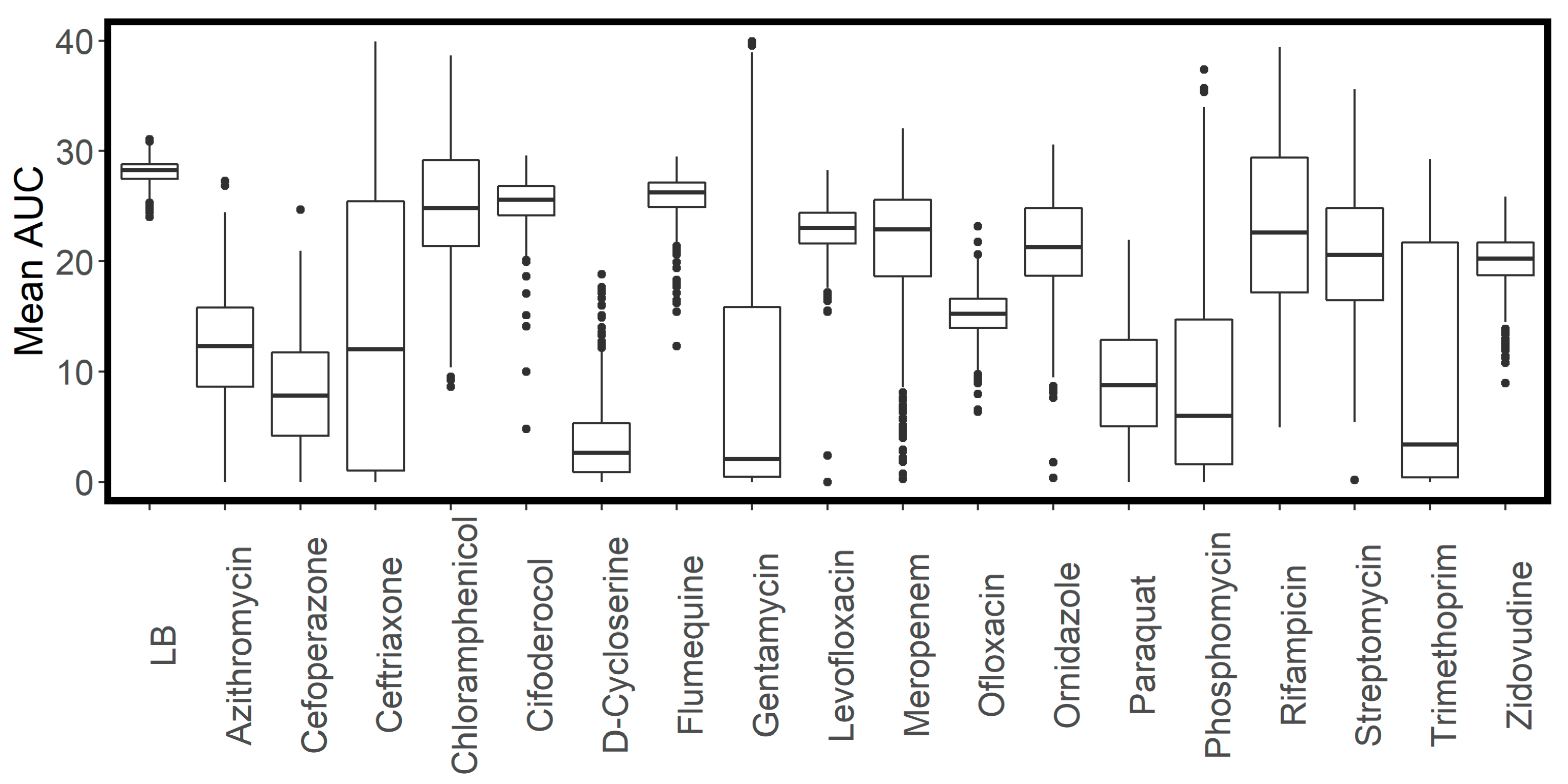
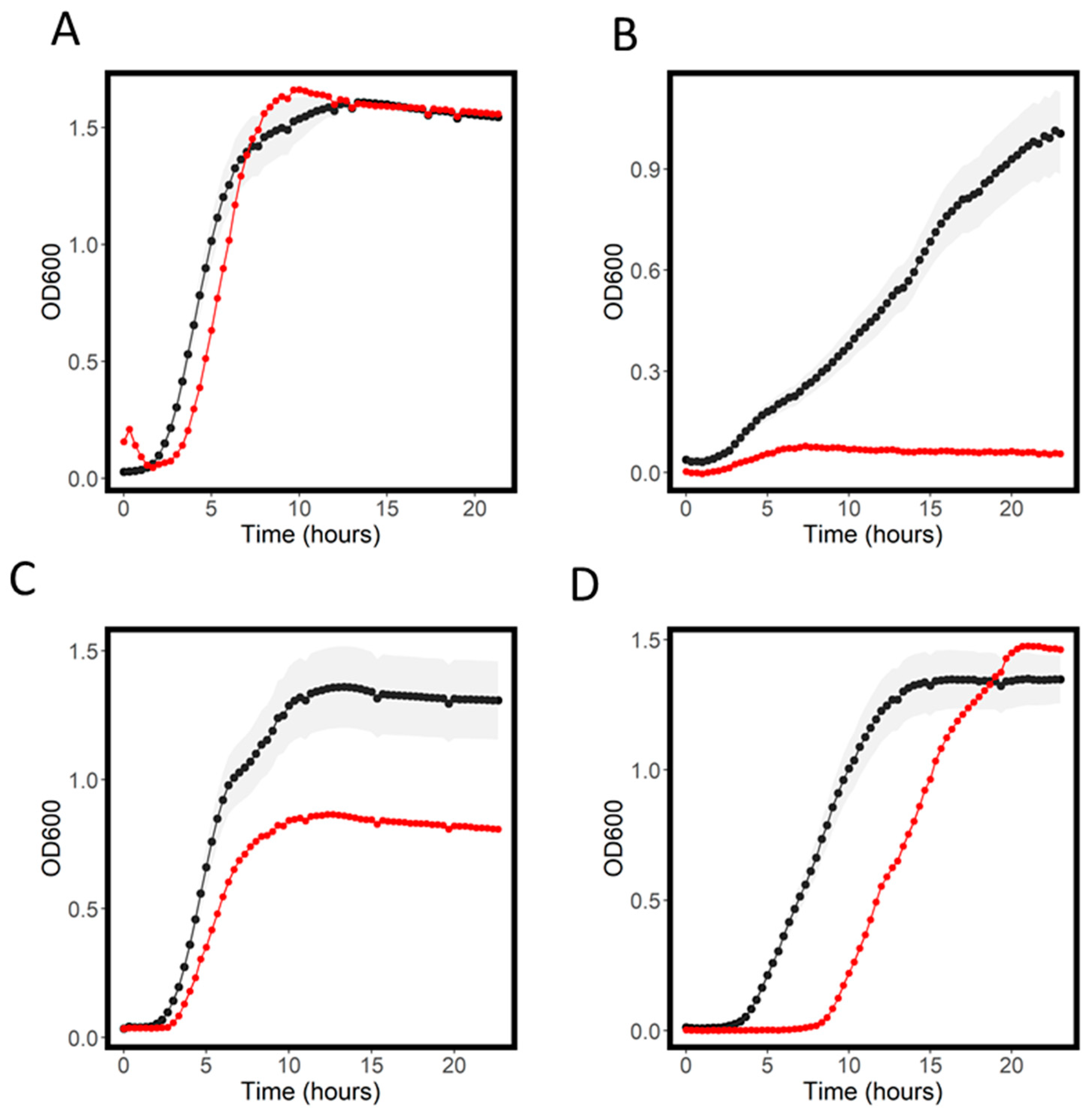
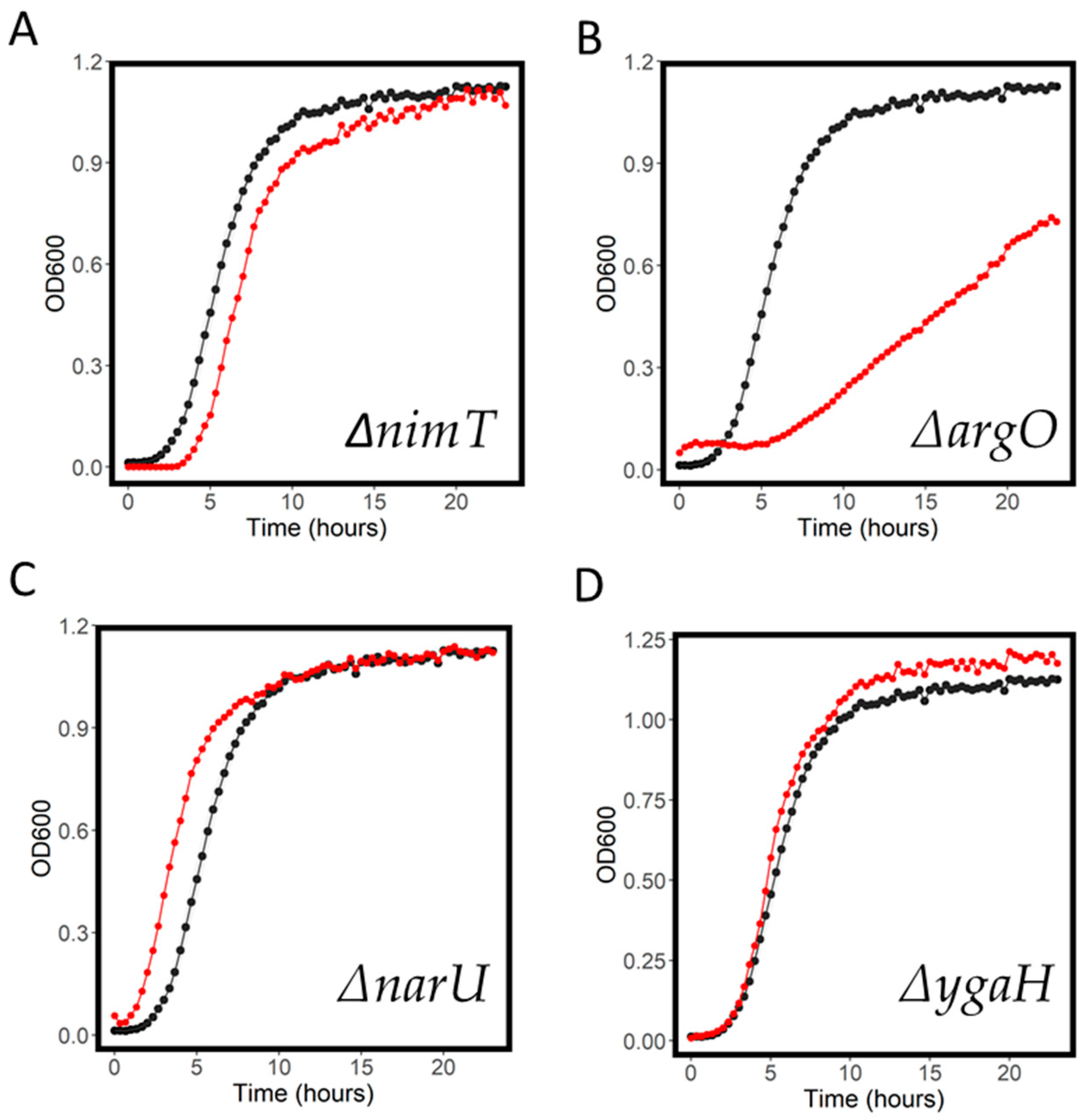

| Antibiotic | WT MIC (mg/L) | Screening Concentration (mg/L) |
|---|---|---|
| Azithromycin | 30 | 7.5 |
| Cefiderocol | 1 | 0.25 |
| Cefoperazone | 0.5 | 0.25 |
| Chloramphenicol | 2.5 | 1.25 |
| Ceftriaxone | 2.5 | 1 |
| D-Cycloserine | 12.5 | 6.25 |
| Flumequine | >1.5 * | 1.5 |
| Gentamycin | 4 | 2 |
| Levofloxacin | 0.25 | 0.125 |
| Meropenem | 0.0625 | 0.03125 |
| Ofloxaxin | 0.125 | 0.0625 |
| Ornidazole | 800 | 400 |
| Paraquat | >268 * | 67 |
| Phosphomycin | 50 | 25 |
| Rifampicin | 25 | 12.5 |
| Streptomycin | 25 | 6.25 |
| Trimethoprim | 2.3 | 0.25 |
| Zidovudine | >100 * | 100 |
| Strain | Max OD | Rate | AUC | Normalized Max OD | Normalized Rate | Normalized AUC |
|---|---|---|---|---|---|---|
| argO | 0.74 | 0.35 | 6.36 | 0.29 | 0.45 | 0.23 |
| ydjE | 0.58 | 0.71 | 6.55 | 0.71 | 0.37 | 0.26 |
| tolQ | 0.60 | 1.05 | 7.98 | 1.16 | 0.38 | 0.29 |
| glvB | 0.75 | 0.57 | 9.78 | 0.54 | 0.44 | 0.34 |
| sapB | 0.64 | 0.73 | 9.05 | 0.74 | 0.41 | 0.35 |
| yebQ | 0.83 | 0.59 | 8.96 | 0.89 | 0.53 | 0.36 |
| ddpF | 0.76 | 0.80 | 11.43 | 0.64 | 0.44 | 0.37 |
| uup | 0.72 | 0.85 | 10.07 | 0.81 | 0.45 | 0.37 |
| cysW | 0.64 | 1.62 | 10.22 | 1.40 | 0.41 | 0.38 |
| ydcS | 0.65 | 0.54 | 9.25 | 0.70 | 0.39 | 0.38 |
| ydeE | 0.78 | 0.89 | 11.00 | 0.74 | 0.48 | 0.39 |
| yicJ | 0.81 | 0.84 | 11.78 | 0.64 | 0.49 | 0.39 |
| guaB | 0.69 | 1.08 | 11.22 | 1.18 | 0.42 | 0.40 |
| gntU | 0.87 | 1.04 | 12.13 | 0.83 | 0.52 | 0.40 |
| ynfA | 0.77 | 0.93 | 9.97 | 1.31 | 0.52 | 0.41 |
| ybhF | 1.15 | 0.95 | 19.30 | 0.96 | 0.66 | 0.65 |
| arnE | 1.14 | 1.03 | 17.97 | 1.14 | 0.70 | 0.66 |
| sapC | 1.11 | 0.74 | 17.12 | 0.77 | 0.67 | 0.66 |
| cycA | 1.17 | 1.07 | 19.65 | 0.83 | 0.69 | 0.66 |
| narU | 1.14 | 0.86 | 19.88 | 0.67 | 0.67 | 0.66 |
| lacY | 1.19 | 0.87 | 20.61 | 0.71 | 0.68 | 0.67 |
| kup | 1.16 | 0.90 | 18.67 | 0.68 | 0.70 | 0.67 |
| mdtH | 1.24 | 0.80 | 18.59 | 0.89 | 0.75 | 0.67 |
| pitA | 1.25 | 1.01 | 18.19 | 0.78 | 0.77 | 0.67 |
| yadG | 1.09 | 0.84 | 17.65 | 0.66 | 0.66 | 0.67 |
| clcB | 1.14 | 0.86 | 18.96 | 0.67 | 0.69 | 0.68 |
| trkH | 1.20 | 0.76 | 18.90 | 0.56 | 0.74 | 0.68 |
| exbD | 1.22 | 0.92 | 20.52 | 0.79 | 0.70 | 0.70 |
| wt | 1.04 | 1.10 | 21.76 | 0.77 | 0.58 | 0.70 |
| dhaM | 1.34 | 0.79 | 23.18 | 0.63 | 0.80 | 0.78 |
| Strain | MaxOD | Rate | AUC | Normalized MaxOD | Normalized Rate | Normalized AUC |
|---|---|---|---|---|---|---|
| betT | 0.06 | 0.02 | 0.30 | 0.02 | 0.04 | 0.01 |
| glvB | 0.11 | 0.03 | 0.32 | 0.03 | 0.07 | 0.01 |
| glnP | 0.11 | 0.02 | 0.33 | 0.02 | 0.06 | 0.01 |
| citT | 0.06 | 0.04 | 0.37 | 0.04 | 0.04 | 0.01 |
| tyrP | 0.24 | 0.01 | 0.56 | 0.01 | 0.15 | 0.02 |
| corA | 0.14 | 0.03 | 0.62 | 0.02 | 0.08 | 0.02 |
| tolR | 0.11 | 1.69 | 0.67 | 1.46 | 0.07 | 0.02 |
| mdtN | 0.67 | 0.50 | 0.70 | 0.40 | 0.40 | 0.02 |
| ydjN | 0.34 | 0.44 | 0.76 | 0.38 | 0.20 | 0.03 |
| yibH | 0.20 | 0.01 | 0.77 | 0.00 | 0.12 | 0.03 |
| torT | 0.18 | 0.04 | 0.80 | 0.03 | 0.11 | 0.03 |
| acrB | 0.09 | 0.66 | 1.35 | 0.54 | 0.05 | 0.05 |
| kdgT | 2.16 | 0.92 | 1.48 | 0.75 | 1.33 | 0.05 |
| tolQ | 0.74 | 1.04 | 1.76 | 1.16 | 0.47 | 0.06 |
| ydcS | 0.31 | 0.28 | 1.61 | 0.36 | 0.18 | 0.07 |
| ddpD | 1.51 | 0.39 | 19.71 | 0.30 | 0.92 | 0.71 |
| ydiN | 1.53 | 0.44 | 20.19 | 0.36 | 0.96 | 0.71 |
| ydjX | 1.58 | 0.43 | 20.37 | 0.35 | 0.96 | 0.72 |
| yddB | 1.59 | 0.40 | 19.66 | 0.28 | 1.00 | 0.72 |
| atpB | 1.57 | 0.44 | 21.09 | 0.35 | 0.96 | 0.73 |
| yccA | 1.54 | 0.43 | 19.30 | 0.37 | 0.99 | 0.73 |
| ygaZ | 1.50 | 0.58 | 18.50 | 0.56 | 1.09 | 0.73 |
| cmtA | 1.54 | 0.41 | 20.84 | 0.36 | 0.95 | 0.74 |
| kgtP | 1.52 | 0.47 | 21.29 | 0.38 | 0.88 | 0.74 |
| pitA | 1.51 | 0.46 | 20.38 | 0.36 | 0.93 | 0.75 |
| ydfJ | 1.58 | 0.55 | 23.16 | 0.41 | 0.93 | 0.76 |
| yhdW | 1.58 | 0.48 | 22.07 | 0.34 | 0.95 | 0.76 |
| yraQ | 2.07 | 0.81 | 24.46 | 0.75 | 1.23 | 0.87 |
| yiaM | 2.51 | 0.51 | 26.88 | 0.42 | 1.50 | 0.90 |
| argO | 1.92 | 0.94 | 27.30 | 0.78 | 1.17 | 0.97 |
| Strain | MaxOD | Rate | AUC | Normalized MaxOD | Normalized Rate | Normalized AUC |
|---|---|---|---|---|---|---|
| ydcZ | 0.11 | 1.55 | 0.39 | 1.35 | 0.06 | 0.01 |
| potH | 0.74 | 0.57 | 1.79 | 0.50 | 0.44 | 0.06 |
| fieF | 1.46 | 1.20 | 7.66 | 0.92 | 0.85 | 0.26 |
| yihO | 1.35 | 1.07 | 8.11 | 0.88 | 0.79 | 0.29 |
| kdgT | 2.22 | 0.49 | 8.51 | 0.40 | 1.36 | 0.29 |
| cusA | 0.69 | 0.49 | 8.66 | 0.50 | 0.41 | 0.29 |
| yfdV | 0.78 | 0.60 | 8.42 | 0.64 | 0.47 | 0.30 |
| uhpC | 2.15 | 0.55 | 9.50 | 0.48 | 1.27 | 0.32 |
| uacT | 2.40 | 0.50 | 10.09 | 0.40 | 1.42 | 0.33 |
| atoS | 1.47 | 0.26 | 11.42 | 0.25 | 0.84 | 0.38 |
| yihN | 1.11 | 0.64 | 11.91 | 0.55 | 0.65 | 0.41 |
| ygjI | 0.97 | 0.53 | 11.79 | 0.41 | 0.58 | 0.42 |
| ygaH | 1.41 | 1.01 | 13.21 | 0.85 | 0.82 | 0.42 |
| mdtN | 0.96 | 0.46 | 12.28 | 0.37 | 0.57 | 0.43 |
| yidK | 1.03 | 1.04 | 12.84 | 0.75 | 0.60 | 0.43 |
| aroP | 2.15 | 0.60 | 28.71 | 0.51 | 1.30 | 1.03 |
| gsiC | 2.21 | 0.68 | 28.48 | 0.64 | 1.37 | 1.03 |
| yaaU | 2.15 | 0.55 | 28.40 | 0.47 | 1.33 | 1.03 |
| copA | 2.15 | 0.57 | 28.30 | 0.50 | 1.30 | 1.03 |
| araJ | 2.16 | 0.64 | 28.94 | 0.52 | 1.32 | 1.03 |
| dinF | 2.13 | 0.60 | 29.45 | 0.47 | 1.27 | 1.04 |
| codB | 2.14 | 0.58 | 29.43 | 0.47 | 1.27 | 1.04 |
| sbp | 2.01 | 0.67 | 27.00 | 0.53 | 1.27 | 1.04 |
| amtB | 2.29 | 0.60 | 29.30 | 0.53 | 1.41 | 1.05 |
| kdpA | 2.34 | 0.60 | 29.41 | 0.50 | 1.42 | 1.05 |
| yfeO | 2.01 | 0.64 | 28.17 | 0.49 | 1.22 | 1.06 |
| uidB | 2.25 | 0.55 | 30.25 | 0.44 | 1.36 | 1.06 |
| caiT | 2.34 | 0.57 | 30.46 | 0.45 | 1.41 | 1.06 |
| acrB | 2.15 | 0.52 | 29.25 | 0.43 | 1.28 | 1.07 |
| focA | 2.26 | 0.61 | 30.58 | 0.52 | 1.37 | 1.09 |
| Transporter 1 | Transporter 2 | Correlation | p Value |
|---|---|---|---|
| yihP | yhdX | 0.91 | 2.46 × 10−7 |
| ydjK | mdtB | 0.91 | 2.36 × 10−7 |
| uup | gsiC | 0.91 | 2.36 × 10−7 |
| yicL | yhdX | 0.91 | 2.26 × 10−7 |
| btuC | araJ | 0.91 | 2.22 × 10−7 |
| mdtG | copA | 0.91 | 2.04 × 10−7 |
| rarD | adeQ | 0.91 | 2.02 × 10−7 |
| mscK | ccmC | 0.91 | 1.98 × 10−7 |
| cynX | btuC | 0.91 | 1.72 × 10−7 |
| ybbA | cynX | 0.91 | 1.65 × 10−7 |
| mdfA | chaA | 0.91 | 1.49 × 10−7 |
| yjfF | gdx | 0.91 | 1.24 × 10−7 |
| ynjB | gdx | 0.91 | 1.18 × 10−7 |
| nhaB | ddpB | 0.91 | 1.16 × 10−7 |
| ydjE | sapB | 0.91 | 1.14 × 10−7 |
| yphD | gudP | 0.91 | 1.07 × 10−7 |
| ybaT | btuC | 0.92 | 9.3 × 10−8 |
| yjfF | adeQ | 0.92 | 8.62 × 10−8 |
| ypjA | fryA | 0.92 | 5.86 × 10−8 |
| rarD | gdx | 0.92 | 5.63 × 10−8 |
| malX | amtB | 0.92 | 5 × 10−8 |
| araJ | acrB | 0.92 | 4.29 × 10−8 |
| gsiC | fetA | 0.93 | 3.67 × 10−8 |
| yaaU | sapA | 0.93 | 3.54 × 10−8 |
| uidB | mdtG | 0.93 | 3.53 × 10−8 |
| yphD | ascF | 0.93 | 2.77 × 10−8 |
| yegT | frwC | 0.93 | 2.61 × 10−8 |
| ytfT | kup | 0.93 | 1.47 × 10−8 |
| copA | ccmC | 0.93 | 1.46 × 10−8 |
| yddB | ddpD | 0.94 | 7.5 × 10−9 |
| ybbA | mscK | 0.95 | 2.46 × 10−9 |
| mscK | fetA | 0.95 | 8.93 × 10−10 |
| cynX | araJ | 0.96 | 3.92 × 10−10 |
| gdx | adeQ | 0.97 | 5.67 × 10−11 |
| yihP | yicL | 0.97 | 7.51 × 10−12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munro, L.J.; Kell, D.B. Analysis of a Library of Escherichia coli Transporter Knockout Strains to Identify Transport Pathways of Antibiotics. Antibiotics 2022, 11, 1129. https://doi.org/10.3390/antibiotics11081129
Munro LJ, Kell DB. Analysis of a Library of Escherichia coli Transporter Knockout Strains to Identify Transport Pathways of Antibiotics. Antibiotics. 2022; 11(8):1129. https://doi.org/10.3390/antibiotics11081129
Chicago/Turabian StyleMunro, Lachlan Jake, and Douglas B. Kell. 2022. "Analysis of a Library of Escherichia coli Transporter Knockout Strains to Identify Transport Pathways of Antibiotics" Antibiotics 11, no. 8: 1129. https://doi.org/10.3390/antibiotics11081129
APA StyleMunro, L. J., & Kell, D. B. (2022). Analysis of a Library of Escherichia coli Transporter Knockout Strains to Identify Transport Pathways of Antibiotics. Antibiotics, 11(8), 1129. https://doi.org/10.3390/antibiotics11081129





