Characterization of Carbapenem-Resistant K. Pneumoniae Isolated from Intensive Care Units of Zagazig University Hospitals
Abstract
1. Introduction
2. Results
2.1. Identification of the Bacterial Isolates
2.2. Characteristics of K. pneumoniae Isolates
2.3. Antimicrobial Susceptibility Testing
2.4. Confirmation of Carbapenem Resistance
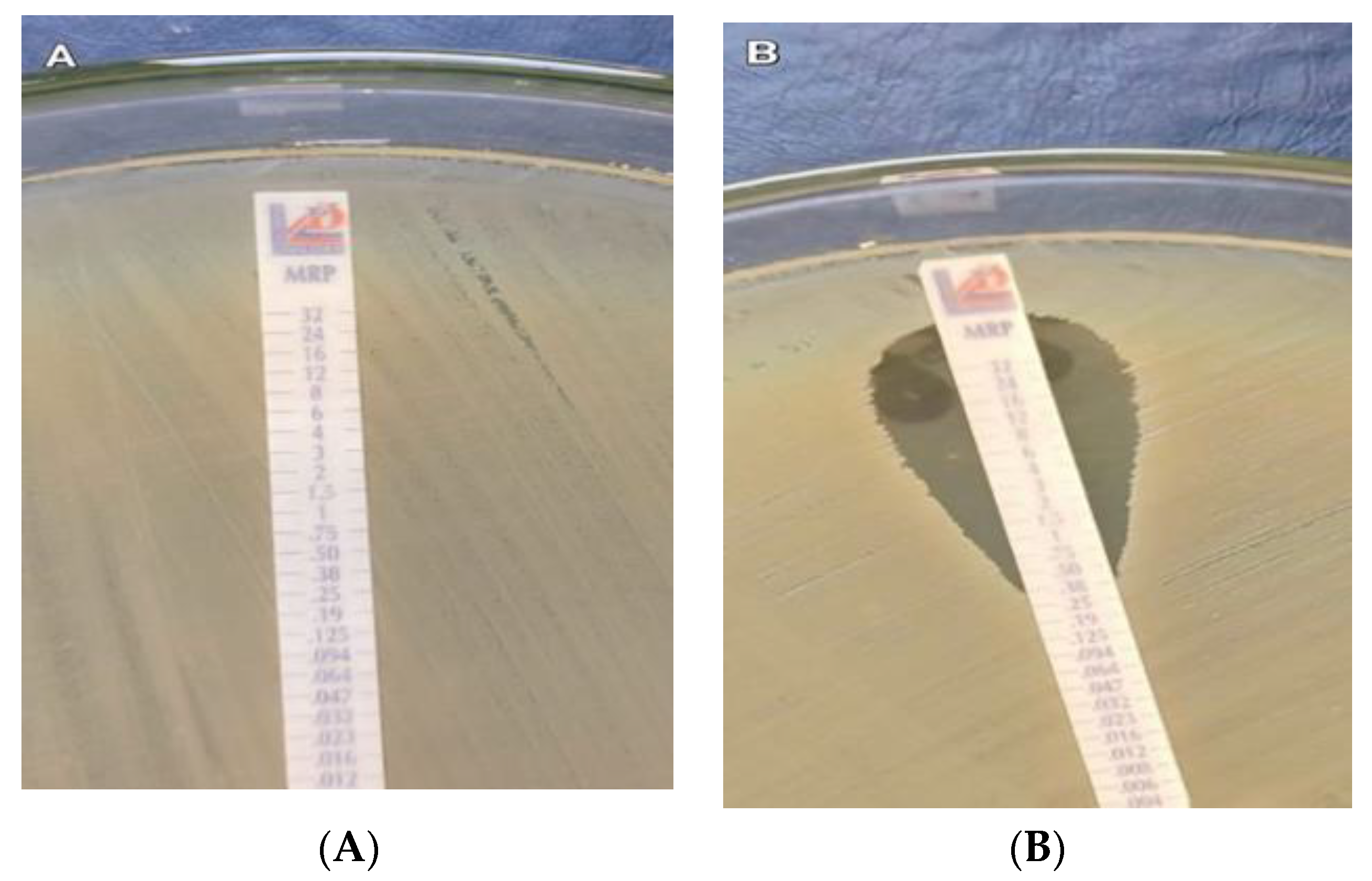
2.5. Phenotypic Carbapenemase Detection
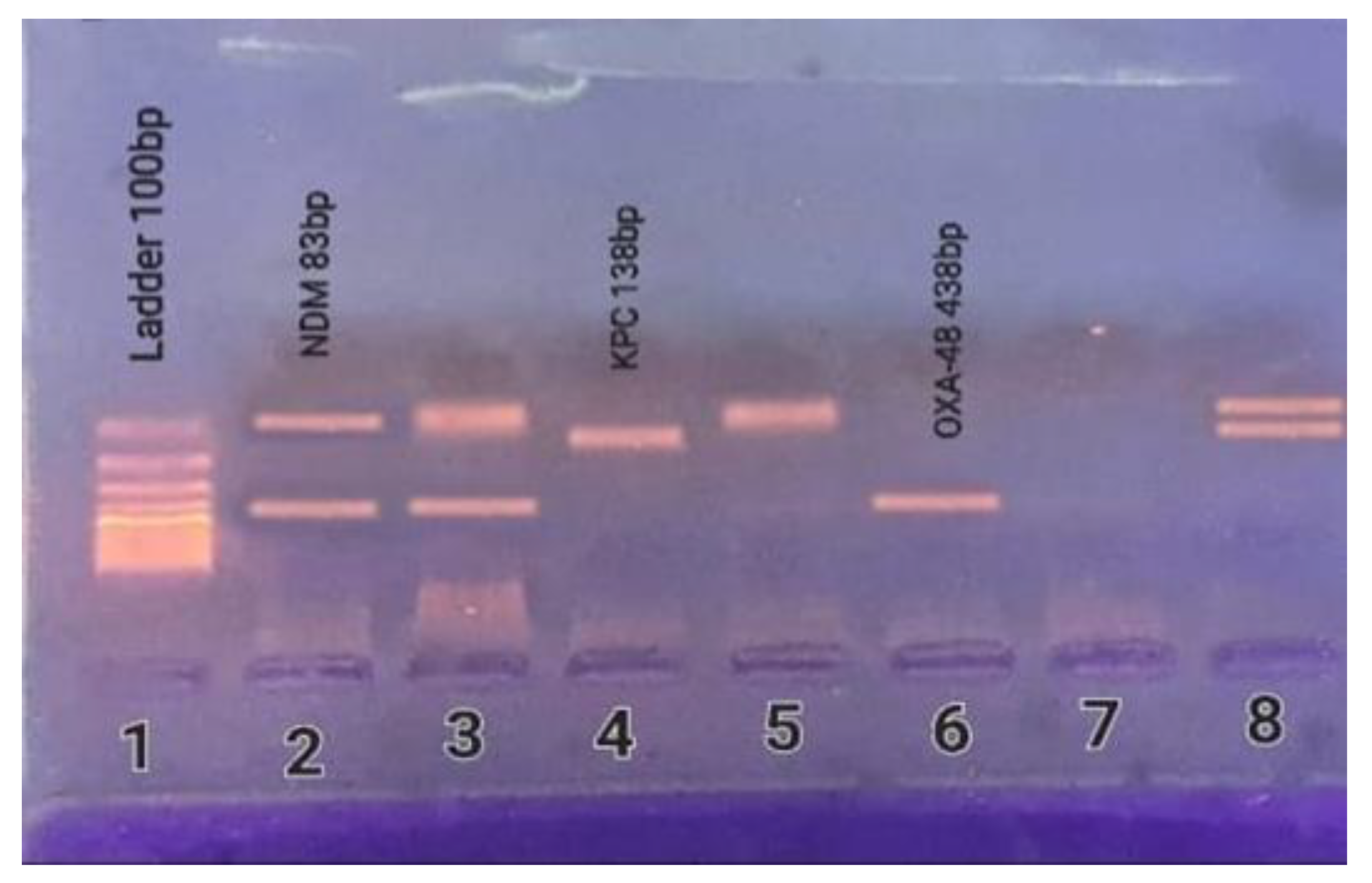
2.6. Detection of Carbapenemase-Encoding Genes
2.7. Demographic Characteristics of CRKP Patients According to the Detected Carbapenemase-Encoding Genes
2.8. Correlation of the Phenotype and Genotype of Carbapenem Resistance
2.9. Susceptibility Testing to New Therapeutic Agent (CZA)
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Ethical Approval
4.3. Case Diagnosis
4.4. Data Collection and Bacterial Strains
4.5. Microbiological Identification
4.6. Antimicrobial Susceptibility Testing
4.7. Confirmation of Carbapenem Resistance
4.8. Phenotypic Carbapenemase Detection
4.8.1. Modified Hodge Test (MHT)
4.8.2. Modified Carbapenem Inactivation Method (mCIM)
4.9. Molecular Detection of Carbapenemase-Encoding Genes
4.10. Susceptibility Testing to New Therapeutic Agent (CZA)
5. Conclusions
6. The Limitations of This Study
7. Recommendations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, Y.T.; Hsueh, P.R. Antimicrobial drug resistance in Taiwan. Int. J. Antimicrob. Agents 2008, 32 (Suppl. 3), S174–S178. [Google Scholar] [CrossRef]
- Moemen, D.; Masallat, D.T. Prevalence and characterization of carbapenem-resistant Klebsiella pneumoniae isolated from intensive care units of Mansoura University hospitals. Egypt J. Basic Appl. Sci. 2017, 4, 37–41. [Google Scholar] [CrossRef][Green Version]
- Pournaras, S.; Poulou, A.; Voulgari, E.; Vrioni, G.; Kristo, I.; Tsakris, A. Detection of the new metallo-beta-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 2010, 65, 1604–1607. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Infect Cont. 2006, 34 (Suppl. 1), S20–S28. [Google Scholar] [CrossRef]
- Abdelaziz, S.M.; Aboshanab, K.M.; Yahia, I.S.; Yassien, M.A.; Hassouna, N.A. Correlation between the Antibiotic Resistance Genes and Susceptibility to Antibiotics among the Carbapenem-Resistant Gram-Negative Pathogens. Antibiotics 2021, 10, 255. [Google Scholar] [CrossRef]
- Moquet, O.; Bouchiat, C.; Kinana, A.; Seck, A.; Arouna, O.; Bercion, R.; Breurec, S.; Garin, B. Class D OXA-48 carbapenemase in Multidrug-resistant Enterobacteria, Senegal. Emerg. Infect. Dis. 2011, 17, 143–144. [Google Scholar] [CrossRef]
- Mushi, M.F.; Mshana, S.E.; Imirzalioglu, C.; Bwanga, F. Carbapenemase Genes among Multidrug Resistant Gram Negative Clinical Isolates from a Tertiary Hospital in Mwanza, Tanzania. Biomed. Res. Int. 2014, 2014, 303104. [Google Scholar] [CrossRef]
- Munoz-Price, L.S.; Poirel, L.; A Bonomo, R.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Jean, B.; Patel, J.; Kamile, R.; Brandon, K. Carbapenemases in Enterobacteriaceae: Activity, Epidemiology, and Laboratory Detection. J. Clin. Microbial. 2009, 31, 55–62. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile beta-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef] [PubMed]
- Walther-Rasmussen, J.; Høiby, N. OXA-type carbapenemases. J. Antimicrob. Chemother. 2006, 57, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Rana, A.P.; Wenzler, E.; Ozer, E.A.; Krapp, F.; Bulitta, J.B.; Hauser, A.R.; Bulman, Z.P. Aminoglycoside-resistance gene signature are predictive of aminoglycoside MICs for carbapenem-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2022, 77, 356–363. [Google Scholar] [CrossRef]
- Ahmed El-Domany, R.; El-Banna, T.; Sonbol, F.; Abu-Sayedahmed, S.H. Co-existence of NDM-1 and OXA-48 genes in Carbapenem Resistant Klebsiella pneumoniae clinical isolates in Kafrelsheikh, Egypt. Afr. Health Sci. 2021, 21, 489–496. [Google Scholar] [CrossRef]
- El-Sweify, M.A.; Gomaa, N.I.; El-Maraghy, N.N.; Mohamed, H.A. Phenotypic Detection of Carbapenem Resistance among Klebsiella pneumoniae in Suez Canal University Hospitals, Ismailiya, Egypt. Int. J. Curr. Microbial. Appl. Sci. 2015, 4, 10–18. [Google Scholar]
- Doi, Y. Treatment options for Carbapenem-resistant Gram-negative Bacterial Infections. Clin. Infect. Dis. 2019, 69, S565–S575. [Google Scholar] [CrossRef]
- Girmenia, C.; Serrao, A.; Canichella, M. Epidemiology of Carbapenem Resistant Klebsiella pneumoniae Infections in Mediterranean Countries. Mediterr. J. Hematol. Infect. Dis. 2016, 8, e2016032. [Google Scholar] [CrossRef]
- Zafer, M.M.; El-Mahallawy, H.A.; Abdulhak, A.; Amin, M.A.; Al-Agamy, M.H.; Radwan, H.H. Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 40. [Google Scholar] [CrossRef]
- Gupta, N.; Limbago, B.M.; Patel, J.B.; Kallen, A.J. Carbapenem-resistant Enterobacteriaceae: Epidemiology and prevention. Clin. Infect. Dis. 2011, 53, 60–67. [Google Scholar] [CrossRef]
- Kandeel, A. Epidemiology of carbapenemase producing Enterobacteriaceae in a general hospital. J. Microbial. Infect. Dis. 2015, 5, 57–62. [Google Scholar] [CrossRef]
- Kotb, S.; Lyman, M.; Ismail, G.; El Fattah, M.A.; Girgis, S.A.; Etman, A.; Hafez, S.; El-Kholy, J.; Zaki, M.E.S.; Rashed, H.-A.G.; et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in Egyptian intensive care units using National Healthcare-associated Infections Surveillance Data, 2011–2017. Antimicrob. Resist. Infect. Control 2020, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Khairy, R.M.M.; Mahmoud, M.S.; Shady, R.R.; Esmail, M.A.M. Multidrug-resistant Klebsiella pneumoniae in hospital-acquired infections: Concomitant analysis of antimicrobial resistant strains. Int. J. Clin. Pract. 2020, 74, e13463. [Google Scholar] [CrossRef]
- Karuniawati, A.; Saharman, Y.R.; Lestari, D.C. Detection of carbapenemase encoding genes in Enterobacteriace, Pseudomonas aeruginosa, and Acinetobacter baumanii isolated from patients at Intensive Care Unit Cipto Mangunkusumo Hospital in 2011. Acta Med. Indones. 2013, 45, 101–106. [Google Scholar] [PubMed]
- Tian, B.; Fadhil, N.H.; Powell, J.E.; Kwong, W.K.; Moran, N.A. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 2012, 3, e00377-12. [Google Scholar] [CrossRef]
- Liu, C.; Wu, Y.; Huang, L.; Zhang, Y.; Sun, Q.; Lu, J.; Zeng, Y.; Dong, N.; Cai, C.; Shen, Z.; et al. The Rapid Emergence of Ceftazidime-Avibactam Resistance Mediated by KPC Variants in Carbapenem-Resistant Klebsiella pneumoniae in Zhejiang Province, China. Antibiotics 2022, 11, 731. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- He, T.; Wang, R.; Liu, D.; Walsh, T.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Poirel, L.; Jayol, A.; Nordmann, P. Polymixins: Antibacterial Activity, Susceptibility Testing, and Resistance Mechanisms Encoded by Plasmid or Chromosomes. Clin. Microbiol. Rev. 2017, 30, 557–596. [Google Scholar] [CrossRef]
- Miriagou, V.; Cornaglia, G.; Edelstein, M.; Galani, I.; Giske, C.; Gniadkowski, M.; Malamou-Lada, E.; Martinez-Martinez, L.; Navarro, F.; Nordmann, P.; et al. Acquired carbapenemases in Gram-negative bacterial pathogens: Detection and surveillance issues. Clin. Microbiol. Infect. 2010, 16, 112–122. [Google Scholar] [CrossRef]
- Yamada, K.; Kashiwa, M.; Arai, K.; Nagano, N.; Saito, R. Comparison of the Modified-Hodge test, Carba NP test, and carbapenem inactivation method as screening methods for carbapenemase-producing Enterobacteriaceae. J. Microbiol. Methods 2016, 28, 48–51. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Poirel, L.; Nordmann, P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 2012, 50, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Carvalhaes, C.G.; Picão, R.C.; Nicoletti, A.G.; Xavier, D.E.; Gales, A.C. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: Be aware of false positive results. J. Antimicrob. Chemother. 2010, 65, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Simner, P.J. Phenotypic Detection of Carbapenemase-Producing Organisms from Clinical Isolates. J. Clin. Microbiol. 2018, 56, e01140-18. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014, 20, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, P.; Garcia-Perdomo, H.A.; Karpinski, T.M.; Issakhanian, L. Metallo-β-lactamases: A review. Molecul. Biol. Rep. 2020, 47, 6281–6294. [Google Scholar] [CrossRef]
- Pierce, V.M.; Simner, P.J.; Lonsway, D.R.; Roe-Carpenter, D.E.; Johnson, J.K.; Brasso, W.B.; Bobenchik, A.M.; Lockett, Z.C.; Charnot-Katsikas, A.; Ferraro, M.J.; et al. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2321–2333. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.S.; Borghi, M.; de Araújo, C.F.M.; Aires, C.A.M.; Oliveira, J.C.R.; Asensi, M.D.; Carvalho-Assef, A.P.D. Clonal Dissemination of OXA-370-Producing Klebsiella pneumoniae in Rio de Janeiro, Brazil. Antimicrob. Agents Chemother. 2015, 59, 4453–4456. [Google Scholar] [CrossRef]
- Clinical & Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 25th ed.; Wayne: Delaware County, PA, USA, 2015; Available online: https://clsi.org/standards/products/microbiology/documents/m100 (accessed on 30 January 2021).
- Georgios, M.; Egki, T.; Effrosyni, S. Phenotypic, and Molecular Methods for the Detection of Antibiotic Resistance Mechanisms in Gram-Negative Nosocomial Pathogens. In Trends in Infectious Diseases; Saxena, S.K., Ed.; IntechOpen: Rijeka, Croatia, 2014; pp. 139–162. [Google Scholar] [CrossRef]
- Baran, I.; Aksu, N. Phenotypic and genotypic characteristics of carbapenem-resistant Enterobacteriaceae in a tertiary-level reference hospital in Turkey. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 20. [Google Scholar] [CrossRef]
- Chmielewska, S.; Leszczyńska, K. Carbapenemase of intestinal rods- the beginning of post-antibiotc era? Postȩpy Mikrobiol. Adv. Micrbiol. 2019, 58, 271–289. [Google Scholar] [CrossRef]
- Emira, A.S.; Madkour, L.A.; Seif, N.E.; Dwedar, R.A. Expressed and Silent Carbapenemase Genes Detected by Multiplex PCR in both Carbapenem-Resistant and Phenotypically-Susceptible Gram Negative Bacilli. Alex. J. Med. 2020, 56, 181–188. [Google Scholar] [CrossRef]
- Khalil, M.A.F.; Elgaml, A.; El-Mowafy, M. Emergence of Multidrug-Resistant New Delhi Metallo-β-Lactamase-1-Producing Klebsiella pneumoniae in Egypt. Microb. Drug Resist. 2017, 23, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G.; Chatzidimitriou, D.; Malisiovas, N. Double- and multi-carbapenemase-producers: The excessively armored bacilli of the current decade. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1487–1493. [Google Scholar] [CrossRef] [PubMed]
- Rea-Neto, A.; Youssef, N.C.; Tuche, F.; Brunkhorst, F.; Ranieri, V.M.; Reinhart, K.; Sakr, Y. Diagnosis of ventilator-associated pneumonia: A systematic review of the literature. Crit. Care. 2008, 12, R56. [Google Scholar] [CrossRef]
- Mohamed, E.R.; Halby, H.M.; Ali, M.Y.; El-Baky, R.M.A.; Waly, N.G.F.M. Spreading of NDMI-Producing Klebsiella Pneumoniaein Different Wards at Assiut University Hospital. Am. J. Infect. Dis. Microbiol. 2020, 8, 24–28. [Google Scholar] [CrossRef]
- Rezai, M.S.; Ahangarkani, F.; Rafiei, A.; Hajalibeig, A.; Bagheri-Nesami, M. Extended-spectrum beta-lactamases producing Pseudomonas aeruginosa isolated from patients with ventilator associated nosocomial infection. Arch. Clin. Infect. Dis. 2018, 13. [Google Scholar] [CrossRef]
- Liao, C.-H.; Lee, N.-Y.; Tang, H.-J.; Lee, S.S.-J.; Lin, C.-F.; Lu, P.-L.; Wu, J.-J.; Ko, W.-C.; Lee, W.-S.; Hsueh, P.-R. Antimicrobial activities of ceftazidime-avibactam, ceftolozane-tazobactam, and other agents against Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa isolated from intensive care units in Taiwan: Results from the surveillance of multicenter antimicrobial resistance in Taiwan in 2016. Infect. Drug Resist. 2019, 12, 545–552. [Google Scholar] [CrossRef]
- de Jonge, B.L.; Karlowsky, J.A.; Kazmierczak, K.M.; Biedenbach, D.J.; Sahm, D.F.; Nichols, W.W. In vitro susceptibility to ceftazidime-avibactam of carbapenem non susceptible Enterobacteriaceae isolates collected during the INFORM Global Surveillance Study (2012 to 2014). Antimicrob. Agents Chemother. 2016, 60, 3163–3169. [Google Scholar] [CrossRef]
- Mauri, C.; Maraolo, A.E.; Di Bella, S.; Luzzaro, F.; Principe, L. The revival of aztreonam in combination with avibactam against metallo-β-lactamase-producing gram-negatives: A systemic review of in vitro studies and clincal cases. Antibiotics 2021, 10, 1012. [Google Scholar] [CrossRef]
- Iwu-Jaja, C.J.; Jaca, A.; Jaja, I.F.; Jordan, P.; Bhengu, P.; Iwu, C.D.; Okeibunor, J.; Karamagi, H.; Tumusiime, P.; Fuller, W.; et al. Preventing and managing antimicrobial resistance in the African region: A scoping review protocol. PLoS ONE 2021, 16, e0254737. [Google Scholar] [CrossRef]
- Chokshi, A.; Sifri, Z.; Cennimo, D.; Horng, H. Global contributors to antibiotic resistance. J. Global. Infect. Dis. 2019, 11, 36–42. [Google Scholar] [CrossRef]
- Clinical & Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; Wayne: Delaware County, PA, USA, 2019; Available online: https://clsi.org/standards/products/microbiology/documents/m100 (accessed on 30 January 2021).
- Babaei, S.; Haeili, M. Evaluating the performance characteristics of different antimicrobial susceptibility testing methodologies for testing susceptibility of gram-negative bacteria to tigecycline. BMC Infect. Dis. 2021, 21, 709. [Google Scholar] [CrossRef] [PubMed]
- Clinical & Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 24th ed.; Wayne: Delaware County, PA, USA, 2014; Available online: https://clsi.org/standards/products/microbiology/documents/m100 (accessed on 30 January 2021).
- Van der Zee, A.; Roorda, L.; Bosman, G.; Fluit, A.d.C.; Hermans, M.; Smits, P.H.M.; Van der Zanden, A.G.M.; Witt, R.; Coppenraet, L.E.S.B.; Stuart, J.C.; et al. Multi-Centre Evaluation of Real-Time Multiplex PCR for Detection of Carbapenemase Genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect. Dis. 2014, 14, 27. Available online: https://www.biomedcentral.com/1471-2334/14/27 (accessed on 14 February 2021). [CrossRef] [PubMed]
- Mohamed, A.; Daef, E.; Nafie, A.; Shaban, L.; Ibrahim, M. Characteristics of Carbapenem Resistant Gram-Negative Bacilli in Patients with Ventilator-Associated Pneumonia. Antibiotics 2021, 10, 1325. [Google Scholar] [CrossRef]
| n | % | |
|---|---|---|
| Gender | ||
| Female | 61 | 51.3% |
| Male | 58 | 48.7% |
| Age (year): | ||
| Mean ± SD | 43.98 ± 22.94 | |
| Range | 44 years (2 days–98 years) | |
| Age groups: | ||
| Infant (<2 years) | 4 | 3.4% |
| Children&Adolescents | 15 | 12.6% |
| Adult | 66 | 55.5% |
| Old Age | 34 | 28.6% |
| Department (ICU): | ||
| Anesthesia | 44 | 37% |
| Chest | 4 | 3.4% |
| Internal Medicine | 32 | 26.9% |
| Neurosurgery | 8 | 6.7% |
| Oncology | 2 | 1.7% |
| Pediatric | 10 | 8.4% |
| Stroke | 8 | 6.7% |
| Surgical | 9 | 7.6% |
| Tropical | 2 | 1.7% |
| Type of Sample: | ||
| Blood | 22 | 18.5% |
| CSF | 3 | 2.5% |
| CVP | 5 | 4.2% |
| Peritoneal Fluid | 3 | 2.5% |
| Pus | 10 | 8.4% |
| Sputum | 47 | 39.5% |
| Urine | 29 | 24.4% |
| Sensitive | Intermediate | Resistant | |
|---|---|---|---|
| n (%) | n (%) | n (%) | |
| Piperacillin/Tazobactam | 0 (0.0) | 0 (0.0) | 119 (100) |
| Piperacillin | 0 (0.0) | 0 (0.0) | 119 (100) |
| Tobramycin | 6 (5.0) | 0 (0.0) | 113 (95.0) |
| Cefepime | 0 (0.0) | 0 (0.0) | 119 (100) |
| Imipenem | 0 (0.0) | 0 (0.0) | 119 (100) |
| Gentamicin | 15 (12.6) | 5 (4.2) | 99 (83.2) |
| Ceftazidime | 0 (0.0) | 0 (0.0) | 119 (100) |
| Meropenem | 0 (0.0) | 0 (0.0) | 119 (100) |
| Amikacin | 14 (11.8) | 0 (0.0) | 105 (88.2) |
| Sulfamethoxazole-Trimethoprim | 26 (21.8) | 0 (0.0) | 93 (78.2) |
| Azithromycin | 0 (0.0) | 0 (0.0) | 119 (100) |
| Minocycline | 7 (5.9) | 5 (4.2) | 107 (89.9) |
| Ticarcillin | 0 (0.0) | 0 (0.0) | 119 (100) |
| Pefloxacin | 0 (0.0) | 6 (5.0) | 113 (95.0) |
| Ciprofloxacin | 0 (0.0) | 2 (1.7) | 117 (98.3) |
| Tigecycline | 66 (55.5) | 10 (8.4) | 43 (36.1) |
| Colistin | 106 (89.1) | 0 (0.0) | 13 (10.9) |
| VITEK2 Compact | Kappa Agreement | p-Value | ||
|---|---|---|---|---|
| Susceptible (n = 61) | Resistant (n = 119) | |||
| MEM E-test | ||||
| Susceptible (n = 70) | 49 (80.3%) | 21 (17.6%) | 0.605 | <0.0001 |
| Resistant (n = 110) | 12 (19.7%) | 98 (82.4%) | ||
| CRKP Isolates | No = 119 | |
|---|---|---|
| No | % | |
| Carbapenmase Gene Presence | 105 | 88.2% |
| No Gene Presence | 14 | 11.8% |
| No = 105 | ||
|---|---|---|
| Carbapenemase-Encoding Gene | No | % |
| blaNDM | 40 | 38.1% |
| blaOXA-48 | 25 | 23.8% |
| blaKPC | 18 | 17.1% |
| blaNDM + blaOXA-48 + blaKPC | 9 | 8.6% |
| blaNDM + blaKPC | 4 | 3.8% |
| blaOXA-48 + blaKPC | 3 | 2.9% |
| blaNDM + blaOXA-48 | 6 | 5.7% |
| KPC | NDM | OXA-48 | ||||
|---|---|---|---|---|---|---|
| Positive n = 34 | Negative n = 85 | Positive n = 50 | Negative n = 69 | Positive n = 43 | Negative n = 76 | |
| Age | ||||||
| Mean ± (SD) | 42.4 ± (19) | 44.4 ± (23.8) | 41.8 ± (24.1) | 45.3 ± (21.2) | 46.6 ± (22.4) | 42.3 ± (22.5) |
| Mann–Whitney | −0.5 | −0.75 | −0.92 | |||
| p-value | 0.62 | 0.46 | 0.36 | |||
| Sex | ||||||
| Male | 13 | 45 | 28 | 30 | 18 | 40 |
| Female | 21 | 40 | 22 | 39 | 25 | 36 |
| Chi-square (χ2) | 2.1 | 1.82 | 1.28 | |||
| p-value | 0.15 | 0.18 | 0.26 | |||
| PCR Genes | Kappa Agreement | p-Value | ||
|---|---|---|---|---|
| Negative (Susceptible) n = 14 (%) | Positive (Resistant) n = 105 (%) | |||
| MHT | ||||
| Susceptible (n = 61) | 10 (71.4) | 51 (48.6) | 0.093 | 0.108006 |
| Resistant (n = 58) | 4 (28.6) | 54 (51.4) | ||
| mCIM | ||||
| Susceptible (n = 13) | 10 (71.4) | 3 (2.6) | 0.328 | <0.00001 |
| Resistant (n = 106) | 4 (28.6) | 102 (97.4) | ||
| CZA | |||
|---|---|---|---|
| Sensitive No (%) | Intermediate No (%) | Resistanat No (%) | |
| CRKP (119) | 28 (23.5) | - | 91 (76.5) |
| -Carbapenemase-encoding genes negative (14) | 8 (57.1) | - | 6 (42.9) |
| -Carbapenemase-encoding genes positive (105) | 20 (19.0) | - | 85 (81) |
| ● blaNDM positive (40) | 0 (0.0) | - | 40 (100) |
| ● blaOXA-48 positive (25) | 11 (44.0) | - | 14 (56.0) |
| ● blaKPC positive (18) | 8 (44.4) | - | 10 (55.6) |
| ● blaNDM + blaOXA-48 + blaKPC positive (9) | 0 (0.0) | - | 9 (100) |
| ● blaNDM + blaOXA-48 positive (6) | 0 (0.0) | - | 6 (100) |
| ● blaNDM + blaKPC positive (4) | 0 (0.0%) | - | 4 (100%) |
| ● blaOXA-48 + blaKPC positive (3) | 1 (33.3%) | - | 2 (66.7%) |
| Amplicon Size (bp) | Nucleotide Sequence (5′-3′) | Primers |
|---|---|---|
| 438 | F-(GCGTGGTTAAGGATGAACAC) | blaOXA-48 |
| R-(CATCAAGTTCAACCCAACCG) | ||
| 83 | F-(CATTAGCCGCTGCATTGATG) | blaNDM |
| R-(GTCGCCAGTTTCCATTTGCT) | ||
| 138 | F-(TGCAGAGCCCAGTGTCAGTTT) | blaKPC |
| R-(CGCTCTATCGGCGATACCA) | ||
| 740 | F-(TGAGCAAGTTATCTGTATTC) | blaIMP |
| R-(TTAGTTGCTTGGTTTTGATG) | ||
| 747 | F-(TCTACATGACCGCGTCTGTC) | blaVIM |
| R-(TGTGCTTTGACAACGTTCGC) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gandor, N.H.M.; Amr, G.E.-S.; Eldin Algammal, S.M.S.; Ahmed, A.A. Characterization of Carbapenem-Resistant K. Pneumoniae Isolated from Intensive Care Units of Zagazig University Hospitals. Antibiotics 2022, 11, 1108. https://doi.org/10.3390/antibiotics11081108
Gandor NHM, Amr GE-S, Eldin Algammal SMS, Ahmed AA. Characterization of Carbapenem-Resistant K. Pneumoniae Isolated from Intensive Care Units of Zagazig University Hospitals. Antibiotics. 2022; 11(8):1108. https://doi.org/10.3390/antibiotics11081108
Chicago/Turabian StyleGandor, Nessma Hessin Mohamed, Ghada EL-Sayed Amr, Sahar Mohamed Saad Eldin Algammal, and Alshymaa Abdullah Ahmed. 2022. "Characterization of Carbapenem-Resistant K. Pneumoniae Isolated from Intensive Care Units of Zagazig University Hospitals" Antibiotics 11, no. 8: 1108. https://doi.org/10.3390/antibiotics11081108
APA StyleGandor, N. H. M., Amr, G. E.-S., Eldin Algammal, S. M. S., & Ahmed, A. A. (2022). Characterization of Carbapenem-Resistant K. Pneumoniae Isolated from Intensive Care Units of Zagazig University Hospitals. Antibiotics, 11(8), 1108. https://doi.org/10.3390/antibiotics11081108






